Abstract
Background
Despite advances in treatments for Hodgkin's lymphoma, about 20% of patients still die from progressive disease. Current prognostic models predict the outcome of treatment with imperfect accuracy, and clinically relevant biomarkers have not been established to improve on the International Prognostic Score.
Methods
Using gene-expression profiling, we analyzed 130 frozen samples obtained from patients with classic Hodgkin's lymphoma during diagnostic lymph-node biopsy to determine which cellular signatures were correlated with treatment outcome. We confirmed our findings in an independent cohort of 166 patients, using immunohistochemical analysis.
Results
Gene-expression profiling identified a gene signature of tumor-associated macrophages that was significantly associated with primary treatment failure (P = 0.02). In an independent cohort of patients, we found that an increased number of CD68+ macrophages was correlated with a shortened progression-free survival (P = 0.03) and with an increased likelihood of relapse after autologous hematopoietic stem-cell transplantation (P = 0.008), resulting in shortened disease-specific survival (P = 0.003). In multivariate analysis, this adverse prognostic factor outperformed the International Prognostic Score for disease-specific survival (P = 0.003 vs. P = 0.03). The absence of an elevated number of CD68+ cells in patients with limited-stage disease defined a subgroup of patients with a long-term disease-specific survival of 100% with the use of current treatment strategies.
Conclusions
An increased number of tumor-associated macrophages was strongly associated with shortened survival in patients with classic Hodgkin's lymphoma and provides a new biomarker for risk stratification.
Current therapies do not cure at least 20% of patients with classic Hodgkin's lymphoma, and a similar proportion of patients are overtreated.1 It remains a challenge to identify patients whose disease will not be eradicated by standard therapies. Currently, most patients receive at least four cycles of polychemotherapy and, if indicated, radiotherapy.2 Autologous hematopoietic stem-cell transplantation can rescue about 50% of patients in whom primary therapy has failed.
Initial clinical decisions and risk stratification for patients with Hodgkin's lymphoma are largely based on clinical variables that distinguish those who are at high risk from those at standard risk. Classic Hodgkin's lymphoma, which constitutes about 95% of all cases of Hodgkin's lymphoma and is distinguished by the presence of Reed– Sternberg cells with characteristic features, is usually managed on the basis of Ann Arbor staging as follows: limited disease (stages I and IIA without constitutional symptoms) or advanced disease (stages IB and IIB with bulky disease [largest deposit, ≥10 cm in diameter] and stages III and IV either A or B [with or without constitutional symptoms]).3 The International Prognostic Score (on a scale of 0 to 7, with higher scores indicating increased risk) is the standard that is used for risk stratification of advanced-stage Hodgkin's lymphoma,4 but it does not apply to limited stages, and none of the published prognostic-factor systems can reliably identify patients in whom treatment is likely to fail. Moreover, neither the International Prognostic Score nor its individual clinical components are suitable for accurately predicting the outcome of autologous hematopoietic stem-cell transplantation in patients with Hodgkin's lymphoma. For these reasons, reliable biomarkers for predicting long-term survival at diagnosis are needed for such patients.
In Hodgkin's lymphoma, unlike most other cancers, the malignant Reed–Sternberg cells are outnumbered by non-neoplastic cells in the microenvironment of the tumor. The frequency and distribution of these cellular components and Reed–Sternberg cells vary considerably among individual patients and among subtypes of Hodgkin's lymphoma.5 Several studies have focused on the prediction of outcomes by means of markers expressed predominantly by Reed–Sternberg cells6-9 or the microenvironment.10-13 However, most of these markers require validation in independent cohorts.
Methods
Samples from Patients
For gene-expression profiling, we selected fresh-frozen lymph-node specimens, which had been obtained at the time of diagnosis from 130 patients with Hodgkin's lymphoma, from the tissue archives at the British Columbia Cancer Agency (BCCA) and the University of Nebraska Medical Center. The criteria that we used included a primary diagnosis of classic Hodgkin's lymphoma after central review, representative lymph-node tissue (at least 1 cm2 in tissue sections), negative status for human immunodeficiency virus infection, and first-line treatment with ABVD chemotherapy (doxorubicin, bleomycin, vinblastine, and dacarbazine) or an ABVD-like regimen and, if indicated, radiation therapy (including wide-field radiation for patients with limited-stage disease).
Primary treatment was defined as a failure if the lymphoma had progressed at any time after the initiation of therapy; treatment success was defined as the absence of progression or relapse. The median follow-up time for living patients in the treatment-success group was 3.9 years (range, 0.5 to 21.0). The gene-expression cohort was stratified according to treatment outcome (failure or success) in order to analyze differences between the two outcomes (see Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Advanced-stage disease was defined with the use of Ann Arbor staging criteria.3
On the basis of the availability of formalin-fixed, paraffin-embedded diagnostic lymph-node specimens, we also selected samples from 166 independent cases of classic Hodgkin's lymphoma for immunohistochemical testing on a tissue microarray. To increase the statistical power for observations linked to progression-free and disease-specific survival, this cohort was enriched for all available cases of treatment failure that were identified through the Centre for Lymphoid Cancer database at the BCCA. The number of cases of treatment failure was roughly matched to the number of cases of treatment success. For the independent cohort, we also recorded the outcome of secondary therapy delivered with curative intent in 61 patients; these secondary therapies included autologous stem-cell transplantation in 55 patients, CVPP chemotherapy (cyclophosphamide, vinblastine, procarbazine, and prednisone) plus involved-field radiation in 5 patients, and GDP chemotherapy (gemcitabine, dexamethasone, and cisplatin) plus extended-field radiation in 1 patient. In this cohort, the median follow-up time was 4.0 years (range, 0.5 to 20.8).
The study was approved by institutional review boards at the University of British Columbia–BCCA and the University of Nebraska Medical Center. Written informed consent was obtained from all patients in accordance with the principles of the Declaration of Helsinki.
Gene-Expression Analysis
Total RNA was extracted from multiple (10 to 20) 20-μm freshly cut tissue sections after mechanical homogenization. Expression profiles were obtained with the use of GeneChip Human Genome U133 Plus 2.0 arrays (Affymetrix). (Data are available at www.ncbi.nlm.nih.gov/geo/query/acc.cgi [accession number, GSE17920].) RNA preparation, array hybridization, and washing were performed according to routine protocol with modifications. All 130 microarrays met homogeneous criteria for quality control (for details on data processing and statistical analysis of array data, see the Supplementary Appendix).
Immunohistochemical Analysis
To confirm the findings of the gene-expression analysis, we performed immunohistochemical analysis on a tissue microarray that was constructed from duplicate 1.5-mm cores of 166 independent samples enriched for cases of treatment failure. Markers that were used included CD3, CD20, CD30, CD68, and MMP11. Immunohistochemical scoring ranged from 1 to 3 for CD68, from 1 to 4 for CD20, and from 0 to 3 for MMP11, with higher scores indicating a greater proportion of positive cells (for details, see the Supplementary Appendix). Scores were stratified as high (3 or 4) or low (1 or 2) for CD20 staining.
Predictive Models
In brief, we used the gene-expression data from the 130 patients for whom all prognostic factors of the International Prognostic Score were recorded and constructed a multidimensional classifier on the basis of feature selection, using sparse multinomial logistic regression (SMLR) and leave-one-out cross-validation.14 Our aim was to build a robust discriminative model that was predictive of treatment failure in addition to identifying a small set of features (genes) that could be used as the basis for separating these data into the respective outcome groups. We determined the relative importance of variables by means of a decision-tree–based algorithm (random forest),15 using the SMLR selected features. The relative importance of variables was defined with the use of a standard method, which is based on randomization of the variable values and measurement of the resultant decline in the accuracy of the model. We measured classification accuracy using a receiver-operating-characteristic (ROC) curve and the area under the curve (AUC).
Treatment failure was assigned as the positive class, and treatment success as the negative class. Thus, an accurate prediction of failure was a true positive result, and an inaccurate prediction of failure was a false positive result; an accurate prediction of success was a true negative result, and an inaccurate prediction of success was a false negative result. (For additional details on the predictive models, see the Supplementary Appendix.)
Statistical Analysis
Group comparisons were performed by means of the chi-square test and Student's t-test. For time-to-event analyses, we used two primary end points: progression-free survival (based on the time from initial diagnosis to progression at any time, relapse from complete response, or initiation of new, previously unplanned treatment) and disease-specific survival (based on the time from initial diagnosis to death from lymphoma or its treatment, with data for patients who died of unrelated causes censored at the time of death). Cox proportional-hazards models and time-to-event analyses with the use of the Kaplan–Meier method were performed with SPSS software, version 11.0. Two-sided P values of less than 0.05 were considered to indicate statistical significance.
Results
Gene-Expression Analysis
Unsupervised hierarchical clustering of the gene-expression results did not identify clusters that were significantly associated with the outcome of treatment or any other reported clinical variable (data not shown). However, using Globaltest, with which we tested all prefiltered genes, we found a significant correlation between the gene-expression profile and the outcome of first-line treatment (P = 0.02) (Table S1 in the Supplementary Appendix).
Table 1 shows the clinical characteristics of the gene-expression cohort. On the basis of supervised analyses with the data set stratified according to the failure or success of primary treatment (Fig. S1 in the Supplementary Appendix), we identified 271 differentially expressed genes in the two outcome groups (Tables S2 and S3 in the Supplementary Appendix). Unsupervised hierarchical clustering of all 130 expression profiles with the use of these differentially expressed genes identified two clusters, one of which was associated with treatment success (cluster A) and the other with both success and failure (cluster B) (Fig. 1A). In the treatment-failure group, pathway analyses identified the functions of cell-mediated immune response, cell-to-cell signaling and interaction, and up-regulation of pathway genes involved in interleukin-12 signaling and production in macrophages and apoptosis (Table S4 in the Supplementary Appendix). Down-regulated pathways in the treatment-failure group included genes involved in CTLA4 signaling in cytotoxic T lymphocytes and G-protein–coupled signaling.
Table 1. Demographic and Clinical Characteristics of the Two Cohorts of Patients.
| Variable | Gene-Expression Profiling (N = 130) | Immunohistochemical Analysis (N = 166) | ||||
|---|---|---|---|---|---|---|
| Treatment Success (N = 92) | Treatment Failure (N = 38) | P Value | Treatment Success (N = 87) | Treatment Failure (N = 79) | P Value | |
| Median age (range) — yr | 37 (8–80) | 46 (12–84) | 0.03 | 33 (16–80) | 36 (15–82) | 0.21 |
| Male sex — % | 51 | 68 | 0.08 | 51 | 53 | 0.74 |
| Histologic subtype — % | 0.15 | 0.19 | ||||
| Nodular sclerosis | 82 | 63 | 89 | 80 | ||
| Mixed cellularity | 12 | 26 | 6 | 8 | ||
| Lymphocyte-rich | 2 | 3 | 0 | 4 | ||
| Lymphocyte-depleted | 1 | 5 | 0 | 3 | ||
| Not classifiable | 3 | 3 | 6 | 6 | ||
| Stage — % | 0.003 | 0.01 | ||||
| I | 14 | 8 | 11 | 3 | ||
| II | 60 | 32 | 53 | 39 | ||
| III | 16 | 37 | 20 | 34 | ||
| IV | 10 | 24 | 16 | 24 | ||
| Presence of constitutional symptoms — % | 36 | 50 | 0.14 | 40 | 53 | 0.10 |
| Tumor size | 0.39 | 0.27 | ||||
| Median (range) — cm† | 6 (2–17) | 7 (2–26) | 6 (0–28) | 7 (0–19) | ||
| ≥10 cm — % | 18 | 26 | 33 | 30 | ||
| IPS ≥4 (high risk) — %‡ | 14 | 18 | 0.05 | 14 | 20 | 0.27 |
| Primary treatment — % | 1.00 | 0.32 | ||||
| ABVD chemotherapy with or without radiation | 96 | 95 | 99 | 100 | ||
| Extended-field radiation alone | 4 | 5 | 1 | 0 | ||
| Secondary treatment — % | ||||||
| Autologous stem-cell transplantation | ND | ND | NA | 70 | ||
| Other therapy with curative intent (CVPP or GDP plus radiation) | ND | ND | NA | 8 | ||
| Palliative treatment (including single-agent chemotherapy or radiation) | ND | ND | NA | 22 | ||
ABVD denotes doxorubicin, bleomycin, vinblastine, and dacarbazine, CVPP cyclophosphamide, vinblastine, procarbazine, and prednisone, GDP gemcitabine, dexamethasone, and cisplatin, IPS International Prognostic Score, NA not applicable, and ND not done.
The tumor size was calculated as the longest diameter of the largest involved area.
The IPS ranges from 0 to 7, with higher scores indicating increased risk.
Figure 1. Hierarchical Clustering of Gene-Expression Profiles in Hodgkin's Lymphoma, True and False Positive and Negative Rates for Three Models of Outcome Prediction, and the Importance of Individual Genes for Outcome Prediction.
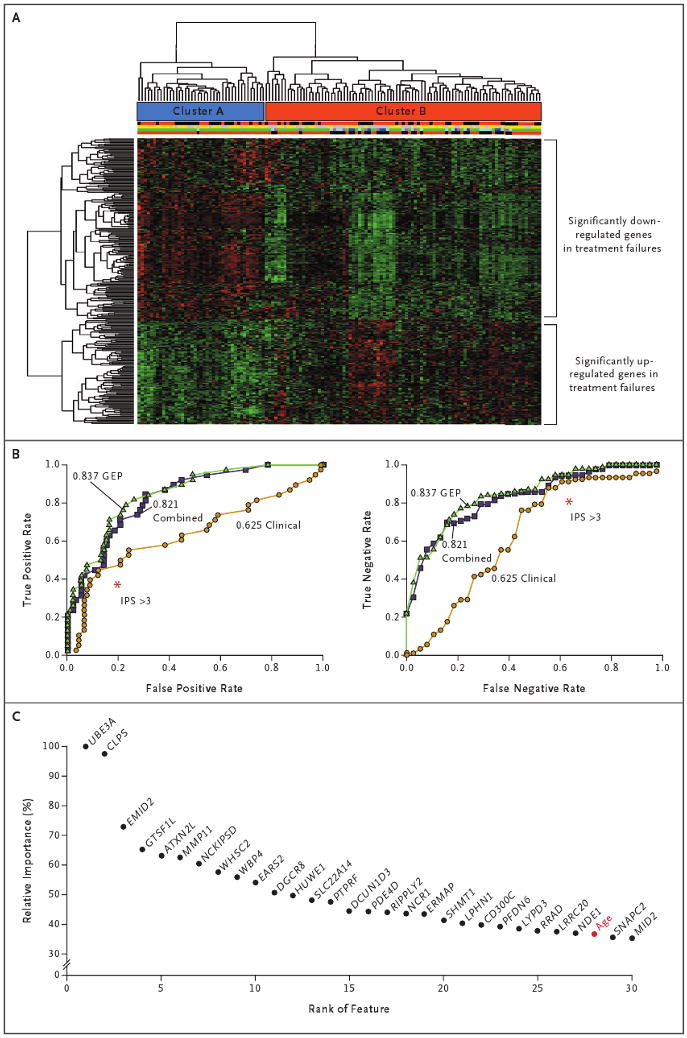
Panel A shows hierarchical clustering of 130 gene-expression profiles for patients with classic Hodgkin's lymphoma. Cluster A has been enriched with primary treatment successes, and Cluster B with both primary treatment successes and failures. Immediately below the cluster bars, the first multicolored bar indicates sex (red for male and black for female), the second bar indicates stage (yellow for limited disease and gray for advanced disease), the third bar indicates the type of treatment failure (green for no treatment failure, purple for refractory, dark blue for early relapse, and light blue for late relapse), and the fourth bar indicates the primary treatment outcome (black for failure and red for success). (For details, see Tables S2 and S3 in the Supplementary Appendix, available with the full text of this article at NEJM.org.) Panel B shows plots of true positive and false positive rates and true negative and false negative rates (receiver-operating-characteristic curves) for three models that were used for feature selection: gene-expression profiling (GEP), a model based on the International Prognostic Score (IPS) for clinical variables, and a model combining these two features. This comparison showed that the value for the area under the curve was highest for the GEP model, as compared with the clinical and combined models (0.837 vs. 0.625 and 0.821, respectively). For comparison with the established IPS, red asterisks indicate an IPS of more than 3, as calculated with the use of IPS thresholds.4 Panel C shows the relative importance of individual genes for outcome prediction. Relative importance is shown for 30 annotated probe sets (selected with the use of sparse multinomial logistic regression) that were more influential than Ann Arbor staging. Among the 27 individual genes exceeding the importance of the best clinical variable, age (shown in red), was MMP11.
These results prompted an investigation of the association between microenvironment gene signatures and outcome. Using Globaltest, we performed associative testing to identify previously described cellular and pathway gene signatures that were differentially expressed in the two outcome groups.16 In the treatment-failure group, there was overexpression of gene signatures of tumor-associated macrophages (P = 0.02)17 and monocytes (P = 0.01),18 findings that are in agreement with the reported overexpression of macrophage-signaling–associated genes according to pathway analysis (Table S1 and Fig. S2 in the Supplementary Appendix). There was also overexpression of gene signatures for angiogenic cells (P = 0.04),19 adipocytes (P = 0.01),20 and Reed–Sternberg cells (P = 0.047)21 and underexpression of a signature for germinal center B cells (P = 0.01)22 in the treatment-failure group (Table S1 in the Supplementary Appendix). Furthermore, previously described genes that are associated with an unfavorable outcome in Hodgkin's lymphoma,23 such as lysozyme (LYZ) and cathepsin L1 (CTSL1), were overexpressed in the treatment-failure group (P = 0.04).
To test the overall power of expression profiles for outcome prediction, we constructed a classifier by means of sparse multinomial logistic regression.14 This algorithm identified 86 non-redundant annotated genes (Table S5 in the Supplementary Appendix), age, and Ann Arbor stage by a cross-validation approach. Figure 1C shows the 30 features with the highest discriminative power, as determined by a random-forest algorithm.15 Among the 27 individual genes with discriminative power exceeding that of the best clinical variable (age) was MMP11 (probe set 235908_at), which was overexpressed in the treatment-failure group (P = 0.03 after adjustment for the false discovery rate). We selected this gene for further immunohistochemical testing, since previous studies have shown that the gene family of matrix metallopeptidases are overexpressed in patients in whom treatment has failed24 and that MMP11 in particular is expressed by tumor-associated macrophages.25
We compared the three data sources for feature selection and settled on a gene-expression profiling model for gene-expression probe sets only, a clinical model based on the International Prognostic Score only, and a combination model including both features. This comparison showed that the AUC value was highest for the gene-expression model, as compared with the clinical and combination models (0.837 vs. 0.625 and 0.821, respectively) (Fig. 1B). The differences in accuracy among the models were most prominent at low false negative rates, with the gene-expression model and the combination model yielding higher true negative rates than the clinical model.
Immunohistochemical Analysis
Our gene-expression study and previous studies by other investigators suggested that a predominance of tumor-infiltrating macrophages, a lack of small B cells, and overexpression of matrix metallopeptidases were correlated with the failure of primary treatment. For these reasons, we selected the markers CD68 (macrophages), CD20 (B cells), and MMP11 for immunohistochemical analysis of a tissue array containing samples from lymph-node biopsies in 166 patients with classic Hodgkin's lymphoma (who were unrelated to the patients in the gene-expression analysis), including 79 patients in whom treatment had failed (Table 1). Of these markers, CD68 and CD20 antibodies are routinely used in the diagnosis of lymphoma. The tissue microarray was also stained for CD30 (a marker for Reed–Sternberg cells) and CD3 (a marker for T cells), but neither the number of CD30+ cells nor the number of CD3+ cells was correlated with outcome (data not shown).
Of these immunohistochemical markers, CD68 stood out because of its significant correlation with primary and secondary treatment outcomes (Fig. 2). Using univariate analysis, we found a significant correlation between the number of CD68+ tumor-infiltrating macrophages and shortened progression-free survival (P = 0.03) (Table 2). Patients with a high number of CD68+ cells (an immunohistochemical score of 3) had a median progression-free survival of 2.7 years, whereas during an observation period of 16.4 years, the median survival was not reached in patients with a score of 1 (Fig. 3A). In a multivariate Cox regression model that included factors with respect to the International Prognostic Score and immunohistochemical scores for CD68, CD20, and MMP11, CD68 was not an independent factor for an association with progression-free survival. In contrast, an increased number of CD68+ macrophages correlated with disease-specific survival in both univariate and multivariate analysis (P = 0.003 for both comparisons) and outperformed the International Prognostic Score (P = 0.03) (Table 2).
Figure 2. Representative Immunohistochemical Analyses for CD68 in Lymph-Node Biopsy Samples from Two Patients.
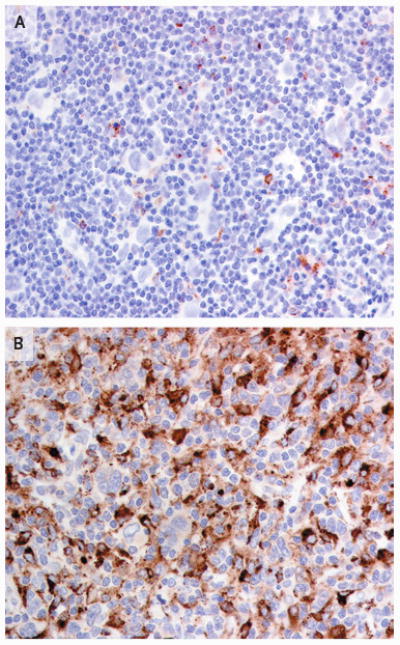
Panel A shows a sample obtained from a patient in the treatment-success group, with few CD68+ macrophages, and Panel B shows a sample from a patient in the treatment-failure group, with many CD68+ macrophages. Both patients had the nodular sclerosis subtype of Hodgkin's lymphoma.
Table 2. Progression-free and Disease-Specific Survival in a Validation Cohort of 166 Patients.*.
| Variable | Patients with Characteristic | P Value for Progression-free Survival | P Value for Disease-Specific Survival | ||
|---|---|---|---|---|---|
| no. (%) | Univariate Analysis | Multivariate Analysis | Univariate Analysis | Multivariate Analysis | |
| Demographic data | |||||
| Male sex | 86 (51.8) | 0.90 | 0.63 | ||
| Age >44 yr | 54 (32.5) | 0.32 | 0.05 | ||
| Immunohistochemical data† | |||||
| ≥5% CD68+ cells (IHC score, >1) | 120 (72.3) | 0.03 | 0.003 | 0.003 | |
| CD20+ cells | |||||
| ≤10% Background B cells (IHC score, <3) | 85 (51.2) | 0.02 | 0.02 | ||
| Reed–Sternberg cells | 20 (12.0) | 0.95 | 0.59 | ||
| Lymphoid follicles | 44 (26.5) | 0.24 | 0.34 | ||
| ≥1% MMP11+ (IHC score, >1)‡ | 65 (40.6) | 0.008 | 0.009 | 0.09 | |
| Laboratory data | |||||
| Albumin <40 g/liter | 68 (41.0) | 0.047 | 0.03 | ||
| Hemoglobin <10.5 g/dl | 29 (17.5) | 0.004 | 0.11 | ||
| White-cell count >15,000/mm3 | 23 (13.9) | 0.52 | 0.23 | ||
| Lymphocyte count <600/mm3 or <8% | 25 (15.1) | 0.13 | 0.15 | ||
| Clinical data | |||||
| IPS >3 (high risk)§ | 28 (16.9) | 0.38 | 0.004 | 0.03 | |
| Advanced-stage disease | 125 (75.3) | 0.002 | 0.001 | 0.05 | |
| Constitutional symptoms | 76 (45.8) | 0.08 | 0.48 | ||
| Bulky tumor (≥10 cm in diameter) | 53 (31.9) | 0.82 | 0.57 | ||
P values are for the correlation between each factor and survival. Univariate analyses were calculated with the use of a Cox proportional-hazards regression model, and multivariate analyses were performed with a Cox proportional-hazards regression model (forward stepwise likelihood ratio).
Immunohistochemical (IHC) scores range from 1 to 3 for CD68, from 1 to 4 for CD20, and from 0 to 3 for MMP11, with higher scores indicating a greater proportion of positive cells.
Data regarding immunohistochemical staining were missing for six patients.
The International Prognostic Score (IPS) ranges from 0 to 7, with higher scores indicating increased risk.
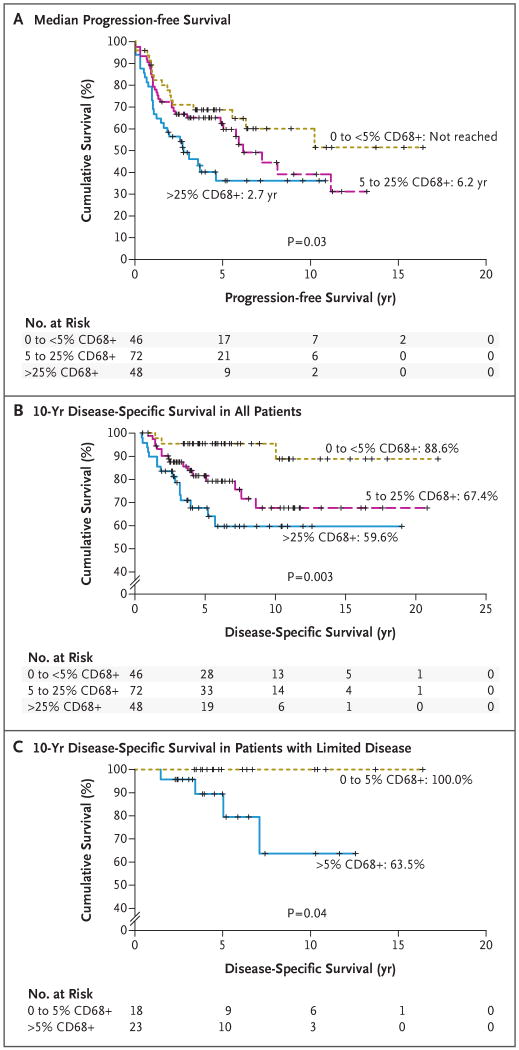
Figure 3. Survival in a Validation Cohort of 166 Patients, According to the Number of Infiltrating CD68+ Macrophages in Pretreatment Lymph-Node Biopsy Specimens.
The graphs show progression-free survival in all patients (Panel A) and disease-specific survival in all patients (Panel B) and in 41 patients with limited-stage disease (Panel C). According to the immunohistochemical scoring system that was used, a score of 1 indicates less than 5% CD68+ cells, a score of 2 indicates 5 to 25%, and a score of 3 indicates more than 25%. Clinically relevant biomarkers for predicting the outcome of treatment in patients with Hodgkin's disease have not been established. In this study, gene profiling and immunohistochemical analysis were used to find such a marker. A strong association was found between a poor outcome of treatment and an increased number of CD68+ cells in the microenvironment of Reed–Sternberg cells. CD68, a marker of macrophages, outperformed the conventional International Prognostic Score and can be immunohistochemically stained in diagnostic samples of Hodgkin's lymphoma.
The 10-year disease-specific survival rate was significantly lower among patients with a CD68 immunohistochemical score of 3 (59.6%) than among those with a score of 2 (67.4%) or 1 (88.6%) (P = 0.003 for all comparisons) (Fig. 3B). The number of tumor-infiltrating macrophages was also correlated with the outcome after secondary treatment. Secondary treatment that was administered with curative intent failed in only 12.5% of patients with a CD68 immunohistochemical score of 1, as compared with failure in 51.7% of those with a score of 2 and in 62.5% of those with a score of 3 (P = 0.009 for all comparisons). In particular, there was a significant correlation between the failure of autologous hematopoietic stem-cell transplantation and a CD68 score of more than 1 (P = 0.008). A high-risk International Prognostic Score (>3)4 was not significantly associated with the number of CD68+ macrophages in diagnostic biopsy samples (8.7% for a score of 1, as compared with 18.1% for a score of 2 and 22.9% for a score of 3; P = 0.17 for all comparisons). When the analysis was restricted to limited-stage disease, a CD68 score of 1 was associated with a long-term disease-specific survival rate of 100% (P = 0.04) (Fig. 3C).
In agreement with the findings of the gene-expression study, MMP11 immunohistochemical staining showed a significant correlation with progression-free survival in both univariate analysis (P = 0.008) and multivariate analysis (P = 0.009), although not with disease-specific survival (Table 2). Among the cells with positive staining were Reed–Sternberg cells, macrophages, and endothelial cells (data not shown).
Using univariate analysis, we also found that an increased number of CD20+ small B cells (immunohistochemical score, >2) was significantly associated with prolonged progression-free survival (P = 0.02) and disease-specific survival (P = 0.02) (Table 2). However, the number of CD20+ small B cells was not an independent predictor of survival and was strongly correlated with advanced-stage disease (P = 0.002). Neither the number of CD20+ Reed–Sternberg cells nor the presence or absence of primary or secondary lymphoid follicles was associated with the outcome.
Discussion
We found that the overexpression of a macrophage signature in expression-profile studies of diagnostic lymph-node specimens obtained from patients with classic Hodgkin's lymphoma was associated with the failure of primary treatment. Using immunohistochemical analysis, we also found that an increased number of CD68+ cells (a marker of benign macrophages) in the diagnostic sample was associated with a poorer outcome in an independent set of samples from 166 patients. Multivariate analysis revealed that the number of CD68+ cells was also associated with the outcome of secondary treatment, independently of the International Prognostic Score.
Three previous studies that have used expression profiles of the microenvironment in Hodgkin's lymphoma have been reported.23,24,26 Of these, one identified a gene signature of macrophages but did not show the clinical value of tumor-associated macrophages assessed by means of a common immunohistochemical marker.23 The association between the number of macrophages and treatment outcome has also been studied in other B-cell cancers.19,27,28 Differences in survival among patients with various lymphoma subtypes, which are linked to macrophage content, might be explained by the variable presence of macrophages with M1 or M2 differentiation in biopsy samples, indicating distinct biologic features of the tumors.29 However, in these lymphoma subtypes, including Hodgkin's lymphoma, the functional link between macrophage numbers and the contribution of these cells to the treatment outcome remains unclear.23,30 Our gene-expression classifier for the outcome of primary treatment outcome revealed MMP11, a gene that has been found by other investigators to be expressed in tumor-associated macrophages involved in remodeling of apoptotic lymphatic tissue.25 Using immunohistochemical analysis, we were able to confirm the correlation between the number of MMP11+ cells and progression-free survival in an independent cohort of patients. However, MMP11 stained many different cell types, including macrophages, and thus did not allow us to identify the particular cells that were responsible for the production of the protein.
Our findings also validate the recent report of a correlation between an increased number of small B cells and a favorable outcome.26 The recently described correlation between the number of CD20-positive B cells and survival in patients with classic Hodgkin's lymphoma31 needs to be reassessed in the context of clinical studies showing successful treatment with the addition of rituximab to standard chemotherapy.32,33
We report the clinical value of a single marker, CD68, in the identification of tumor-associated macrophages by immunohistochemical analysis, an analytic method that can be easily incorporated into a routine diagnostic approach. The use of such markers in combination with well-established clinical risk factors could improve on the predictive value of a single biomarker used alone.
We focused on tumor-associated macrophages because of the strong signal from the gene-expression data and the recently renewed interest in these non-neoplastic cells as major contributors to the biologic features of lymphoma and outcome prediction.19 The value of assessing the number of tumor-associated macrophages as a biomarker is highlighted by the association between these cells and the outcome after secondary therapy with autologous stem-cell transplantation, a widely used treatment option. Accurate prediction of the outcome after secondary treatments with curative intent would provide better risk stratification for these therapeutic options. Clinical predictors of the outcome after autologous stem-cell transplantation have been of limited value.34 In addition, our finding that in the tissue-microarray cohort, none of the patients with limited-stage disease and a low number of macrophages died is encouraging, since the applicability of the International Prognostic Score is restricted to advanced-stage disease. Thus, the CD68+ macrophage content represents a biomarker with clinical applicability in all stages of classic Hodgkin's lymphoma, both at the time of diagnosis and at the time of relapse.
In summary, our study showed the value of enumerating CD68+ macrophages in diagnostic lymph-node samples for prediction of the outcome after primary treatment and secondary treatment (in particular, autologous stem-cell transplantation). The absence of an increased number of CD68+ cells in patients with limited-stage disease defines a subgroup of patients for whom the rate of long-term disease-specific survival is 100% with the use of available treatments.
Supplementary Material
Acknowledgments
Supported by grants from Deutsche Forschungsgemeinschaft, the Cancer Research Society, and the Lymphoma Research Foundation (to Dr. Steidl), the Michael Smith Foundation for Health Research (to Drs. Steidl and Shah), Roche Molecular Systems, the Canadian Institutes of Health Research (178536, to Dr. Gascoyne), the National Cancer Institute (UO1-CA114778-01, to Dr. Chan), and the Intramural Research Program of the National Cancer Institute (NCI) and by an NCI Strategic Partnering to Evaluate Cancer Signatures (SPECS) grant (UO1-CA 114778) to the Lymphoma/Leukemia Molecular Profiling Project consortium.
Dr. Gascoyne reports receiving consulting fees from Genentech, Roche Canada, and Eli Lilly and research support from Roche Canada. No other potential conflict of interest relevant to this article was reported.
We thank Adele Telenius, Bruce Woolcock, Lorraine May, and the Center for Translational and Applied Genomics for their technical support.
Footnotes
Presented in part at the Seventh International Symposium on Hodgkin Lymphoma, Cologne, Germany, November 5, 2007, and the 50th American Society of Hematology Annual Meeting, San Francisco, December 8, 2008.
References
- 1.Björkholm M, Axdorph U, Grimfors G, et al. Fixed versus response-adapted MOPP/ABVD chemotherapy in Hodgkin's disease: a prospective randomized trial. Ann Oncol. 1995;6:895–9. doi: 10.1093/oxfordjournals.annonc.a059356. [DOI] [PubMed] [Google Scholar]
- 2.Canellos GP, Anderson JR, Propert KJ, et al. Chemotherapy of advanced Hodgkin's disease with MOPP, ABVD, or MOPP alternating with ABVD. N Engl J Med. 1992;327:1478–84. doi: 10.1056/NEJM199211193272102. [DOI] [PubMed] [Google Scholar]
- 3.Diehl V, Stein H, Hummel M, Zollinger R, Connors JM. Hodgkin's lymphoma: biology and treatment strategies for primary, refractory, and relapsed disease. Hematology Am Soc Hematol Educ Program. 2003:225–47. doi: 10.1182/asheducation-2003.1.225. [DOI] [PubMed] [Google Scholar]
- 4.Hasenclever D, Diehl V, Armitage JO, et al. A prognostic score for advanced Hodgkin's disease. N Engl J Med. 1998;339:1506–14. doi: 10.1056/NEJM199811193392104. [DOI] [PubMed] [Google Scholar]
- 5.Swerdlow SH, Campo E, Harris NL, et al., editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 6.Sup SJ, Alemany CA, Pohlman B, et al. Expression of bcl-2 in classical Hodgkin's lymphoma: an independent predictor of poor outcome. J Clin Oncol. 2005;23:3773–9. doi: 10.1200/JCO.2005.04.358. [DOI] [PubMed] [Google Scholar]
- 7.Doussis-Anagnostopoulou IA, Vassilakopoulos TP, Thymara I, et al. Topoisomerase IIalpha expression as an independent prognostic factor in Hodgkin's lymphoma. Clin Cancer Res. 2008;14:1759–66. doi: 10.1158/1078-0432.CCR-07-1395. [DOI] [PubMed] [Google Scholar]
- 8.Natkunam Y, Lossos IS, Taidi B, et al. Expression of the human germinal center-associated lymphoma (HGAL) protein, a new marker of germinal center B-cell derivation. Blood. 2005;105:3979–86. doi: 10.1182/blood-2004-08-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diepstra A, van Imhoff GW, Karim-Kos HE, et al. HLA class II expression by Hodgkin Reed-Sternberg cells is an independent prognostic factor in classical Hodgkin's lymphoma. J Clin Oncol. 2007;25:3101–8. doi: 10.1200/JCO.2006.10.0917. [DOI] [PubMed] [Google Scholar]
- 10.Alvaro T, Lejeune M, Salvado MT, et al. Outcome in Hodgkin's lymphoma can be predicted from the presence of accompanying cytotoxic and regulatory T cells. Clin Cancer Res. 2005;11:1467–73. doi: 10.1158/1078-0432.CCR-04-1869. [DOI] [PubMed] [Google Scholar]
- 11.Alvaro-Naranjo T, Lejeune M, Salvadó -Usach MT, et al. Tumor-infiltrating cells as a prognostic factor in Hodgkin's lymphoma: a quantitative tissue microarray study in a large retrospective cohort of 267 patients. Leuk Lymphoma. 2005;46:1581–91. doi: 10.1080/10428190500220654. [DOI] [PubMed] [Google Scholar]
- 12.Kelley TW, Pohlman B, Elson P, Hsi ED. The ratio of FOXP3+ regulatory T cells to granzyme B+ cytotoxic T/NK cells predicts prognosis in classical Hodgkin lymphoma and is independent of bcl-2 and MAL expression. Am J Clin Pathol. 2007;128:958–65. doi: 10.1309/NB3947K383DJ0LQ2. [DOI] [PubMed] [Google Scholar]
- 13.Tzankov A, Meier C, Hirschmann P, Went P, Pileri SA, Dirnhofer S. Correlation of high numbers of intratumoral FOXP3+ regulatory T cells with improved survival in germinal center-like diffuse large B-cell lymphoma, follicular lymphoma and classical Hodgkin's lymphoma. Haematologica. 2008;93:193–200. doi: 10.3324/haematol.11702. [DOI] [PubMed] [Google Scholar]
- 14.Krishnapuram B, Carin L, Figueiredo MA, Hartemink AJ. Sparse multinomial logistic regression: fast algorithms and generalization bounds. IEEE Trans Pattern Anal Mach Intell. 2005;27:957–68. doi: 10.1109/TPAMI.2005.127. [DOI] [PubMed] [Google Scholar]
- 15.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 16.Shaffer AL, Wright G, Yang L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 17.Duff MD, Mestre J, Maddali S, Yan ZP, Stapleton P, Daly JM. Analysis of gene expression in the tumor-associated macrophage. J Surg Res. 2007;142:119–28. doi: 10.1016/j.jss.2006.12.542. [DOI] [PubMed] [Google Scholar]
- 18.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–23. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urs S, Smith C, Campbell B, et al. Gene expression profiling in human preadipocytes and adipocytes by microarray analysis. J Nutr. 2004;134:762–70. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
- 21.Karube K, Ohshima K, Suzumiya J, Kawano R, Kikuchi M, Harada M. Gene expression profile of cytokines and chemokines in microdissected primary Hodgkin and Reed-Sternberg (HRS) cells: high expression of interleukin-11 receptor alpha. Ann Oncol. 2006;17:110–6. doi: 10.1093/annonc/mdj064. [DOI] [PubMed] [Google Scholar]
- 22.Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–42. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Aguilera A, Montalbán C, de la Cueva P, et al. Tumor microenvironment and mitotic checkpoint are key factors in the outcome of classic Hodgkin lymphoma. Blood. 2006;108:662–8. doi: 10.1182/blood-2005-12-5125. [DOI] [PubMed] [Google Scholar]
- 24.Devilard E, Bertucci F, Trempat P, et al. Gene expression profiling defines molecular subtypes of classical Hodgkin's disease. Oncogene. 2002;21:3095–102. doi: 10.1038/sj.onc.1205418. [DOI] [PubMed] [Google Scholar]
- 25.Odaka C, Izumiyama S. Expression of stromelysin-3 (matrix metalloproteinase-11) in macrophages of murine thymus following thymocyte apoptosis. Cell Immunol. 2005;235:21–8. doi: 10.1016/j.cellimm.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 26.Chetaille B, Bertucci F, Finetti P, et al. Molecular profiling of classical Hodgkin lymphoma tissues uncovers variations in the tumor microenvironment and correlations with EBV infection and outcome. Blood. 2009;113:2765–3775. doi: 10.1182/blood-2008-07-168096. [DOI] [PubMed] [Google Scholar]
- 27.Dave SS, Wright G, Tan B, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–69. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 28.Farinha P, Masoudi H, Skinnider BF, et al. Analysis of multiple biomarkers shows that lymphoma-associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL) Blood. 2005;106:2169–74. doi: 10.1182/blood-2005-04-1565. [DOI] [PubMed] [Google Scholar]
- 29.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–55. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 30.Küppers R. The biology of Hodgkin's lymphoma. Nat Rev Cancer. 2009;9:15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 31.Jones RJ, Gocke CD, Kasamon YL, et al. Circulating clonotypic B cells in classic Hodgkin lymphoma. Blood. 2009;113:5920–6. doi: 10.1182/blood-2008-11-189688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Younes A, Romaguera J, Hagemeister F, et al. A pilot study of rituximab in patients with recurrent, classic Hodgkin disease. Cancer. 2003;98:310–4. doi: 10.1002/cncr.11511. [DOI] [PubMed] [Google Scholar]
- 33.Falchi L, Capello D, Palumbo B, et al. A case of nodular sclerosis Hodgkin's lymphoma repeatedly relapsing in the context of composite plasma cell-hyaline vascular Castleman's disease: successful response to rituximab and radiotherapy. Eur J Haematol. 2007;79:455–61. doi: 10.1111/j.1600-0609.2007.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perz JB, Giles C, Szydlo R, et al. LACE-conditioned autologous stem cell transplantation for relapsed or refractory Hodgkin's lymphoma: treatment outcome and risk factor analysis in 67 patients from a single centre. Bone Marrow Transplant. 2007;39:41–7. doi: 10.1038/sj.bmt.1705544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.