Abstract
Geographic distribution among members of the Sigmodon hispidus complex (Sigmodon hirsutus, S. hispidus, and S. toltecus) were examined using DNA sequences from the mitochondrial cytochrome-b gene. Geographic distribution of each taxon was defined based on DNA sequences obtained from 69 samples (19 newly obtained and 50 from previous studies) collected from North, Central, and South America. These data indicated that S. hispidus is restricted to the southern one-half of the United States and northeastern Mexico (Nuevo León and Tamaulipas), S. toltecus occupies the eastern one-third of Mexico (central Tamaulipas) to northern Honduras, and S. hirsutus is distributed from central Chiapas and southeastern Oaxaca to northern South America (Venezuela). The newly collected data extend distributions of S. hispidus from the southern United States southward into northeastern Mexico and that of S. toltecus from Chiapas, Mexico, southward to Honduras. Genetic divergence and patterns of phylogeography were examined within each taxon.
DNA-sequence data from the mitochondrial cytochrome-b gene (Peppers and Bradley, 2000; Peppers et al., 2002; Carroll et al., 2005) and intron 7 of the beta-fibrinogen gene (Carroll and Bradley, 2005) indicated that the taxon, historically referred to as S. hispidus, was a composite and paraphyletic assemblage that consisted of three distinct species (S. hirsutus, S. hispidus, and S. toltecus). Peppers and Bradley (2000) applied the term cryptic species to these taxa to illustrate the lack of morphological divergence accompanying the high level of genetic divergence among the three species. To date, genetic data provide the only means of identifying the three species. Carroll et al. (2005) further attempted to describe distributions and hypothesized in general that S. hispidus was restricted to southern portions of the United States north of the Rio Grande, S. toltecus was restricted to regions south of the Rio Grande along the eastern coastal regions of Mexico to central Chiapas, and S. hirsutus occupied regions of southern Mexico (Chiapas and Oaxaca) to Venezuela. Additionally, Carroll et al. (2005) considered possible explanations (habitat preferences and phylogeography) for the proposed distributions of these three species.
Concerning separation of S. hispidus and S. toltecus along boundaries of the United States and Mexico, individuals from Brownsville, Texas, and Calabazas, Tamaulipas, represented peripheral samples of each respective taxon. These individuals were separated by ca. 400 km and genetically differed by genetic-distance values that were typical for any comparison between S. hispidus or S. toltecus. Carroll et al. (2005) offered two hypotheses to explain the distribution of these two species. First, the Rio Grande historically has been considered to be a formidable barrier separating various species (Toledo and Ordonez, 1993; Edwards et al., 2001) and may be responsible for limiting the distribution of these two species in this geographic region. Second, S. hispidus from southern Texas occupied a more arid-plains-grassland habitat (Arid/Semi Arid Terrestrial Ecological Zones; Toledo and Ordonez, 1993), whereas S. toltecus appeared to occupy more humid grasslands of the coastal lowlands in eastern and southeastern Mexico (Subhumid Tropical and Humid Tropical Terrestrial Ecological Zones; Toledo and Ordonez, 1993). Unfortunately, Carroll et al. (2005) could not determine whether the Rio Grande played a role in the distributional limits of S. hispidus and S. toltecus, or if habitat was a more influential component.
Concerning the separation of S. hirsutus and S. toltecus in southern Mexico, Carroll et al. (2005) proposed the distribution of S. toltecus to include the coastal grasslands east of the Sierra Madre Oriental, south to grasslands associated with the Yucatan Peninsula and lowlands of Chiapas (Subhumid Tropical and Humid Tropical Ecological Zones; Toledo and Ordonez, 1993); whereas, S. hirsutus occupied medium- to high-elevation-grassland habitats associated with the Guatemalan Highlands. In central Chiapas (near Ocozocoautla, Chiapas) the two species were reported as being sympatric (Carroll et al., 2005), with two individuals of S. toltecus and three individuals of S. hirsutus being collected in the same trapline. In reference to habitat, this locality represents an interface between lowland grasslands and more montane grasslands associated with the Guatemalan Highlands. However, an individual of S. hirsutus from the coastal lowlands of southern Mexico (La Blanca, Oaxaca) complicated this view.
In this study, DNA sequences from the mitochondrial cytochrome-b gene (Cytb) were obtained from 19 additional specimens of the three species of Sigmodon to increase sample sizes in geographic regions where taxa were parapatric or sympatric. These data were combined with data from Carroll et al. (2005) and Peppers and Bradley (2000) to assess phylogenetic patterns within taxa, evaluate habitat associations and preferences, and determine species boundaries.
Materials And Methods
Samples
Nineteen individuals were collected from natural populations through-out the ranges of S. hirsutus, S. hispidus, and S. toltecus (Fig. 1). Animal care and use procedures followed guidelines approved by the American Society of Mammalogists (Animal Care and Use Committee 1998). In addition, 50 DNA sequences reported in Carroll et al. (2005), Peppers and Bradley (2000), Peppers et al. (2002), and Smith and Patton (1999) were included in this study. Initial identification of specimens as S. hispidus-like was confirmed by morphology and karyology following diagnostic characteristics listed in Bailey (1902), Zimmerman (1970), Elder (1980), Hall (1981), Elder and Lee (1985), and Carleton et al. (1999); however, final identification was based on DNA sequences. Specimen numbers and collection localities are listed in Appendix I.
Fig. 1.
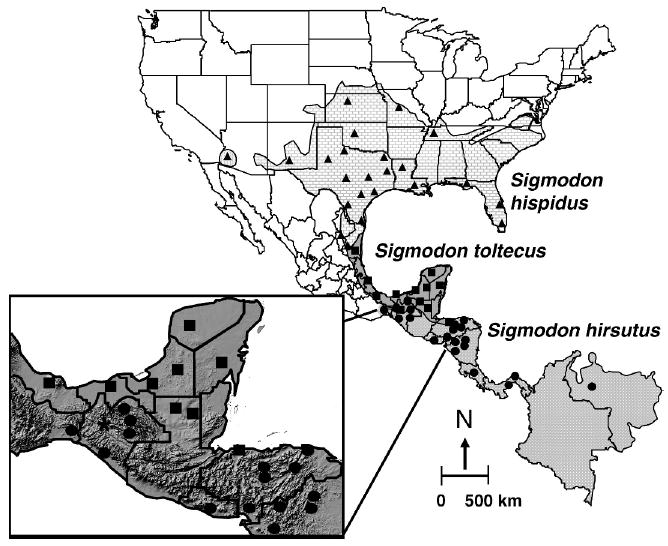
Distribution of members of the hispidus species group (Sigmodon hirsutus, S. hispidus, and S. toltecus). Localities from which samples were obtained are indicated as follows: S. hirsutus (circles), S. hispidus (triangles), and S. toltecus (squares). An asterisk indicates locality where samples of S. hirsutus and S. toltecus were sympatric.
Sequence Data
Genomic DNA was extracted from frozen samples of liver (0.1 g) using the Qiagen DNeasy Tissue Kit (Qiagen, Valencia, California). The complete Cytb gene (1,143 base pairs) was amplified using the following polymerase-chain-reaction (PCR) parameters modified from Saiki et al. (1988): 35 cycles of 95°C denaturation (1 min), 50°C annealing (1 min), 72°C extension (2 min), and 1 final 72°C extension cycle (7 min). Primers used in the PCR reactions were MVZ05 (5′ end; Smith and Patton, 1993) or H15915 (3′ end; Irwin et al., 1991) and P3′ (Tiemann-Boege et al., 2000) or MVZ14 (3′ end; Smith and Patton, 1993). The resulting PCR product was purified using the QIAquick PCR purification kit (Qiagen, Valencia, California). The PCR primers and following five primers were used in cycle sequencing reactions to amplify three fragments (ca. 400 base pairs each) on the forward and reverse strands, respectively: F1 (Whiting et al., 2003), 400R (Tiemann-Boege et al., 2000), 700H and 700L (Peppers and Bradley, 2000), and 870R (Peppers et al., 2002). Cycle sequencing was conducted using the ABI Big Dye terminator v3.1 reaction mix (PE Applied Biosystems, Foster City, California) and samples were analyzed on an ABI 3100-Avant automated sequencer (PE Applied Biosystems, Foster City, California). Sequencher 3.0 software (Gene Codes, Ann Arbor, Michigan) was used to align and proof nucleotide sequences. All Cytb sequences obtained in this study were deposited in GenBank and are listed in Appendix I.
Data Analyses
Based on phylogenetic relationships presented in Peppers et al. (2002), S. alstoni was used as the outgroup taxon in all analyses. In addition, representatives of S. alleni (n = 1), S. arizonae (n = 2), S. fulviventer (n = 2), S. leucotis (n = 1), S. mascotensis (n = 5), S. ochrognathus (n = 2), and S. peruanus (n = 2), were included as references. These taxa were identified by Peppers et al. (2002) as being either sister taxa or as being closely related to certain members of the ingroup taxa. Bayesian and parsimony models (PAUP*; Swofford, 2002; MrBayes 2.01; Huelsenbeck and Ronquist, 2001) were used to generate hypotheses concerning phylogenetic relationships of taxa.
Parsimony analyses were constructed using equally weighted characters and the heuristic search option with tree-bisection-reconnection, random stepwise addition of taxa, and 100 repetitions to obtain the most-parsimonious tree-set. Phylogenetically uninformative characters were excluded from analyses and variable nucleotide positions within the dataset were treated as unordered, discrete characters with four possible states; A, C, G, or T. Due to computational limitation, a Fast-step-wise-addition bootstrap analysis (PAUP*; Swofford, 2002) with 1,000 iterations was used to assess nodal support.
A Bayesian analysis (MrBayes 2.01; Huelsenbeck and Ronquist, 2001) using a GTR+I+G model with rates set equal to the proportion of invariable sites and sites partitioned by codon was used with the following options: 4 Markov-chains (one cold and three heated), 10 million generations, sample frequency = every 1,000th generation. S. alstoni was designated as the outgroup and two runs were conducted simultaneously. The GTR+I+G model was chosen so that data analyses were comparable to those conducted by Carroll et al. (2005). After a visual inspection of likelihood scores, convergence statistics, and potential scale-reduction factors the first 100,000 trees were discarded and a consensus tree (50% majority rule) was constructed from remaining trees. Clade probabilities were estimated and values ≥95 were used as indication of nodal support.
Kimura two-parameter genetic distances (Kimura, 1980) were estimated for individuals within each of the three respective taxa. The Kimura two-parameter was chosen so that genetic-distance values were comparable to those reported in Peppers and Bradley (2000) and Carroll et al. (2005). These distances were used to evaluate genetic divergences relative to geographic regions based on structure of clade obtained from parsimony and Bayesian analyses.
Results
Complete nucleotide sequences (1,143 base pairs) from the mitochondrial Cytb gene were included from 69 individuals, representing S. hirsutus (n = 29), S. hispidus (n = 25), and S. toltecus (n = 15). Both phylogenetic analyses (parsimony and Bayesian) produced similar topologies. Given the levels of molecular evolution relative to branch lengths, only the topology recovered from the Bayesian analysis was depicted (Fig. 2). Clade probability and bootstrap support values were superimposed on the tree recovered from the Bayesian analysis. DNA sequences were included for S. alleni, S. alstoni, S. arizonae, S. mascotensis, S. ochrognathus, and S. peruanus solely for the purpose of reference; therefore, the phylogenetic relationships of these taxa were not described in detail.
Fig. 2.
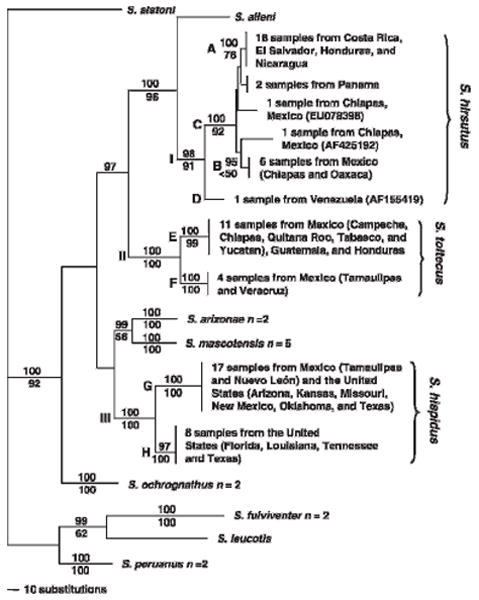
Phylogenetic tree obtained from the Bayesian analysis of 69 taxa. Major clades are depicted by Roman numerals (I–III) and subclades by letters (A–H). Clade probability values are indicated above branches and bootstrap values are indicated below branches. Branch lengths for multiple individuals of a species were collapsed for clarity. Vertical bars at terminal nodes indicate that genetic changes are so small that branch lengths are virtually non-existent.
In the parsimony analysis, 394 phylogenetically informative characters generated 172,798 equally parsimonious trees (1,428 steps, consistency index = 0.401, retention index = 0.864). A strict consensus tree generated a topology (not shown) similar to that obtained in the Bayesian analysis described below. Bootstrap values (Fig. 2) were high for the S. hirsutus clade (91), the S. toltecus clade (100), and the S. hispidus clade (100).
In the Bayesian analyses, the two simultaneous runs generated identical topologies (Fig. 2) and clade probability values differed only slightly; therefore, if support values differed at a node, both were reported. In this analysis, a monophyletic clade (I, clade probability value = 98) was formed containing 29 individuals that were referable to S. hirsutus. Within this clade, four supported subclades were identified (A through D): subclade A contained 20 individuals from Central America (Costa Rica, El Salvador, Honduras, and Nicaragua), B contained six individuals from Mexico (Chiapas and Oaxaca), C was comprised of individuals from clades A and B plus two individuals from Chiapas, Mexico, and two individuals from Panama, and D contained one individual from South America (Venezuela). The S. toltecus clade (II) included 15 individuals that formed a monophyletic clade (clade probability = 100) and two subclades (E and F). Subclade E contained seven individuals from southeastern Mexico (Campeche, Chiapas, Quintana Roo, Tabasco, and Yucatán) and four individuals from Guatemala and Honduras, whereas subclade F contained four individuals from northeastern Mexico (Tamaulipas and Veracruz). The 25 individuals formed a monophyletic clade (III, clade probability = 100) and were referred to as S. hispidus. Two supported subclades (G and H) were present with subclade G contained 15 individuals from the western United States and 2 individuals from northern Mexico (Tamaulipas and Nuevo León); whereas F represented individuals primarily restricted to the southeastern United States.
Kimura two-parameter (Kimura, 1980) genetic distances were estimated within each species and selected clades and are presented in Table 1. Genetic distances between the three species ranged from 12.55% (S. hirsutus and S. hispidus) to 12.90% (S. hirsutus and S. toltecus). Genetic distances within the three species ranged from 2.34% (S. hirsutus) to 2.81% (S. toltecus). Genetic distance values within and between clades ranged from 0.56% (within clade A) to 4.89% (between clades C and D).
Table 1.
Average genetic distances (Kimura, 1980) for selected taxa and clades of Sigmodon.
| Taxa | Average genetic distance (%) |
|---|---|
| S. hirsutus versus S. alleni | 9.77 |
| S. hirsutus versus S. toltecus | 12.90 |
| S. hirsutus versus S. hispidus | 12.55 |
| S. toltecus versus S. hispidus | 12.81 |
| S. hispidus versus S. arizonae | 11.32 |
| S. hispidus versus S. mascotensis | 11.04 |
| S. arizonae versus S. mascotensis | 8.75 |
| Within S. hirsutus | 2.34 |
| Within Clade A | 0.56 |
| Within Clade B | 3.92 |
| Within Clade C | 2.15 |
| Within Clade D | — |
| Clade A versus Clade B | 3.99 |
| Clade C versus Clade D | 4.89 |
| Within S. toltecus | 2.81 |
| Within Clade E | 1.46 |
| Within Clade F | 1.42 |
| Clade E versus Clade F | 4.69 |
| Within S. hispidus | 2.78 |
| Within Clade G | 1.17 |
| Within Clade H | 0.76 |
| Clade G versus Clade H | 4.80 |
Discussion
Phylogenetic analyses (parsimony and Bayesian) support the overall conclusions of Peppers et al. (2002) and Carroll et al. (2005) that S. hirsutus, S. hispidus, and S. toltecus represent valid species. Individuals of S. hirsutus formed a monophyletic group sister to S. alleni. The 15 individuals representing S. toltecus formed a monophyletic clade sister to the S. alleni and S. hirsutus clade. Individuals of S. hispidus formed a single clade; however, the sister taxon to S. hispidus was not resolved with this dataset, nor has it been resolved in other molecular studies (Carroll and Bradley, 2005; Carroll et al., 2005). Phylogenetic relationships of other members of the genus Sigmodon are depicted in Fig. 2, but are not discussed herein as they are beyond the scope of this study. Below, the geographic distribution and biogeographic parameters for each taxon are discussed in light of the newly examined samples.
Sigmodon hirsutus
The distribution of S. hirsutus remains relatively unchanged from that described by Carroll et al. (2005). Additional samples examined herein document the presence of S. hirsutus in Honduras and Nicaragua. These samples provide evidence for a distribution from southern Mexico (Chiapas and Oaxaca) into northern South America (Venezuela); however, four observations prevent this from being a simple and straightforward geographic range. First, as discussed by Carroll et al. (2005), S. hirsutus primarily occurs in medium-to-highelevation grasslands of the Central America Highlands or Guatemalan Highlands (Ryan, 1963; Carleton et al., 2002), whereas S. toltecus appears to be restricted to the coastal regions (Subhumid Tropical and Humid Tropical Terrestrial Ecological Zones; Toledo and Ordonez, 1993) of eastern Mexico (discussed below). However, both species were recorded in sympatry in north-central Chiapas, Mexico (near Ocozocoautla), where lowland grasslands of the Depression Central de Chiapas transition into those occurring at more moderate elevations in the Sierra Madre de Chiapas (Ferrusquía-Villafranca, 1993). Therefore, it is possible that one or both species may be encountered in the foothills in central Chiapas. Second, samples of S. hirsutus from southeastern Oaxaca (La Blanca) and southwestern Chiapas (Mapastepec) are from coastal grassland habitats, more typically associated with S. toltecus. Third, samples of S. hirsutus from Capiro y Calenturo, Honduras, were collected within 1 km of the coast; however, these were collected from a foothill region (a few hundred meters in elevation). Fourth, although our study included only two samples from Panama and did not include samples from Costa Rica, other authors (Goodwin, 1946; Handley, 1966; McPherson, 1985) have reported cotton rats from low-elevation scrubland and savanna habitats. These cotton rats formerly were assigned to S. hispidus, but given more recent interpretations of this species complex they presumably are referable to S. hirsutus.
In general, it appears that the key to interpreting the distribution of S. hirsutus lies between central Chiapas and northern Honduras. In Chiapas, two mountain ranges (Montañas del Norte de Chiapas and Sierra Madre de Chiapas) provide suitable habitat (moderate-elevation grasslands) for S. hirsutus and presumably restrict the distribution of S. toltecus to the northeast along the coastal regions of the Yucatan Peninsula. However, in north-central Chiapas, a lowelevation trough (Meseta Central de Chiapas and Depression Central de Chiapas) is formed between the two mountain regions and allows for an interdigitation between the coastal grasslands and the grasslands associated with medium-to-high elevations as described for the area of sympatry between S. hirsutus and S. toltecus near Ocozocoautla. The southern boundary of this trough (Sierra Madre de Chiapas) presumably prevents S. toltecus from occupying the lowelevation grasslands of southwestern Chiapas and southern Oaxaca, but provided an opportunity for S. hirsutus to expand its distribution from more southern regions; although it is not clear whether this would have been a recent or older invasion.
Sigmodon hirsutus appears to be the primary inhabitant of Central America except for the occurrence of S. toltecus along the northern coast of Honduras. The presence of S. toltecus in this region depends upon availability of suitable habitat (coastal or lowland grasslands) along the Yucatan Peninsula and Belize to about Tela and La Ceiba in northern Honduras. The Cordillera Nombre de Dios range may restrict the distribution of S. toltecus to the east and provides suitable habitat for S. hirsutus to the south. This hypothesis is supported by the presence of S. toltecus at Tela (Jardin Botanico Lancetilla) and samples of S. hirsutus 150 km E Tela (Trujillo, Parque Nacional Capiro y Calentura) and 100 km S Tela (La Guama).
Specimens comprising clades A, B, and D conform to three general geographic regions discussed above. Clade A contained 18 individuals from the Central American Highlands (Costa Rica, El Salvador, Honduras, and Nicaragua), clade B contained 6 individuals from the foothills and low-elevation grasslands of southern Mexico (southeastern Oaxaca and central and western Chiapas), whereas clade D contained the single individual from South America (Venezuela). However, two caveats hinder this simplistic interpretation. First, the two individuals from Panama were not included in clade A and second, two individuals from Chiapas were not contained in clade B, although, these four individuals are part of the larger clade C, which contains all specimens from Mexico and Central America.
The delineation and divergence between clades C and D may be explained by the influences of an emerging and disappearing Panamanian Land Bridge along the Isthmus of Panama, which may have acted to isolate the more northern forms from those to the south. Average genetic distances between those two clades (4.89%, Table 1) indicated a molecular divergence of the two clades ca. 1.5–2 million years ago (based on an approximation of 2.5–3.5%/million year divergence rate in the mammalian mitochondrial Cytb gene; Arbogast and Slowinski, 1998). If these rates and estimates are valid, then clades C and D may represent independent evolutionary trajectories. Voss (1992) examined the available specimens of S. hirsutus from South America and concluded that they are not morphologically different from more northern samples. Unfortunately, lack of sufficient genetic data from South America precludes a satisfactory test of this hypothesis.
Phylogenetic differentiation between clades A and B is more complex. Genetic distances (Table 1) between these two clades (3.99%) suggest a substantial divergence, although geographically some members of each group are in proximity. It is further complicated if genetic variation within clades is considered. For example, within clade A average genetic distance among individuals was low (0.56%) compared to that for members of clade B (3.92%). It appears that individuals from the Central American Highlands are genetically homogenous, whereas individuals from foothills and lowlands of Oaxaca and Chiapas may have been isolated by factors associated with establishment of the Isthmus of Tehuantepec.
Sigmodon toltecus
Evidence obtained from this study alters the conclusions of Carroll et al. (2005) concerning the distribution of S. toltecus and refutes the possibility that the Rio Grande forms a barrier between S. toltecus and S. hispidus. Based on newly acquired samples, S. toltecus appears to be distributed from northeastern Mexico (Tamaulipas and Nuevo León) to the northern coast of Honduras. In Mexico, S. toltecus typically is found in the coastal grasslands east of the Sierra Madre Oriental range. As discussed previously, the distribution of S. toltecus in Chiapas is complex and an area of sympatry with S. hirsutus has been described (Carroll et al., 2005). In southeastern Mexico, S. toltecus occurs throughout the Yucatan Peninsula to Belize and the northern coast of Honduras (see above). The distribution of S. toltecus in northern Mexico is difficult to discern, as few samples were available for study and the fact that individuals of S. hispidus were collected in proximity to individuals of S. toltecus (La Carbonera, Tamaulipas, and Doctor Arroyo, Nuevo León). To date, the northern-most sample of S. toltecus is from Calabazas, Tamaulipas (near Cuidad Victoria), located to the east of the Sierra Madre Oriental range. Doctor Arroyo, Nuevo León, is located ca. 100 km W of Calabazas and is located on the western side of the Sierra Madre Oriental range. It is plausible that this mountain range serves to isolate S. hispidus and S. toltecus. The individual of S. hispidus from La Carbonera is located ca. 16 km NE Calabazas and is on the coast of central Tamaulipas. It may be that some of the smaller mountain ranges (Sierra de San José de las Rusias, Sierra de Tamaulipas, and Sierra de San Carlos) affect the distribution of S. hispidus and S. toltecus in east-central Tamaulipas.
Individuals of S. toltecus formed two clades that correspond to a southern geographical group (E) and a northern group (F). Clade E consists of a genetically homogenous series of individuals (average genetic distance between samples = 1.46%, Table 1) from coastal lowlands of southern Mexico and northeastern Central America. Clade F was comprised of individuals from Tamaulipas and Veracruz, Mexico, that also were genetically similar (average genetic distance between samples = 1.42%). However, the two clades differed by a value of 4.69% indicating some degree of historical isolation that may correspond to the Isthmus of Tehuantepec.
Sigmodon hispidus
As discussed above, data collected from newly acquired samples of S. hispidus refute the hypothesis of Carroll et al. (2005) concerning the possibility of the Rio Grande forming a barrier to these two taxa. As discussed above, it now appears that S. hispidus is distributed from central-southern United States into northern Mexico (Tamaulipas), although there are few available samples from northern Mexico to assist in a thorough characterization of the southern portion of its range. Samples from Coahuila, Nuevo León, San Luis Potosí, and Tamaulipas will be crucial in determining the ranges of S. hispidus and S. toltecus. This especially will be true where the interdigitation of mountain ranges and intervening valleys may complicate the issue, or even provide opportunities for sympatry.
Carroll et al. (2005) and Phillips et al. (2007) proposed that samples of S. hispidus genetically could be separated into eastern and western clades. Based on genetic-distance values (4.80%, Table 1), these two clades are divergent, whereas there is little divergence within each clade (1.17% western, 0.76% eastern). Carroll et al. (2005) postulated that boundaries of the two genetic forms could be tentatively explained by habitat preferences with western clade members being associated with plains grasslands and eastern clade members being associated with southeast hardwood grasslands. Alternatively, the eastern and western forms may have been isolated as a result of a some vicariant event and rejoined at a latter date. Phillips et al. (2007) provided evidence that the two genetic lineages hybridized in eastern Texas and that the contact zone coincided with the interface of the post oak savannah and piney woods. The possibility of two ecological species (eastern and western) remains to be resolved.
Based on data from this study, individuals of S. hispidus from northern Mexico (Nuevo León and Tamaulipas) phylogenetically are more similar to individuals from the western United States. It is not clear whether samples in northern Mexico represent an extension of populations across the Rio Grande from southern Texas or are they connected, via the Mesa Central, to samples from farther west. More sampling is needed in northern Mexico where S. hispidus and S. toltecus are in proximity. Also, a broader sampling scheme is needed in southern Mexico and northern Central America to determine the distribution and habitat affiliation of S. toltecus and S. hirsutus, as well as the possible relationship of S. zanjonensis (Carleton et al., 1999; Musser and Carleton, 2005). Comparison of genetic-distance values between major clades (A and B, C and D, E and F, and G and H) produced high levels of intraspecific genetic divergence that should be further investigated in light of the possibility of genetic isolation without substantial morphological differentiation (genetic species concept, sensu Baker and Bradley, 2006). Genetic-distance values (Table 1) between these clades were ca. 4–5% indicating a substantial divergence time. In addition, clades corresponded to either different habitats or regions potentially separated by geographic barriers.
Acknowledgments
We thank M. J. Hamilton, R. A. Van Den Bussche, and the 2001, 2004, 2005, and 2006 Field Methods class at Texas Tech University for assistance in collecting specimens. Tissue loans were provided by T. Lee (Abilene Christian University Natural History Collection), R. Dowler (Angelo State Natural History Collections), D. S. Rogers (Monte L. Bean Life Science Museum), M. Hafner (Louisiana State University Museum of Natural History), T. L. Yates (Museum of Southwestern Biology), R. J. Baker (Museum of Texas Tech University), J. L. Patton (Museum of Vertebrate Zoology), Karen McBee (Oklahoma State University), M. D. Engstrom (Royal Ontario Museum), C. F. Fulhorst (University of Texas Medical Branch at Galveston), and R. Tesh (University of Texas Medical Branch at Galveston). This research was supported in part by the National Institutes of Health for projects entitled “Ecology of Emerging Arenaviruses in the Southwestern U.S.” (DHHS A141435-01) and “West Texas Rural EXPORT Center” (R24MD001097) and J. B. Sowell (Sowell Expeditions).
Appendix I
Specimens Examined
The specimens examined are listed below by museum acronym and catalogue number (Hafner et al., 1997) and GenBank accession number. In some cases, no museum catalogue number was available so a collector number was reported. All localities are in the United States unless otherwise specified. Abbreviations for museum catalogue or collector numbers are as follows: Abilene Christian University Natural History Collection (ACUNHC); Angelo State Natural History Collections (ASU); Brigham Young University (BYU); Centro Interdisciplinario de Investigacion para el Desarrollo Integral Regional Unidad Durango (CRD), Collection of Tissues, Oklahoma State University (OSU); John C. Patton (JCP); Midwestern State University (MWSU), Museum of Texas Tech University (TTU and TK); Museum of Natural Science, Louisiana State University (LSUMZ); Museum of Vertebrate Zoology (MVZ); Royal Ontario Museum (ROM), Museum of Southwestern Biology, (MSB); and University of Texas Medical Branch at Galveston (T and FSH). In some cases, localities are provided with Universal Transverse Mercator (UTM) coordinates. Sequences generated in this study are identified by GenBank accession numbers EU073164-EU073182 and EU078398; all others were reported by Peppers and Bradley (2000), Peppers et al. (2002), Carroll et al. (2005), or were obtained from GenBank.
Sigmodon alleni vulcani
MEXICO: Michoacán; 3.5 km N Tancotaro, 2,353 m (TK45276, AF155425).
Sigmodon alstoni
VENEZUELA: Portuguesa; Municipality Guanare, Gato Negro (just S Guanare) (T2140, AF293396).
Sigmodon arizonae major
MEXICO: Durango; San Juan de Camarones, UTM 13-385011E-2775718N (TTU81699, AF155423); Sonora; 20 km W Alamos (by road) (MSB55566, AF293398).
Sigmodon fulviventer dalquesti
Texas; Jeff Davis County, 2.4 km W Point of Rocks Park (MWSU17910, AF293399).
Sigmodon fulviventer fulviventer
MEXICO: Durango; 2.2 km S, 2.5 km E Vicente Guerrero (TK48915, AF293400).
Sigmodon hirsutus borucae
COSTA RICA: Puntarenas; Finca Mamos, Chomes, 60 m (BYU15259, AF108702).
Sigmodon hirsutus chiriquensis
PANAMA: Chiriquí; Hotel La Siesta, by airport (TTU39163, AF155416); Canal Zone; Gamboa (ROM104228, AF425191).
Sigmodon hirsutus griseus
EL SALVADOR: La Pax; 4.8 km NW San Luis Talpa (TK34810, AF155417). HONDURAS: Colon; Trujillo, Parque Nacional Capiro y Calentura, UTM 16-612297E-1758742N (TTU104184, EU073174; TTU104185, EU073176; TTU103971, EU073175); Cortez; La Guama, UTM 16-395886E- 1649092N (TTU104420, EU073170; TTU104422, EU073171); Francisco Morazan; El Picacho Zoological Parque, UTM 16-497451E-1561275N (TTU83741, AY517528); Olancho; 4 km E Catacamas, Escuela de Sembrador, UTM 16-624523E-1637511N (TTU84767, AY517529; TTU84634, EU073172); Valle; 3 km N, 9 km SW San Lorenzo, UTM 16-442952E-1486788N (TTU83788, AY517530); 3 km N, 12.5 km SW San Lorenzo, UTM 16-441040E-1484097N (TTU83794, EU073173). NICARAGUA: Nueva Segovia; El Balsamo (TTU101442, EU073164; TTU100560, EU073168); Boaco; El Paraiso (TTU100570, EU073166; TTU100579, EU073165); Matagalpa; El Tigre (TTU105136, EU073169); Jinotega; El Cua (TTU108154, EU073167).
Sigmodon hirsutus hirsutus
VENEZUELA: Lara; Finca Santa María Sarano (T4632, AF155419).
Sigmodon hirsutus ssp
MEXICO: Chiapas; 14.4 km N Ocozocoaut la, UTM 15 -451772E-1864243N (TTU82801, AF425193; TTU82798, AF425196; TTU82799, AF425192; TTU82798, AF425196); Ixtapa, 12 km SE of Ixtapa (ROM97578, AF425197); Soyalo (ROM97584, AF425195); Mapastepec, Tutuan, Rancho El Trebol, UTM 15-491419E-1703719N (TK138046, EU078398); 2.1 km SW La Sombra (TTU108157, AF425198); Oaxaca; 2 km S La Blanca UTM 15-318523E-1833293N (TTU82796, AF425194).
Sigmodon hispidus berlandieri
MEXICO: Tamaulipas; 12.8 km S la Carbonera, UTM 14-627591E-2711834N (TTU108155, EU073177); Nuevo León; 8.7 km W Doctor Arroyo, UTM 14-369819E-2622301N (TTU108156, EU073178). New Mexico; Otero County, Holloman Air Force Base (TTU79008, AF188198). Texas; Cameron County, Brownsville (TK32481, AF425199); Dimmit County, Chaparral Wildlife Management Area UTM 14-458437E-3134279N (TTU80759, AF425200); Lubbock County, Lubbock Lake Landmark State Historical Park (TTU76231, AF435110); McMullen County, James Daughtrey Wildlife Management Area UTM 14-555436E-3153833 (TTU82847, AF425201); Refugio County, Guadalupe Delta Wildlife Management Area UTM 14-709976E-3146702N (TTU75154, AF155415).
Sigmodon hispidus eremicus
Arizona; Yuma County, 3.2 km S, 1 km W Gadsden UTM 11-707004E-3600561N (TTU78493, AF155421).
Sigmodon hispidus hispidus
Florida; Walton County, 16 km E Destin, Topsail Hill State Recreation Area (TTU77517, AF425206). Louisiana; Bossier Parish, Wright Island (LSUMZ23836, AF425205); East Baton Rouge Parish, (LSUMZ28519, AF425204). Missouri; Howard County, Booneville (TK28256, AF425203). Tennessee; Shelby County, Meeman Biological Station (TTU79181, AF425202).
Sigmodon hispidus littoralis
Florida; Brevard County, Sebastion Inlet State Park (TTU97859, AF425207; TTU97861, AF425208).
Sigmodon hispidus spadicipygus
Florida; Dade County, Homestead Airforce Reserve Base golf course (FSH33, AF155420).
Sigmodon hispidus texianus
Kansas; Ellis County, Hays (OSU13231, AF425213; OSU13227, AF425209). Okla Oklahoma; Caddo County, 3.2 km S, 1.6 km W Anadarko (TTU54962, AF155414). Texas; Cottle County, Matador Wildlife Management Area UTM 14-374841E- 3778551N (TTU78822, AF425212); Freestone County, Richland Creek Wildlife Management Area UTM 14-775688E-3542219N (TTU75347, AF425210); Kimble County, Junction, Texas Tech University Center UTM 14-425829E-3373218N (TTU40477, AF425211); Lamar County, Pat Mayse Wildlife Management Area (TTU80626, AF425227); Nacogdoches County, Alazon Bayou Wildlife Management Area (ASU4996, AF425214).
Sigmodon leucotis leucotis
MEXICO: Durango; 12 km E Ojitos, UTM 13-385011E-2775718 (TTU81692, AF293401).
Sigmodon mascotensis inexoratus
MEXICO: Jalisco; 2 km NW Mesconcitos UTM 13-808753E-2375677N (TTU82793, AF425215).
Sigmodon mascotensis mascotensis
MEXICO: Colima; 6 km W Colima (JCP1020, AF155424). MEXICO: Michoacán; Apatzingán (TK45737, AF425216); Patzcuaro (Lake) (JCP1061, AF296188); Oaxaca; Las Minas (TTU82794, AF425217).
Sigmodon ochrognathus
MEXICO: Durango; 24 km N Las Herreras (CRD1033, AF155422). Arizona; Cochise County, 3.2 km W Portal (ACUNHC500, AF155592).
Sigmodon peruanus
EQUADOR: El Oro; Progreso (MSB140112, AF293395); Guayas, Manglares Churute, Cerro Cimalon UTM 17-650092E-9732559N (TTU103494, EU073179).
Sigmodon toltecus furvus
HONDURAS: Atlantida; Jardin Botanico Lancetilla, UTM 16-451012E-1740282N (TTU84378, EU073180; TTU103840, EU073181). MEXICO: Quintana Roo; 6 km S, 1.5 km W Tres Garantias (ASNHC7480, AF293402).
Sigmodon toltecus microdon
MEXICO: Campeche; Escárcega (ASNHC7474, AF425218); LaValeta (ROM95219, AF425221); Tabasco; Jonuta (ROM96256, AF425220); Yucatán; Labna (ROM96474, AF425219).
Sigmodon toltecus saturatus
GUATEMALA: El Petén; El Remate (ROM99644, AF425223); 10 km N Tikal (ROM99640, AF425222). MEXICO: Chiapas; 14.4 km N Ocozocoautla, UTM 15-451772E-1864243N (TTU108152, AF425228; TTU108153, AF425224).
Sigmodon toltecus toltecus
MEXICO: Tamaulipas; 3.2 km W Calabazas, Rancho Calabazas (TTU44946, AF425225); Veracruz; Estación Biológica Morro de la Mancha, 19°35′23.80″ N, 96°22′49.2″ W, 8 m (MSB75587, AF155418); 30 km N, 3 km E Cardel, Estación Biologica La Mancha (FN29909, AF425226); Paso del Patel , UTM 14-665394E-2282430N (TTU105062, EU073182).
Contributor Information
Robert D. Bradley, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409-3131; Museum of Texas Tech University, Lubbock, TX 79409-3191.
Dallas D. Henson, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409-3131
Nevin D. Durish, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409-3131
Literature Cited
- Animal Care And Use Committee. Guidelines for the capture, handling, and care of mammals as approved by the American Society of Mammalogists. Journal of Mammalogy. 1998;79:1416–1431. [Google Scholar]
- Arbogast BS, Slowinski JB. Pleistocene speciation and the mitochondrial DNA clock. Science. 1998;282:1955a. [Google Scholar]
- Bailey V. Synopsis of the North American species of Sigmodon. Proceedings of the Biological Society of Washington. 1902;15:101–116. [Google Scholar]
- Baker RJ, Bradley RD. Speciation in mammals and the genetic species concept. Journal of Mammalogy. 2006;87:643–662. doi: 10.1644/06-MAMM-F-038R2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton MD, Fisher RD, Gardner AL. Identification and distribution of cotton rats, genus Sigmodon (Muridae: Sigmodontinae), of Nayarit, Mexico. Proceedings of the Biological Society of Washington. 1999;112:813–856. [Google Scholar]
- Carleton MD, Sánchez O, Urbano Vidales G. A new species of Habromys (Muridae: Neotominae) from México, with generic review of species definitions and remarks on diversity patterns among Mesoamerican small mammals restricted to humid montane forests. Proceedings of the Biological Society of Washington. 2002;115:488–533. [Google Scholar]
- Carroll DS, Bradley RD. Systematics of the genus Sigmodon: DNA sequences from beta-fibrinogen and cytochrome-b. Southwestern Naturalist. 2005;50:342–349. [Google Scholar]
- Carroll DS, Peppers LL, Bradley RD. Molecular systematics and phylogeography of the Sigmodon hispidus species group. In: ánchez-Cordero VS, Medellín RA, editors. Contribuciones mastozoológicas en homenaje a Bernardo Villa. Instituto de Biología e Instituto de Ecología, Instituto de Biología e Instituto de Ecología, Universidad Nacional Autónoma de México y Comision Nacional el Conocimiento y Uso de la Bioversidad; México: 2005. pp. 87–100. [Google Scholar]
- Edwards CW, Fulhorst CF, Bradley RD. Molecular phylogenetics of the Neotoma albigula species group: further evidence of a paraphyletic assemblage. Journal of Mammalogy. 2001;82:267–279. [Google Scholar]
- Elder FFB. Tandem fusion, centric fusion, and chromosomal evolution in the cotton rats, genus Sigmodon. Cytogenetics and Cell Genetics. 1980;26:199– 210. doi: 10.1159/000131441. [DOI] [PubMed] [Google Scholar]
- Elder FFB, Lee MR. The chromosomes of Sigmodon ochrognathus and S. fulviventer suggest a realignment of Sigmodon species groups. Journal of Mammalogy. 1985;66:511–518. [Google Scholar]
- Ferrusquía-Villafranca I. Biodiversity scenario: a review of terrestrial habitats. In: Ramamoorthy TP, Bye R, Lot A, Fa J, editors. Biological diversity of Mexico: origins and distribution. Oxford University Press; New York: 1993. pp. 3–107. [Google Scholar]
- Goodwin GG. Mammals of Costa Rica. Bulletin of the American Museum of Natural History. 1946;87:271–471. [Google Scholar]
- Hafner MS, Gannon WL, Salazar-Bravo J, Alvarez-Castañeda ST. Mammal collections in the Western Hemisphere: a survey and directory of existing collections. Allen Press; Lawrence, Kansas: 1997. [Google Scholar]
- Hall ER. The mammals of North America. Second edition. John Wiley & Sons; New York: 1981. [Google Scholar]
- Handley CO., Jr . Checklist of the mammals of Panama. In: Wenzel RL, Tipton VJ, editors. Ectoparasites of Panama. Field Museum of Natural History; Chicago, Illinois: 1966. pp. 753–795. [Google Scholar]
- Huelsenbeck JP, Ronquist FR. MRBAYES: Bayesian inference of phylogeny. Biometrics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Irwin DM, Kocher TD, Wilson AC. Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution. 1991;32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- McPherson AB. A biogeographical analysis of factors influencing the distribution of Costa Rican rodents. Brenesia. 1985;23:97–273. [Google Scholar]
- Musser GG, Carleton MD. Superfamily Muroidea. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. Third edition. Johns Hopkins University Press; Baltimore, Maryland: 2005. pp. 894–1531. [Google Scholar]
- Peppers LL, Bradley RD. Cryptic speciation in Sigmodon hispidus: evidence from DNA sequences. Journal of Mammalogy. 2000;81:332–343. [Google Scholar]
- Peppers LL, Carroll DS, Bradley RD. Molecular systematics of the genus Sigmodon (Rodentia: Muridae): evidence from the mitochondrial cytochrome b gene. Journal of Mammalogy. 2002;83:396–407. [Google Scholar]
- Phillips CD, Henard CA, Pfau RS. Amplified fragment length polymorphism and mitochondrial DNA analyses reveal patterns of divergence and hybridization in the hispid cotton rat (Sigmodon hispidus) Journal of Mammalogy. 2007;88:351–359. [Google Scholar]
- Ryan RM. The biotic provinces of Central America as indicated by mammalian distribution. Acta Zoologica Mexicana. 1963;6:1–54. [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Smith MF, Patton JL. The diversification of South American rodents: evidence from mitochondrial sequence data for the akodontine tribe. Biological Journal of the Linnean Society. 1993;50:149–177. [Google Scholar]
- Smith MF, Patton JL. Phylogenetic relationships and the radiation of sigmodontine rodents in South America: evidence from cytochrome b. Journal of Mammalian Evolution. 1999;6:89–128. [Google Scholar]
- Swofford DL. PAUP: phylogenetic analysis using parsimony (* and other methods). Version 4.0b10. Sinauer Associates, Inc., Publishers; Sunderland, Massachusetts: 2002. [Google Scholar]
- Tiemann-Boege I, Kilpatrick CW, Schmidly DJ, Bradley RD. Molecular phylogenetics of the Peromyscus boylii species group (Rodentia: Muridae) based on mitochondrial cytochrome b sequences. Molecular Phylogenetics and Evolution. 2000;16:366–378. doi: 10.1006/mpev.2000.0806. [DOI] [PubMed] [Google Scholar]
- Toledo VM, Ordonez MJ. Biodiversity scenario: a review of terrestrial habitats. In: Ramamoorthy TP, Bye R, Lot A, Fa J, editors. Biological diversity of Mexico: origins and distribution. Oxford University Press; New York: 1993. pp. 757–777. [Google Scholar]
- Voss RS. A revision of the South American species of Sigmodon (Mammalia: Muridae) with notes on their natural history and biogeography. American Museum Novitates. 1992;3050:1–56. [Google Scholar]
- Whiting AS, Bauer AS, Sites JW., Jr Phylogenetic relationships and limb loss in sub-Saharan African scincine lizards (Squamata: Scincidae) Molecular Phylogenetics and Evolution. 2003;29:583–598. doi: 10.1016/s1055-7903(03)00142-8. [DOI] [PubMed] [Google Scholar]
- Zimmerman EG. Karyology, systematics, and chromosomal evolution in the rodent genus, Sigmodon. Vol. 4. Michigan State University, Publications of the Museum, Biological Series; 1970. pp. 385–454. [Google Scholar]


