Abstract
Background.
Despite the significant improvement in internal medicine and supportive therapy in recent years, liver fibrosis/cirrhosis remains a serious health issue in hepatitis B virus (HBV) infected patients. Invasive liver biopsy is presently the best means of diagnosing cirrhosis, but it carries a significant risk and has well recognised limitations such as sampling error, hence the importance in developing early diagnosis biomarkers. With this aim, we performed a pilot proteomic study to assess this as a strategy for plasma marker detection in patients suffering from HBV-associated liver cirrhosis.
Methods.
Plasma from eight chronic HBV-infection patients and from eight HBV-related cirrhotic patients were selected and proteome profiles were created by two-dimensional electrophoresis. The strategy included the use of ProteoMiner enrichment kit for the reduction of highly abundance proteins (e.g. albumin and IgG) prior to proteomic analyses with the goal to improve detection of novel candidate markers.
Results.
One reproducible spot was found to be completely repressed in plasma samples from cirrhotic patients and mass spectrometry analysis identified this a specific variant of the gelsolin actin-depolymerizing factor. Though further investigations are needed, especially in term of clinical validation, to our knowledge this is the first time that gelsolin is proposed as potential biomarker in HBV-related liver pathologies.
Conclusions.
Our findings confirm the potential utility of gelsolin either as a prognostic marker or a replacement therapeutic agent to alleviate liver injury.
Keywords: hepatitis B virus (HBV), inactive chronic HBV-infection, HBV-associated liver cirrhosis, human plasma, biomarker discovery
Introduction
Hepatitis B virus (HBV) is the prototype member of the family Hepadnaviridae that also includes viruses that can infect higher primates such as chimpanzees, and lower primates such as tupaia1. Approximately 350 million individuals has been infected with HBV and each year, an estimated 1 million persons die from chronic complications of the disease. Although chronic hepatitis B infection has a worldwide distribution, the vast majority of infected persons reside in Asia, the Middle East or Africa2, where there is a concomitant high incidence of hepatocellular carcinoma (HCC)3. HBV is a non-cytopathic virus and chronic hepatitis B is developed when the immune response that normally clears the infection fails to have a function or is too weak to be effective. Thus, infections are almost always chronic following exposure of children younger than 1 year or of immunocompromised individuals4–6. HBV infection may or may not be symptomatic and the outcome of infection to a large extent is determined by the immune status of the individual7. Successful clearance and resolution of infection also depends on the age and immune status of the individual. The complications of chronic HBV infection are well known and include liver cirrhosis, liver cancer as well as liver failure8,9. Cirrhosis is a consequence of chronic liver disease characterized by replacement of liver tissue by fibrous scar tissue as well as regenerative nodules, leading to progressive loss of liver function. Liver cirrhosis could be reversible, and accurate diagnosis is crucial to the management of patients. Pathologic diagnosis with liver biopsy has long been the gold standard for assessing the degree of fibrosis, but it is an invasive procedure with inherent risk and sampling variability. Plasma-based tests of liver cirrhosis have attracted more attention in recent years because plasma sample can be easily obtained from blood collection of patients10. Human blood plasma is one of the most important proteome from a clinical and medical point of view and the discovery of new biomarkers is a very challenging process which has become the basis for preventive medicine. However, plasma is also the most complex human-derived sample for proteomic analysis because it contains the widest dynamic range of cellular protein species in the body. In fact, several plasmatic proteins are synthesized in the liver and the majority of these change their structures and abundance in response to liver disease11. Tens of thousands of proteins, with their cleaved or modified forms, have been estimated to be present in the plasma. A small number of proteins such as albumin, immunoglobulins, α-1-antitrypsin, transferrin, and haptoglobins are present in concentrations in the milligram to tens of milligrams per milliliter range and together account for as much as 90% of the total plasma protein called “highly-abundant proteins” (HAP)12. On the other hand, a large number of proteins, including many that are, or could be, diagnostically significant are comprised into “low-abundant proteins” (LAP) because when such proteins are released into around 6 L of blood, their final concentration becomes extremely low. The presence of “highly-abundant proteins” and “low-abundant proteins” represents the major problem in proteome studies which use plasma and serum samples because the first protein content masks the second one. Depletion of abundant plasma proteins will help in the discovery and detection of less abundant proteins that may prove to be informative disease markers13. To overcome the above-described difficulties, prefractionation methods have been recently developed14 and a novel sample preparation tool is now commercially available under the trade name of ProteoMiner15. This protein enrichment technology is based on the interaction of complex protein sample with large, highly diverse library of hexapeptides bound to a chromatographic support where each unique hexapeptide binds to a unique protein sequence. High-abundance proteins quickly saturate their ligands because of the bead capacity limits binding capacity and excess protein is washed out during the procedure. In contrast, low-abundance proteins are concentrated on their specific ligands, thereby decreasing the dynamic range of sample proteins. When analyzed in downstream applications (e.g., electrophoresis) the number of proteins detected is dramatically increased16. This peculiar property of revealing novel low-abundance species is of extremely interest within biomarker discovery investigations.
In our work, comparison between plasma proteomes from patients with liver cirrhosis associated to hepatitis B infection and chronic HBV-infection patients who were asymptomatic, has been performed to show a possible approach for plasma biomarker discovery and validation. Before proteomic analysis, the sample has been treated by ProteoMiner™ Protein Enrichment Technology and then analysed by 2D-IEF-SDS-PAGE and MS/MS to search for candidate markers of this liver disease.
Materials and methods
Plasma samples
With patients’ consent, we collected plasma samples from a total of 16 HBsAg positive and HBeAg negative subjects attending the Hepatitis Clinic of Shariati Hospital, Tehran University of Medical Sciences. 8 were patients with inactive chronic HBV-infection and 8 with HVB-related liver cirrhosis. Only patients with HBV DNA detected using a non-commercial hemi-nested PCR (sensitivity of approximately 100 copies/mL)17 were recruited for this study, whereas patients with undetectable HBV DNA, or those with evidence of concomitant hepatitis C or D virus infection, HIV infection, autoimmune, drug-induced or other causes of chronic liver disease were excluded. Inactive chronic HBV-infection defined as chronic hepatitis B with persistently normal ALT levels for 6 months prior to enrolment. Cirrhosis was defined as the presence of fibrosis stage 4 or more than 4 in HAI score18. Demographic data and plasma samples were collected at the initial assessment before liver biopsy and stored at −70°C prior to analysis. None of patients received antiviral treatment prior to liver biopsy. HBV DNA was quantified in the Light-Cycler (Roche) using the RealART™ HBV LC PCR (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The protocol for this study was approved by the ethics committee of Shariati Hospital. Detailed information on subjects enrolled in proteomics analyses are shown in Table I.
Table I.
Demographic, biochemical and virologic characteristics of patients with chronic hepatitis B
| Patients with chronic hepatitis B (n = 8) | Patients with cirrhosis (n = 8) | P value | |
|---|---|---|---|
| Age (Years) | 37 ± 7 | 44 ± 7.5 | 0.07 |
| Gender M/F | 4/4 | 6/2 | - |
| Viral load (copies/mL) | 6.9 ± 10.3 E3 | 647 ± 169 E3 | 0.3 |
| ALT (IU/L) | 30 ± 7 | 72 ± 23 | 0.001 |
| AST (IU/L) | 32 ± 6 | 79 ± 28 | 0.002 |
| Platelets × 103 mm−3 | 174 ± 48 | 158 ± 56 | 0.5 |
| Prothrombin time(s) | 12.3 ± 0.4 | 13.9 ± 1.4 | 0.01 |
| Albumin (g/L) | 4.5 ± 0.4 | 4.2 ± 0.3 | 0.1 |
| HAI score | 4.5 ± 1.6 | 9.5 ± 1.4 | 0.001 |
| Bilirubin Total (mg/dL) | 1.0 ± 0.2 | 2.3 ± 0.9 | 0.05 |
ProteoMiner enrichment
Before proteomic analysis, each plasma sample was pre-fractioned using the ProteoMiner™ kit (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s protocol. The protein concentration of sample was 50mg/mL.
The equalized plasma samples were stored at −80 °C until use.
Quantification of plasma protein concentration
The protein concentration of each sample was determined according to Bradford19 using BSA as a standard curve. The protein concentration was estimed on whole plasma sample before and after ProteoMiner treatment.
Two-dimensional electrophoresis
To remove lipids, proteins were precipitated from a desired volume (containing 400 μg of proteins) of each sample with cold (4 °C) acetone (80% v/v) over-night, then centrifuged at 18000 g for 20 min. The supernatant was removed and the pellet was air-dried and then solubilized in the focusing solution 8 M urea, 4% (w/v) CHAPS, 0.5% (w/v) pH 4–7 carrier ampholyte (Bio-lyte; Bio-Rad, Hercules, CA, USA) and 40 mM Tris base with continuous stirring. Proteins were subsequently reduced (10 mM tributylphosphine, 1 h) and alkylated (40 mM IAA, 1 h). To prevent over-alkylation, iodoacetamide (IAA) excess was destroyed by adding 10 mM DTE. IEF was performed using Biorad Multiphore II and Dry Strip Kit (Bio-Rad-Protean-IEF-Cell-System). Seventeen centimeter IPG strips (Bio-Rad, Hercules, CA, USA) pH 4–7 were rehydrated overnight with 345 μL of rehydratation solution containing 8 M urea, 4% (w/v) CHAPS, 0.5% (w/v) pH 4–7 carrier ampholyte (Bio-lyte; Bio-Rad, Hercules, CA, USA), 10 mM DTE and 100μL of sample was loaded using the cup-loading method. The total product time × voltage applied was 80 000 V h for each strip at 20 °C. For the second dimension, IPG strips were incubated in the equilibration solution [6 M urea, 50 mM Tris-HCl (pH 6.8), 30% (v/v) glycerol, 3% (w/v) SDS, 0.002% (w/v) bromophenol blue] for 30 min with gentle agitation. Equilibrated strips were then placed on SDS-polyacrylamide gels, 16 cm × 20 cm, 11% acrylamide, and sealed with 0.5% (w/v) agarose. SDS-PAGE was performed using the Protean II xi Cell, large gel format (Bio-Rad) at constant current (35 mA per gel) at 7 °C until the bromophenol blue tracking dye was approximately 2–3 mm from the bottom of the gel. Protein spots were stained by Coomassie Brilliant Blue G-250 stain20.
Image analysis
Sixteen two-dimension stained gel were digitized and image analysis was performed with Progenesis SameSpots software vers. 2.0 (Nonlinear Dynamics), which allows novel spot detection. The gel image showing the highest number of spots and the best protein pattern was chosen as a reference template, and spots in a standard gel were then matched across all gels. Each gel was analysed for novel spot detection and background subtraction.
In-Gel Digestion
Protein spots were carefully excised from Coomassie stained gels and subjected to in-gel trypsin digestion according to Shevchenko and colleagues21 with minor modifications. The gel pieces were swollen in a digestion buffer containing 50 mM NH4HCO3 and 12.5 ng/μL of trypsin (modified porcine trypsin, sequencing grade, Promega, Madison, WI) in an ice bath. After 30 min the supernatant was removed and discarded, 20 μL of 50 mM NH4HCO3 were added to the gel pieces and digestion allowed to proceed at 37 ° C overnight. The supernatant containing tryptic peptides was dried by vacuum centrifugation. Prior to mass spectrometric analysis, the peptide mixtures were redissolved in 10 μL of 5% FA (Formic Acid).
Protein identification by MS/MS
Peptide mixtures were separated using a nanoflow-HPLC system (Ultimate; Switchos; Famos; LC Packings, Amsterdam, The Netherlands). A sample volume of 10 μL was loaded by the autosampler onto a homemade 2 cm fused silica precolumn (75 μm I.D.; 375 μm O.D.; Reprosil C18-AQ, 3 μm (Ammerbuch-Entringen, DE)) at a flow rate of 2 μL/min. Sequential elution of peptides was accomplished using a flow rate of 200 nL/min and a linear gradient from Solution A (2% acetonitrile; 0.1% formic acid) to 50% of Solution B (98% acetonitrile; 0.1% formic acid) in 40 minutes over the precolumn in-line with a homemade 10–15 cm resolving column (75 μm I.D.; 375 μm O.D.; Reprosil C18-AQ, 3 μm (Ammerbuch-Entringen, Germany)). Peptides were eluted directly into a High Capacity ion Trap (model HCTplus, Bruker-Daltonik, Germany). Capillary voltage was 1.5–2 kV and a dry gas flow rate of 10 L/min was used with a temperature of 230 °C. The scan range used was from 300 to 1800 m/z. Protein identification was performed by searching the National Center for Biotechnology Information non-redundant database (NCBInr, version 20100129, www.ncbi.nlm.nih.gov) using the Mascot program (in-house version 2.2, Matrix Science, London, UK). The following parameters were adopted for database searches: complete carbamidomethylation of cysteines and partial oxidation of methionines, peptide Mass Tolerance ± 1.2 Da, Fragment Mass Tolerance ± 0.9 Da, missed cleavages 2. For positive identification, the score of the result of (−10 x Log(P)) had to be over the significance threshold level (P < 0.05).
Results
Application of ProteoMiner™ Technology
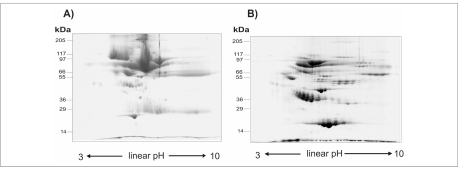
Inactive chronic HBV-infected plasma samples together with cirrhotic samples were analysed by 2D IEF-SDS-PAGE and proteins subsequently visualized by coomassie staining. Before to perform electrophoresis analysis on plasma collected from patients, efficiency of ProteoMiner™ Technology was tested. Figure 1 shows two representative maps of plasma control samples (inactive hepatitis B infection) before and after treatment with ProteoMiner enrichment kit. As expected, in the untreated sample albumin dominated the gel, obscuring signals from less abundance proteins. When an equal amount of protein is pre-treated with ProteoMiner, generated 2D gels showed a dramatically improved resolution and a greater number of spots was detected.
Figure 1.
Small size 2D gels (linear IPGs pH 3–10, 7 cm length) of plasma samples from inactive chronic HBV-infection patients before (A) and after (B) treatment with ProteoMiner enrichment kit (Bio-Rad Laboratories, Hercules, CA, USA). Total protein load: 200 μg.
Comparison between plasma proteome of both inactive HBV infected patients and patients suffering from HBV-associated liver cirrhosis
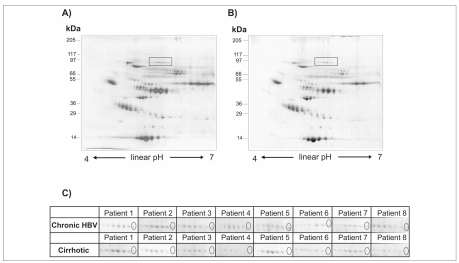
The experimental procedure involved large 2D gel electrophoresis (17 cm, pI 4-7) conducted on 8 plasma samples from inactive HBV-infected patients and 8 plasma samples from patients with hepatitis B virus-related liver cirrhosis. Eight biological and three technical replicates (for a total of 24 maps) per condition were performed and cross-compared by image software analysis Progenesis SameSpot (NonLinear Dynamics, Newcastle, UK) which generated the reference maps shown in Figure 2A–2B. As can be seen, the 2DE pattern is similar between samples. We found 624 spots that were common to all the gels and searching for qualitative changes resulted in only one spot of difference which was detected in 2-DE gels of inactive HBV infected plasma and was completely repressed in 2-DE map of plasma from patients with HBV-associated liver cirrhosis. Figure 2C displays sections of 2DE images showing reproducible detection of the candidate protein in all the inactive HBV infected patients which were analyzed. Interestingly, this protein appeared as the more basic spot of a train pattern, typically due to post-translational modifications (PTMs). The spot of interest was excised from the gels, digested by trypsin and peptide mixtures were then analysed by LC-ESI-MS/MS for protein identification. Mass spectrometry analysis identified this completely repressed protein as gelsolin actin-depolymerizing factor (ADF). Results are summarized in Table II.
Figure 2.
Representative analytic 2D-gels (linear IPGs pH 4–7, 17 cm length) of ProteoMiner pre-fractioned plasma samples. A, inactive HBV-infected patients. B, HBV-related cirrhotic patients. Total protein load: 400 μg. Boxes indicate gel regions which are zoomed in the panel below. C, close-up views of the 2D gel area showing the reproducible difference of the candidate marker protein. Ellipse indicates gelsolin spot totally repressed in patients suffering from HBV-associated liver cirrhosis.
Table II.
Detailed MS/MS peptide sequence analysis of the identified candidate biomarker
| Protein name | Mr, kDa theor/exper a | pI theor/exper a | NCBI Accession No. | Peptides identified by MS/MS | Mascot Ion Score | |||
|---|---|---|---|---|---|---|---|---|
| m/z | charge state | start-end b | sequence | |||||
| 500.76 | 2+ | 33–43 | R.GASQAGAPQGR.V | 42 | ||||
| 418.90 | 3+ | 62–72 | K.AGKEPGLQIWR.V | 36 | ||||
| 425.76 | 2+ | 169–177 | K.KGGVASGFK.H | 51 | ||||
| 361.61 | 2+ | 170–177 | K.GGVASGFK.H | 46 | ||||
| 638.36 | 2+ | 178–188 | K.HVVPNEVVVQR.L | 66 | ||||
| Gelsolin | 441.73 | 2+ | 361–368 | K.TASDFITK.M | 36 | |||
| [Homo sapiens] | 86.04/88.0 | 5.9/5.7 | gi|4504165 | 915.46 | 2+ | 374–390 | K.QTQVSVLPEGGETPLFK.Q | 54 |
| 528.76 | 2+ | 554–564 | R.EGGQTAPASTR.L + pyro-Glu (N-term E) | 31 | ||||
| 378.22 | 2+ | 578–584 | R.AVEVLPK.A | 55 | ||||
| 660.39 | 2+ | 585–597 | K.AGALNSNDAFVLK.T | 66 | ||||
| 919.46 | 2+ | 598–615 | K.TPSAAYLWVGTGASEAEK.T | 93 | ||||
| 444.25 | 2+ | 616–623 | K.TGAQELLR.V | 60 | ||||
| 758.30 | 2+ | 627–648 | R.AQPVQVAEGSEPDGFWEALGGK.A | 55 | ||||
| 488.81 | 2+ | 721–729 | K.TEALTSAKR.Y | 44 | ||||
| 379.22 | 2+ | 742–748 | R.TPITVVK.Q | 46 | ||||
theoretical Mr/pI was calculated with Mr/pI tool on the Expasy web site (http://expasy.org/tools/pi_tool.html)
start-end positions of identified peptides were calculated against complete amino acid sequence of the protein
Discussion
Currently, lack of robust biomarkers still limits evaluation of hepatic fibrosis stages and progression in chronic diseases, especially in HBV infection22,23. Liver biopsy remains the gold standard for assessment of hepatic fibrosis, but it is an invasive procedure with inherent risk and sampling variability24. In addition, the diagnostic accuracy depends on the size of the biopsy specimens25. Furthermore, intra- and inter-observer variation for interpretation of biopsies is 10–20%, even among experienced pathologists26. Serum-based tests of liver fibrosis have attracted more attention in recent years. In particular, comparison of proteomes of disease and control serum samples has been shown to be a possible approach for discovering serum biomarkers of liver diseases associated with hepatitis B infection27–30. Surface enhanced laser desorption/ionization (SELDI) was used to develop fingerprinting models for discriminating different stages of fibrosis and predicting fibrosis, cirrhosis and hepatocellular carcinoma in chronic HBV infections31–33. Although serum/plasma became an important resource for proteome analysis, and several depletion and fractionation technologies have been developed to remove highly abundant proteins such as albumin and immunoglobulin G, little study has been reported on the use of enrichment pre-treatment in biomarker discovery for HBV-associated liver diseases34. Recently, the ProteoMiner technology has been proposed as a promising and powerful alternative to common immuno-subtraction tools and a flurry of applications emerged in the literature15. For the first time, with this study the use of ProteoMiner was tested in searching new potential candidate markers in plasma samples of (HBV)-related cirrhotic patients. Though many differential proteins or discriminatory patterns were demonstrated in previous studies, most of them were focused on end-stage liver diseases, especially on hepatocellular carcinoma35–37. Moreover, these studies looked at differentially expressed proteins and not to absolute protein expression changes (newly expressed or totally repressed spots). Furthermore, to our knowledge, our preliminary study is the first on plasma samples of patients suffering of HBV-associated liver cirrhosis. Interestingly, our findings demonstrated the repression of a gelsolin-containing spot. Gelsolin is a highly conserved, multifunctional actin-binding protein initially described in the cytosol of macrophages and subsequently identified in many vertebrate cells. A unique property of gelsolin is that in addition to its widely recognized function as a cytoplasmic regulator of actin organization, the same gene expresses a splice variant coding for a distinct isoform, plasma gelsolin, which is secreted into extracellular fluids38. The secreted form of gelsolin was implicated in a number of processes such as the extracellular actin scavenging system and the presentation of lysophosphatidic acid and other inflammatory mediators to their receptors, in addition to its function as a substrate for extracellular matrix-modulating enzymes38. Consistent with these proposed functions, blood gelsolin levels decrease markedly in a variety of clinical conditions such as acute respiratory distress syndrome, sepsis, major trauma, prolonged hyperoxia, malaria, and liver injury39–44. Therefore, gelsolin could not be considered as a specific marker of HBV-related cirrhosis. However, our identified spot corresponded to a specific isoform of gelsolin. In fact, gelsolin protein generated spot trains in 2D maps, typically due to post-translational modifications or to small differences in amino acid composition (usually a splice variant). This progressive change in the pI and Mr of protein spots on serum/plasma 2-DE maps was reported before29,30 and detection of specific modified isoforms is a pathological hallmark45. As a rule of thumb, the correlation between blood gelsolin levels and several critical clinical conditions suggests the potential utility of gelsolin as a prognostic marker as well as the possibility for therapeutic replenishment of gelsolin to alleviate the injurious cascades in these settings38. On the other hand, the presence or absence of a particular isoform for this protein may be specific for a certain disease. To this regard, deeper investigations are being carried out in our lab with the intent to further characterize the detected gelsolin isoform which was found to be repressed in plasma samples of patients suffering from liver cirrhosis associated with hepatitis B infection. Additional experimentation is also needed to validate this protein as a real biomarker with approval of clinical efficiency. At any rate, the indication of new candidate markers may help in early diagnosis of HBV-related liver diseases.
Acknowledgments
This work was financially supported by the Italian National Blood Centre (CNS-ISS) and the “GENZOOT” research program funded by the Italian Ministry of Agriculture.
References
- 1.Seeger C, Mason WS. Hepatitis B Virus Biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre F. Hepatitis B epidemiology in Asia, the Middle East and Africa. Vaccine. 2000;18(Suppl 1):S20–22. doi: 10.1016/s0264-410x(99)00456-9. [DOI] [PubMed] [Google Scholar]
- 3.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(Suppl 1):S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Norman G. Hepatitis B: diagnosis, prevention, and treatment. Clin Chem. 1997;43:1500–06. [PubMed] [Google Scholar]
- 5.Villarejos VM, Visoná KA, Gutiérrez A, Rodríguez A. Role of saliva, urine and feces in the transmission of type B hepatitis. N Engl J Med. 1974;291:1375–8. doi: 10.1056/NEJM197412262912602. [DOI] [PubMed] [Google Scholar]
- 6.Karayiannis P, Novick DM, Lok AS, et al. Heatitis B virus DNA in saliva, urine, and seminal fluid of carriers of hepatitis B e antigen. Br Med J. 1985;290:1853–5. doi: 10.1136/bmj.290.6485.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang JJ, Lewin SR. Immunopathogenesis of hepatitis B virus infection. Immunol Cell Biol. 2007;85:16–23. doi: 10.1038/sj.icb.7100009. [DOI] [PubMed] [Google Scholar]
- 8.Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000;275:2247–50. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- 9.Canbay A, Taimr P, Torok N, et al. Apoptotic body engulfment by a human stellate cell line is profibrogenic. Lab Invest. 2003;83:655–63. doi: 10.1097/01.lab.0000069036.63405.5c. [DOI] [PubMed] [Google Scholar]
- 10.Dienstag JL. The role of liver biopsy in chronic hepatitis C. Hepatology. 2002;36:152–60. doi: 10.1053/jhep.2002.36381. [DOI] [PubMed] [Google Scholar]
- 11.Zolla L. Proteomics studies reveal important information on small molecule therapeutics. Drug Discovery Today. 2008;13:1042–51. doi: 10.1016/j.drudis.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson LN, Anderson NG. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol Cell Proteomics. 2002;1:845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Adkins JN, Varnum SM, Auberry KJ, et al. Toward a Human Blood Serum Proteome analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics. 2002;1:947–55. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed FE. Sample preparation and fractionation for proteome analysis and cancer biomarker discovery by mass spectrometry. J Sep Sci. 2009;32:771–798. doi: 10.1002/jssc.200800622. [DOI] [PubMed] [Google Scholar]
- 15.Boschetti E, Righetti PG. The ProteoMiner in the proteomic arena: a non-depleting tool for discovering low-abundance species. J Proteomics. 2008;71:255–264. doi: 10.1016/j.jprot.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Sennels L, Salek M, Lomas L, et al. Proteomic Analysis of Human Blood Serum Using Peptide Library Beads. J Proteome Res. 2007;6:4055–62. doi: 10.1021/pr070339l. [DOI] [PubMed] [Google Scholar]
- 17.Poutchi H, Mohamadkhani A, Bowden S, et al. Clinical significance of precore and core promoter mutations in genotype D hepatitis B-related chronic liver disease. J Viral Hepat. 2008;15:753–60. doi: 10.1111/j.1365-2893.2008.00998.x. [DOI] [PubMed] [Google Scholar]
- 18.Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–9. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 20.Candiano G, Bruschi M, Musante L, et al. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–33. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 21.Shevchenko A, Tomas H, Havlis J, et al. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2007;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 22.Rossi E, Adams LA, Bulsara M, Jaffrey GP. Assessing liver fibrosis whit serum marker models. Clin Biochem Rev. 2007;28:3–10. [PMC free article] [PubMed] [Google Scholar]
- 23.Rockey DC, Bissell DM. Noninvasive measures of liver fibrosis. Hepatology. 2006;43(Suppl 1):S113–20. doi: 10.1002/hep.21046. [DOI] [PubMed] [Google Scholar]
- 24.Grant A, Neuberger J. Guidelines on the use of liver biopsy in clinical practice. British Society of Gastroenterology Gut. 1999;45(Suppl 4):IV1–11. doi: 10.1136/gut.45.2008.iv1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheuer PJ. Liver biopsy size matters in chronic hepatitis: bigger is better. Hepatology. 2003;38:1356–8. doi: 10.1016/j.hep.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Soloway RD, Baggestoss AH, Schoenfield LJ, Summerskill WH. Observer error and sampling variability tested in evaluation of hepatitis and cirrhosis by liver biopsy. Am J Dig Dis. 1971;16:1082–6. doi: 10.1007/BF02235164. [DOI] [PubMed] [Google Scholar]
- 27.Poon TC, Johnson PJ. Proteome analysis and its impact on the discovery of serological tumor markers. Clin Chim Acta. 2001;313:231–9. doi: 10.1016/s0009-8981(01)00677-5. [DOI] [PubMed] [Google Scholar]
- 28.Poon TC, Yip TT, Chan AT, et al. Comprehensive proteomic profiling identifies serum proteomic signatures for detection of hepatocellular carcinoma and its subtypes. Clin Chem. 2003;49:752–60. doi: 10.1373/49.5.752. [DOI] [PubMed] [Google Scholar]
- 29.He QY, Lau GK, Zhou Y, et al. Serum biomarkers of hepatitis B virus infected liver inflammation: a proteomic study. Proteomics. 2003;3:666–74. doi: 10.1002/pmic.200300394. [DOI] [PubMed] [Google Scholar]
- 30.Steel LF, Shumpert D, Trotter M, et al. A strategy for the comparative analysis of serum proteomes for the discovery of biomarkers for hepatocellular carcinoma. Proteomics. 2003;3:601–9. doi: 10.1002/pmic.200300399. [DOI] [PubMed] [Google Scholar]
- 31.Cui J, Kang X, Dai Z, et al. Prediction of chronic hepatitis B, liver cirrhosis and hepatocellular carcinoma by SELDI-based serum decision tree classification. J Cancer Res Clin Oncol. 2007;133:825–34. doi: 10.1007/s00432-007-0224-y. [DOI] [PubMed] [Google Scholar]
- 32.Poon TC, Hui AY, Chan HL, et al. Prediction of liver fibrosis and cirrhosis in chronic hepatitis B infection by serum proteomic fingerprinting: a pilot study. Clin Chem. 2005;51:328–35. doi: 10.1373/clinchem.2004.041764. [DOI] [PubMed] [Google Scholar]
- 33.He QY, Zhu R, Lei T, et al. Toward the proteomic identification of biomarkers for the prediction of HBV related hepatocellular carcinoma. J Cell Biochem. 2008;103:740–52. doi: 10.1002/jcb.21443. [DOI] [PubMed] [Google Scholar]
- 34.Feng JT, Liu YK, Song HY, et al. Heat-shock protein 27: a potential biomarker for hepatocellular carcinoma identified by serum proteome analysis. Proteomics. 2005;5:4581–8. doi: 10.1002/pmic.200401309. [DOI] [PubMed] [Google Scholar]
- 35.Li N, Long Y, Fan X, et al. Proteomic analysis of differentially expressed proteins in hepatitis B virus-related hepatocellular carcinoma tissues. J Exp Clin Cancer Res. 2009;28:122. doi: 10.1186/1756-9966-28-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Na K, Lee EY, Lee HJ, et al. Human plasma carboxylesterase 1, a novel serologic biomarker candidate for hepatocellular carcinoma. Proteomics. 2009;9:3989–99. doi: 10.1002/pmic.200900105. [DOI] [PubMed] [Google Scholar]
- 37.Ren F, Chen Y, Wang Y, et al. Comparative serum proteomic analysis of patients with acute-on-chronic liver failure: alpha-1-acid glycoprotein maybe a candidate marker for prognosis of hepatitis B virus infection J Viral Hepat 2010. DOI 10.1111/j.1365-2893.2009.01242.x. [DOI] [PubMed]
- 38.Bucki R, Levental I, Kulakowska A, Janmey PA. Plasma gelsolin: function, prognostic value, and potential therapeutic use. Curr Protein Pept Sci. 2008;9:541–51. doi: 10.2174/138920308786733912. [DOI] [PubMed] [Google Scholar]
- 39.Lind SE, Smith DB, Janmey PA, Stossel TP. Depression of gelsolin levels and detection of gelsolin-actin complexes in plasma of patients with acute lung injury. Am Rev Respir Dis. 1988;138:429–34. doi: 10.1164/ajrccm/138.2.429. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Cheng B, Chen Q, et al. Time course of plasma gelsolin concentrations during severe sepsis in critically ill surgical patients. Crit Care. 2008;12:R106. doi: 10.1186/cc6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang S, Rhoads SL, DiNubile MJ. Temporal association between serum gelsolin levels and clinical events in a patient with severe falciparum malaria. Clin Infect Dis. 1997;24:951–4. doi: 10.1093/clinids/24.5.951. [DOI] [PubMed] [Google Scholar]
- 42.Suhler E, Lin W, Yin HL, Lee WM. Decreased plasma gelsolin concentrations in acute liver failure, myocardial infarction, septic shock, and myonecrosis. Crit Care Med. 1997;25:594–8. doi: 10.1097/00003246-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 43.Mounzer KC, Moncure M, Smith YR, DiNubile MJ. Relationship of admission plasma gelsolin levels to clinical outcomes in patients after major trauma. Am J Respir Crit Care Med. 1999;160:1673–81. doi: 10.1164/ajrccm.160.5.9807137. [DOI] [PubMed] [Google Scholar]
- 44.Spinardi L, Witke W. Gelsolin and disease. Subcell Biochem. 2007;45:55–69. doi: 10.1007/978-1-4020-6191-2_3. [DOI] [PubMed] [Google Scholar]
- 45.Fernández-Irigoyen J, Santamaría E, Sesma L, et al. Oxidation of specific methionine and tryptophan residues of apolipoprotein A-I in hepatocarcinogenesis. Proteomics. 2005;5:4964–72. doi: 10.1002/pmic.200500070. [DOI] [PubMed] [Google Scholar]