SUMMARY
Regulation of stem cells depends on both tissue-specific transcriptional regulators and changes in chromatin organization, yet the coordination of these events in endogenous niches is poorly understood. In the Drosophila testis, local JAK-STAT signaling maintains germline and somatic stem cells (GSCs and cyst progenitor cells, or CPCs) in a single niche. Here we show that epigenetic regulation via the nucleosome remodeling factor (NURF) complex ensures GSC and CPC maintenance by positively regulating JAK-STAT signaling, thereby preventing premature differentiation. Conversely, NURF is not required in early differentiating daughter cells of either lineage. Since three additional ATP-dependent chromatin remodelers (ACF, CHRAC, and dMi-2/NuRD) are dispensable for stem cell maintenance in the testis, epigenetic regulation of stem cells within this niche may rely primarily on NURF. Thus, local signals cooperate with specific chromatin remodeling complexes in intact niches to coordinately regulate a common set of target genes to prevent premature stem cell differentiation.
Keywords: Stem cell niche; Chromatin remodeler; spermatogonial stem cell; niche; NURF, JAK-STAT; Drosophila testis
INTRODUCTON
Stem cells sustain tissues by dividing asymmetrically to generate both stem cells and differentiating daughter cells. Signals from the local microenvironments (niches) where stem cells reside govern the balance between these opposing fates by activating distinct transcriptional programs (Morrison and Spradling, 2008). However, chromatin structure imposes an additional level of regulation during this process (Clapier and Cairns, 2009). While the roles of both cell signaling and chromatin structure in the regulation of cell fate are under intense investigation, little is known of how these events are coordinately regulated in endogenous niches.
The germline stem cells (GSCs) sustaining gametogenesis within Drosophila gonads are some of the best-understood adult stem cells, and the importance of local signaling in the regulation of stem cell function in these tissues is well established (Fuller and Spradling, 2007; Gregory et al., 2008). Two distinct populations of stem cells reside in the Drosophila testis apex: GSCs, which produce differentiating germ cells, and somatic stem cells (cyst progenitor cells or CPCs), which produce daughter cells (cyst cells) that envelop germ cells and ensure their differentiation (Fig. 1A). The hub, a cluster of non-mitotic somatic cells, creates a stem cell niche by secreting the cytokine Unpaired; local activation of the Janus Kinase-Signal Transducer and Activator of Transcription (JAK-STAT) signaling cascade prevents differentiation within adjacent stem cells (Kiger et al., 2001; Tulina and Matunis, 2001). Although the JAK-STAT pathway is a clear example of stem cell regulation via extrinsic signaling, the role of epigenetic regulation, including the state of chromatin, has not been studied in this niche.
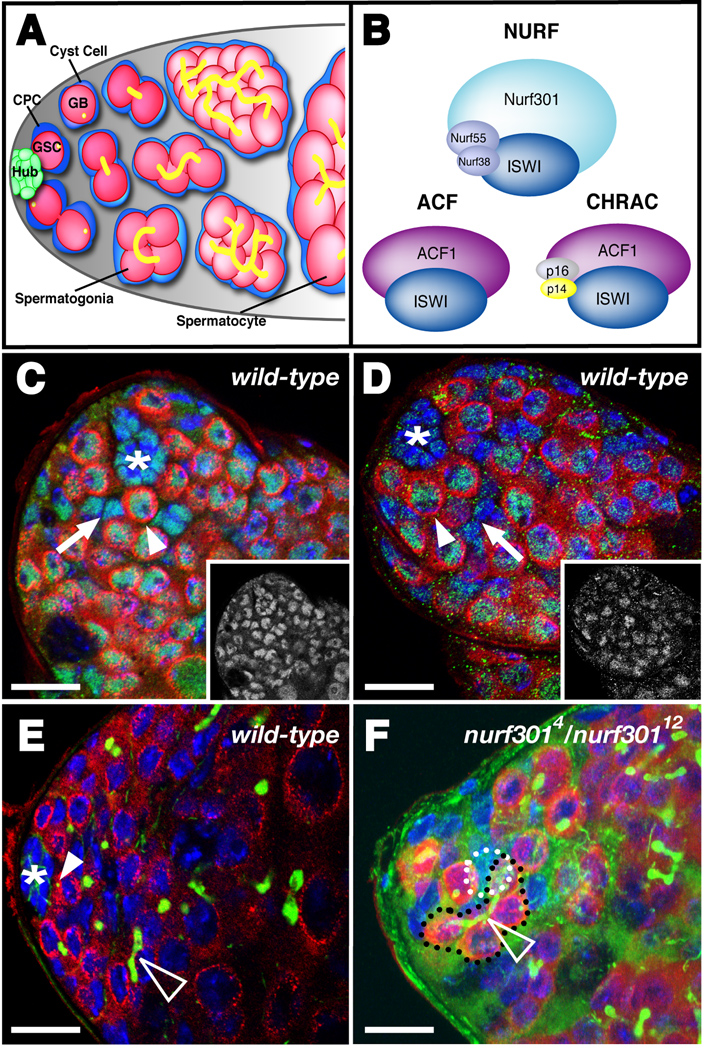
Figure 1. Nurf301 is a unique member of the NURF chromatin remodeling complex and is required for GSC maintenance in the Drosophila testis stem cell niche.
(A) The Drosophila testis apex. Approximately 10 GSCs make broad contact with the hub. GSCs divide asymmetrically to produce gonialblast (GB) daughters that are displaced away from the hub. GBs undergo four rounds of mitosis giving rise to 16-interconnected spermatogonia, which further differentiate into spermatocytes. Approximately two CPCs flank each GSC and contact the hub through thin cytoplasmic extensions. CPCs divide asymmetrically, giving rise to non-mitotic somatic cyst cells that encyst differentiating germ cells. (B) NURF is one of three ISWI-containing ATP-dependent chromatin remodeling complexes in Drosophila. The Nurf301 subunit is unique to the NURF complex and is essential for its function. ACF and CHRAC are distinct from NURF and share a common subunit: ACF1. (C–D) Confocal section of a wild-type testis apex immunostained with germline specific marker anti-Vasa (red), the DNA counterstain DAPI (blue), and (C) anti-ISWI (green, inset) or (D) anti-Nurf301 (green, inset). (C) ISWI is expressed in GSCs (Vasa-positive and contacting the hub, one indicated, arrowhead), CPCs (Vasa-negative and within two cell diameters of the hub, one indicated, arrow) and the hub (*). (D) Nurf301 is expressed in all cell types in the testis apex including GSCs (arrowhead), CPCs (arrow), and the hub (*). (E–F) Testes immunostained with anti-Vasa (red), the hub marker anti-Armadillo (green), the fusome marker anti-1B1 (green) and DAPI (blue). (E) A wild-type testis with a rosette of GSCs surrounding the hub (*). GSCs contain spherical fusomes (green, arrowhead) whereas differentiating spermatogonia contain elongated and branched fusomes (open arrowhead). (F) Projection of three confocal sections of a testis with reduced Nurf301 function (nurf3014/nurf30112) showing differentiating spermatogonia (black outline) containing a branched fusome (open arrowhead) and contacting the hub (white outline). Bars, 10 microns.
Chromatin is highly structured to provide for efficient packaging of DNA and transcriptional regulation (Loden and van Steensel, 2005). Nucleosomes, the fundamental repeating units of chromatin, contain DNA and histones, and are regulated by two main classes of chromatin-remodeling enzymes: those that use ATP hydrolysis to alter histone–DNA contacts, and those that covalently modify histone proteins (Becker and Horz, 2002). Recent work has focused on the role of chromatin in embryonic stem cells (ES cells), which are enriched in euchromatin but accumulate transcriptionally inactive, highly compacted heterochromatin upon differentiation (Arney and Fisher, 2004; Francastel et al., 2000). Consistent with this finding, ATP-dependent chromatin remodelers are found at elevated levels in ES cells (Meshorer and Misteli, 2006). However, the existence of many ATP-dependent chromatin remodelers and few well-characterized niches makes understanding the role of chromatin state in endogenous stem cells challenging.
Nine different ATP-dependent remodelers, grouped into four distinct families, are currently known in Drosophila (Bouazoune and Brehm, 2006). Our previous work indicated that components from one of these families (ISWI) have enriched expression in the Drosophila testis apex (Terry et al., 2006), providing an opportunity to analyze the role of epigenetic regulation in a well-characterized niche.
The Drosophila ISWI ATPase, which is homologous to the yeast SWI2/SNF2 enzyme (Elfring et al., 1998), is found in three distinct chromatin remodeling complexes (Fig. 1B): ACF (ATP-utilizing chromatin assembly and remodeling factor), CHRAC (chromatin accessibility complex), and NURF (nucleosome remodeling factor) (Bouazoune and Brehm, 2006). ACF and CHRAC are involved in chromatin assembly, DNA replication, and transcriptional regulation. NURF regulates higher-order chromatin structure and can act as a transcriptional repressor or activator (Badenhorst et al., 2005). Since expression profiling experiments indicated that three of the four NURF components (ISWI, Nurf55, and Nurf301) are expressed within the testis apex (Terry et al., 2006), we focused on this complex.
Of the four subunits of the Drosophila NURF complex, Nurf301 and ISWI are necessary and sufficient for the accurate and efficient sliding of nucleosomes. Nurf301, the only NURF-specific subunit, is well-characterized biochemically: it is essential to the structural integrity of the complex, interacts directly with sequence-specific transcription factors, and binds tri-methylated lysine 4 on histone H3 tails (H3K4me3) (Wysocka et al., 2006), a histone mark typically found in the promoter of actively transcribed genes (Bernstein et al., 2005; Santos-Rosa et al., 2002). Nurf301 has also been well-characterized genetically in Drosophila: it is required to maintain homeotic gene expression during development (Badenhorst et al., 2002; Xiao et al., 2001), repress JAK-STAT signaling in the immune system (Badenhorst et al., 2002), and promote ecdysone signaling during metamorphosis (Badenhorst et al., 2005). Here, we examined the role of NURF in the Drosophila testis niche.
RESULTS
nurf301 is required for GSC maintenance in the Drosophila testis
To pursue our previous findings that members of the NURF complex are expressed in the Drosophila testis apex (Terry et al., 2006), we immunostained testes with antisera specific for the ISWI and Nurf301 subunits of NURF. As expected, both proteins were expressed in all cells within the testis apex including GSCs, CPCs, and the hub, although Nurf301 levels appeared to be slightly lower in the latter (Fig. 1C, D). Both ISWI and Nurf301 displayed nuclear localization consistent with their well-characterized roles in chromatin remodeling. Since these results were consistent with a role for NURF in the germline and/or somatic stem lineages in this tissue, we next analyzed the testis phenotypes from flies with reduced levels of the essential NURF component, Nurf301. Although nurf301 is required for viability, flies containing a combination of hypomorphic alleles (nurf3014/nurf30112) survive to adulthood (Badenhorst et al., 2005). Testes from 0–3 day old nurf3014/nurf30112 adults contained a wild-type number of GSCs (6.38 ± 0.44, n = 21) compared to heterozygous controls (6.20 ± 0.26, n = 25, p-value > 0.05). However, differentiating spermatogonial cysts were found adjacent to the hub in 25% of nurf3014/nurf30112 testes (n = 20); this phenotype was never observed in heterozygous controls (nurf3014/+, n = 15 and nurf30112/+, n = 10) (Fig. 1E, F), suggesting that nurf301 prevents premature GSC differentiation. Consistent with this finding, larvae with a combination of null alleles (nurf3012/nurf3013), which survive until the third instar (Badenhorst et al., 2005), contained significantly fewer GSCs (4.65 ± 0.51, n = 17) than heterozygous controls (10.66 ± 0.36, n = 32, p-value < 0.0001). Larval testes lacking Nurf301 also appeared to contain fewer differentiating germline cysts than controls (Fig. 2A, B); this is likely due to reduced GSC numbers rather than a reduction in their division rate, since the frequency of GSC mitoses, as assayed by the mitotic marker phospho-histone H3 (PH3), did not vary significantly from wild-type (0.3% of nurf301 GSCs were PH3-positive, n=279; 1.5% of wild-type GSCs were PH3-positive, n=482, p-value = 0.270). We conclude that Nurf301 is required for GSC maintenance.
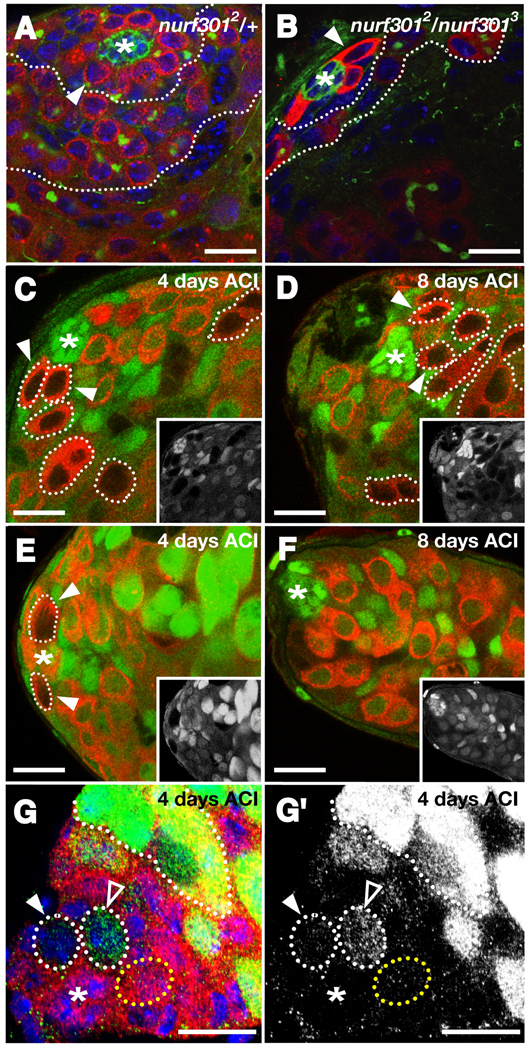
Figure 2. Nurf301 is required cell-autonomously for GSC maintenance. (A–B).
Larval testes immunostained with anti-Vasa (red), anti-Armadillo (green), anti-1B1 (green), and DAPI (blue). (A) Larval testes heterozygous for nurf301 (nurf3012 / +) have a wild-type number of GSCs (one indicated, arrowhead) surrounding the hub (*). Numerous differentiating daughters (outlined) are displaced away from the hub. (B) Larval testes homozygous mutant for nurf301 (nurf3012 / nurf3013) have significantly fewer GSCs (one indicated, arrowhead) surrounding the hub (*) and a smaller zone of differentiating spermatogonia (outlined). (C–F) Confocal sections of testes immunostained with anti-GFP (green, inset) and anti-Vasa (red); hubs denoted with an asterisk. (C–D) Wild-type GSC clones (arrowheads, outlined) and their differentiating daughters (outlined), identified by the absence of GFP and the presence of Vasa, are detected (C) 4 days and (D) 8 days ACI. (E) nurf301 mutant GSC clones are found at the hub 4 days ACI, but are no longer detected 8 days ACI (F). (G) Testis immunostained with anti-β-galactosidase (red) and a differentiation marker bam-GFP (green). bam-GFP is not normally expressed in GSCs (yellow outline) but is detected in differentiating spermatogonia (white outline). Two nurf301 mutant GSCs (β-galactosidase-negative, white outline, arrowheads) surround the hub (*). One nurf301 GSC expresses bam-GFP (outlined, open arrowhead) while the other is bam-GFP-negative (outlined, arrowhead), indicating that nurf301 null GSCs differentiate prematurely in the niche. (G') bam-GFP (green) channel alone. Bars, 10 microns.
nurf301 is required cell-autonomously for GSC maintenance but is dispensable for spermatogonial differentiation
GSCs could directly or indirectly require nurf301 for their maintenance. To distinguish between these possibilities we created negatively marked nurf301 null clones in adult testes using FLP/FRT mediated mitotic recombination (Xu and Rubin, 1993). GSC clones were identified as Vasa-positive cells contacting the hub but lacking GFP. Testes were scored for the presence of one or more negatively marked GSCs at 2, 4, 6, 8, or 10 days after clone induction (ACI). As expected, wild-type control GSC clones were maintained over this time interval. In contrast, GSCs lacking nurf301 were rapidly lost from the niche, indicating that GSCs directly require nurf301 for their maintenance (Fig. 2C-F, Table 1). Interestingly, differentiating germline cysts lacking nurf301 were detected up to the primary spermatocyte stage for several days ACI, suggesting that although nurf301 is required for GSC maintenance, it is not required for spermatogonial differentiation (Supplemental Fig. 1, Supplemental Table 1).
Table 1.
Nurf301, Nurf38, and ISWI are essential for maintaining GSCs in the Drosophila testis.
| Genotype | 2 days ACI | 4 days ACI | 6 days ACI | 8 days ACI | 10 days ACI |
|---|---|---|---|---|---|
| Average number of marked GSCs per testis | |||||
| Wild-type (FRT80B) |
0.193 (11/57) |
0.395 (32/81) |
0.348 (46/132) |
0.344 (21/61) |
0.471 (16/34) |
| nurf3012 | 0.434 (23/53) |
0.222 (16/72) |
0.151 (16/106) |
0 (0/61) |
0.12 (3/25) |
| nurf3013 | 0.359 (23/64) |
0.182 (16/88) |
0.029 (4/138) |
0.053 (4/76) |
0.037 (2/54) |
| Wild-type (FRT42D) |
0.06 (3/50) |
0.109 (5/46) |
ND | 0.262 (11/42) |
0.279 (12/43) |
| nurf38k16102 | 0.308 (8/26) |
0.28 (7/25) |
ND | 0 (0/40) |
0 (0/36) |
| Testes with a marked GSC* | |||||
| Wild-type (FRT42B) |
47.9% (46/96) |
58.1% (43/74) |
50% (43/86) |
51.1% (24/47) |
59.7% (46/77) |
| Iswi2 | 48.1% (25/52) |
16.7% (12/72) |
6.8% (4/59) |
0% (0/41) |
1.7% (1/59) |
The number of testes with one or more marked GSCs/the total number of testes.
Since GSCs lacking nurf301 were depleted from the testis over time, nurf301 could either maintain GSC viability or prevent GSCs from differentiating. To distinguish between these possibilities, a transcriptional reporter revealing the expression of the differentiation factor bag of marbles (bam) (Chen and McKearin, 2003) was analyzed in testes containing nurf301 null clones. Bam is normally detected in spermatogonia but almost never in GSCs (0.4% of wild-type GSCs express Bam, n=263). In contrast, Bam was expressed in 31.4% (n = 35) of nurf3012 GSCs and 40.6% (n = 32) of nurf3013 GSCs at 4 days ACI (Fig. 2G, G'). These results suggest that nurf301 null GSCs are lost from the niche due to premature differentiation. Consistent with this finding, the number of apoptotic cells in testes containing nurf301 null clones did not increase compared to testes containing control clones (1.96 ± 0.2 TUNEL-positive cells, n = 66 for nurf3012; 2.19 ± 0.2, n = 70 for nurf3013; 3.10 ± 0.23, n = 92 for wild-type). Together, these results indicate that nurf301 is not required generally to sustain germ cell viability or spermatogonial differentiation; rather, it is specifically required within GSCs to prevent them from prematurely entering the differentiation pathway.
nurf301 is required cell-autonomously for CPC maintenance but not for cyst cell differentiation
GSCs cohabit the niche with somatic stem cells called CPCs, and both types of stem cells express NURF components (Figure 1C, D). Therefore, we used mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo, 1999) to determine the requirement for nurf301 in the CPC lineage. CPC clones were identified as Vasa-negative, GFP-positive cells contacting the hub. Testes were scored for the presence of one or more CPC clones at 2, 4, 6, 8, 10, or 14 days ACI. As expected, wild-type control CPC clones were readily observed throughout the time course (Fig. 3A, B). By 14 days ACI, the percentage of testes with wild-type CPC clones decreased by about half, but CPC clones were still frequently observed (Fig. 3B, Table 2). This moderate loss of wild-type CPC clones likely reflects the relatively short half-life (5–10 days) that has been reported for CPCs (Voog et al., 2008). In contrast, nurf301 null CPCs were lost much more rapidly than wild-type CPC clones and were rarely detected after 4 days ACI (Fig. 3C, D, Table 2). About 85.5% of nurf3012 and 100% of nurf3013 null CPCs were lost after two weeks, indicating that nurf301 is essential for CPC maintenance (Table 2). The frequency of apoptotic cells did not differ significantly in testes with nurf301 null clones compared to testes with control clones (assayed via TUNEL staining, as above), suggesting that nurf301 null CPCs are lost via differentiation, rather than death. Prior to being lost from the niche, nurf301 null CPCs express the CPC marker zinc-finger-homeodomain protein 1 (Zfh-1) (Leatherman and Dinardo, 2008) (100% of nurf301 null CPCs at 4 days ACI were Zfh-1-positive, n=34) and produce differentiating CPC daughters (cyst cells) that encyst adjacent spermatogonia (Fig. 3D, Supplemental Table 2). Importantly, cyst cells lacking nurf301 extinguish Zfh-1 expression (data not shown), and eventually express the late cyst cell marker Eya (Fig. S1). Thus, cyst cells lacking nurf301 appear to differentiate appropriately. Together, these results indicate that nurf301 is specifically required to autonomously maintain both germline and somatic stem cells in the testis, yet is dispensable for early daughter cell differentiation in each lineage.
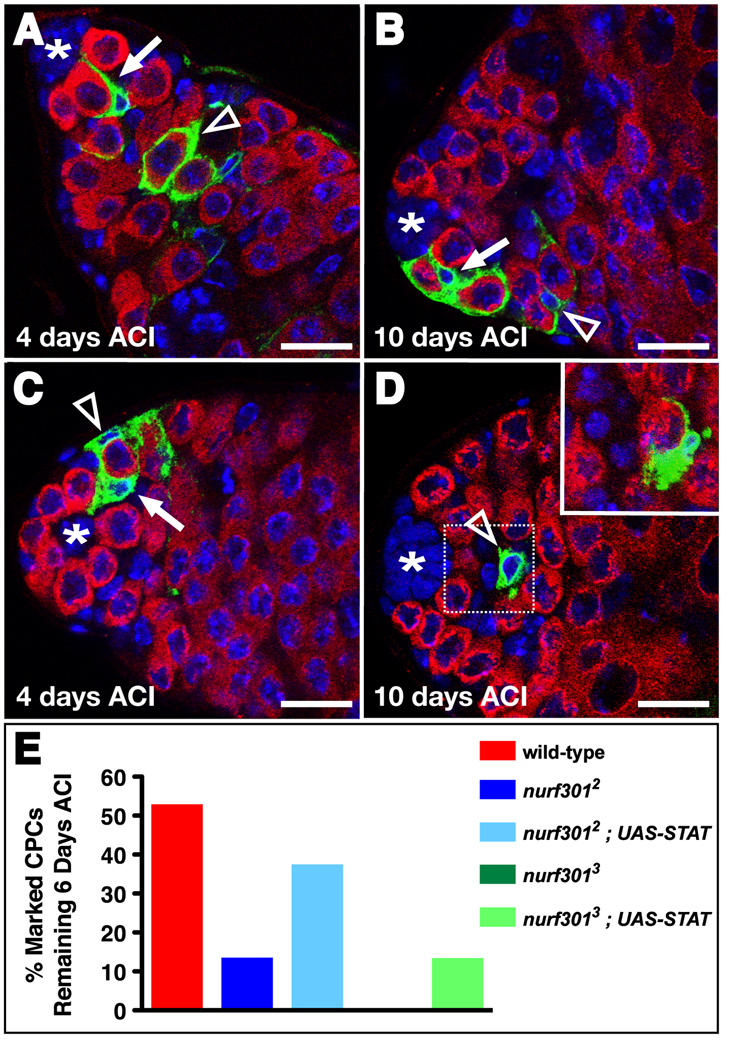
Figure 3. Nurf301 is required cell-autonomously for CPC maintenance. (A-D).
Confocal sections of testes immunostained with anti-GFP (green), anti-Vasa (red), and DAPI (blue) containing positively-marked wild-type (A, B) or nurf301 mutant (C, D) CPC clones (arrow) and their differentiating daughters (open arrowhead) identified by the presence of GFP, absence of Vasa, and their position within the tissue. (A–B) Wild-type CPC clones and their daughters are detected at (A) 4 and (B) 10 days ACI. nurf301 mutant CPC clones are detected 4 days ACI (C), but are not maintained at 10 days ACI (D). Differentiating mutant CPC daughters are occasionally still found 10 days ACI (open arrowhead). Inset; higher magnification and adjacent confocal section of the boxed area in D showing a mutant CPC daughter cell encysting a differentiating germline cell. (E) Restoring STAT92E to nurf301 null CPCs partially rescues their stem cell loss phenotype. Bars, 10 microns.
Table 2.
Nurf301 is essential for maintaining CPCs in the Drosophila testis.
| Genotype | 2 days | 4 days | 6 days | 8 days | 10 days | 14 days |
|---|---|---|---|---|---|---|
| Testes with a marked CPC* | ||||||
| Wild-type (FRT2A) |
62.3% (66/106) |
66.7% (20/30) |
33.3% (14/42) |
20.9% (9/43) |
19.2% (10/52) |
33.3% (8/24) |
| nurf3012 | 33.3% (14/42) |
24.1% (13/54) |
4.7% (2/43) |
4.7% (2/43) |
1.9% (1/54) |
4.8% (1/21) |
| nurf3013 | 36.4% (16/44) |
4.8% (2/42) |
0% (0/39) |
0% (0/41) |
0% (0/38) |
0% (0/20) |
|
nurf3012 UAS-STAT |
34.1% (14/41) |
41.0% (9/22) |
13.0% (3/23) |
12.0% (3/25) |
3.4% (2/59) |
ND |
|
nurf3013 UAS-STAT |
14.3% (6/42) |
14.0% (7/50) |
2.0% (1/51) |
0% (0/51) |
0% (0/43) |
ND |
The number of testes with one or more marked CPCs divided by the total number of testes.
The NURF complex maintains GSCs and CPCs in the Drosophila testis
Since nurf301is a unique subunit of the NURF complex and is essential to its function, our results suggested that the NURF complex is essential for maintaining stem cell fate in the Drosophila testis. Therefore, we analyzed the role of additional members of this complex in GSC maintenance via genetic mosaic analysis as described above. Loss-of-function alleles have not been identified for nurf55, but exist for the inorganic pyrophosphatase nurf38 and the ATPase iswi. Therefore, we created nurf38 and iswi loss-of-function clones as described above for nurf301. We found that nurf38k16102 mutant GSCs were completely lost from the testis by 8 days ACI. Similarly, the number of testes containing iswi2 mutant GSCs declined by about 99% by 10 days ACI (Table 1). Interestingly, the timing of loss of both nurf38 and iswi mutant GSCs was similar to that of nurf301 mutant GSCs. These results indicate that Nurf38 and ISWI are required for GSC maintenance, and support the hypothesis that the NURF complex is required for stem cell maintenance in the testis.
We also wanted to determine if CPCs, like GSCs, require ISWI for their maintenance. We successfully reduced ISWI levels in the CPC lineage by expressing an ISWI-RNAi construct specifically in CPCs and their daughters using the c587-Gal4 driver (Voog et al., 2008). In wild-type testes, ISWI was detected in CPCs and GSCs at comparable levels (Fig. 4A). However, induction of the ISWI-RNAi construct for 7 days at 29°C led to greatly reduced levels of ISWI in CPCs but not GSCs (Fig. 4B). To quantify CPCs before and after ISWI-RNAi induction, we immunostained testes with antibodies against Zfh-1 (Issigonis et al., 2009; Leatherman and Dinardo, 2008). Before RNAi induction, flies carrying the ISWI-RNAi construct contained the same number of CPCs as GFP-RNAi controls (27.8 ± 2.1, n = 8 versus 26.6 ± 1.5, n = 17, p-value > 0.05) (Fig. 4G). However, after RNAi induction, flies carrying the ISWI-RNAi construct contained significantly fewer CPCs (18 ± 1.2, n = 17) (Fig 4D, G) than GFP-RNAi controls (30.4 ± 1.2, n = 22, p-value < 0.0001) (Fig. 4C, G). Thus, ISWI is directly required for CPC maintenance. Following induction of ISWI-RNAi in CPCs and their daughters we also observed a decrease in GSC number (3.80 ± 2.25, n=10) compared to GFP-RNAi controls (8.73 ± 2.66, n=22, p-value 0.0002). This suggests that CPCs with reduced levels of ISWI do not properly signal to the GSCs, thus indirectly causing loss of GSCs from the niche. Signaling between CPCs and GSCs plays an important role in the balance between stem cell self-renewal and differentiation (Kawase et al., 2004; Kiger et al., 2000; Leatherman and Dinardo, 2008), but is poorly understood. It will be interesting to determine if NURF ensures appropriate signaling between stem cell types or if the loss of GSCs following ISWI-RNAi in the CPCs is an indirect effect due to the exit of ISWI-deficient CPCs from the niche. Together our results demonstrate that multiple members of the NURF complex autonomously maintain CPC and GSC fate in the Drosophila testis niche.
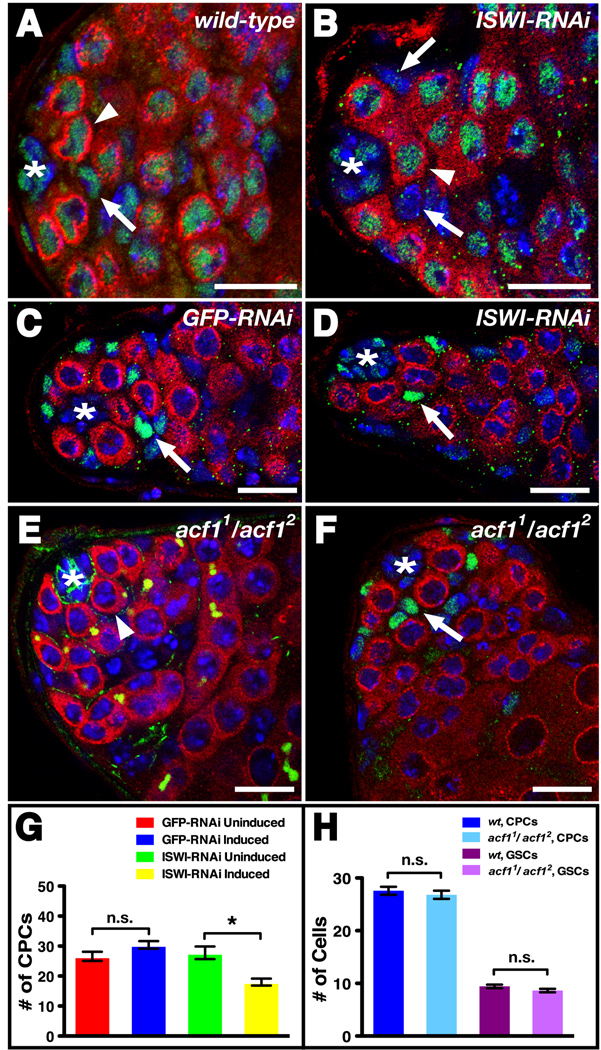
Figure 4. NURF is the sole ISWI-containing chromatin remodeling complex required for stem cell maintenance in the Drosophila testis.
(A–B) Testes immunostained with anti-ISWI (green), anti-Vasa (red), and DAPI (blue). An asterisk denotes the hub. (A) In wild-type testes, ISWI is expressed at comparable levels in CPCs (arrows) and GSCs (arrowhead). (B) ISWI-RNAi knockdown in CPCs and their daughters reduces ISWI expression in CPCs (arrows) compared to GSCs (arrowhead). (C–D) Testes immunostained with anti-Vasa (red), CPC marker anti-Zfh-1 (green), and DAPI (blue) expressing (C) GFP-RNAi or (D) ISWI-RNAi in CPCs (arrows) and their daughters. When compared to controls (C), a reduction in ISWI leads to fewer CPCs around the hub (*) (D). (E) acf1 homozygous mutant testis immunostained with anti-Vasa (red), anti-Armadillo (green), anti-1B1 (green), and DAPI (blue) containing a wild-type number of GSCs (one indicated, arrowhead) surrounding the hub (*). (F) acf1 homozygous mutant testis immunostained with anti-Vasa (red), anti-Zfh-1 (green), and DAPI (blue) containing a wild-type number of CPCs (one indicated, arrow) surrounding the hub (*). (G) ISWI-RNAi knockdown in CPCs and their daughters leads to a significant decrease in CPCs [n.s. (not significant), p-value > 0.05; *p-value < 0.001]. (H) acf1 homozygous mutant testes have the same number of GSCs and CPCs as wild-type (n.s., p-value > 0.05). Data are represented as mean +/− SEM. Bars, 10 microns.
NURF is one of nine unique chromatin-remodeling complexes currently identified in Drosophila, and increasing evidence indicates that chromatin remodelers may play both general and specific roles in regulating cell fate decisions (Clapier and Cairns, 2009). We wondered whether multiple remodelers are required for stem cell maintenance in the testis, or if instead this is a unique feature of NURF. The NURF ATPase ISWI is a component of three distinct remodeling complexes (NURF, ACF and CHRAC), but ACF and CHRAC share a common subunit, ACF1, which is not present in NURF. Therefore, we determined the requirement for ACF and CHRAC in the testis by removing ACF1 function. Two different null alleles of acf1 exist: acf11 and acf12. Since acf1 homozygous null mutants are semi-lethal, but those flies that do survive are fertile (Fyodorov et al., 2004), we asked if acf1 is necessary for maintaining stem cells in the testis by comparing the number of GSCs and CPCs in surviving acf11/acf12 adults to wild-type controls. Testes from acf11/acf12 flies had the same number of GSCs as wild-type controls (8.64 ± 0.64, n = 28 versus 9.43 ± 0.34, n = 21, p-value > 0.05) (Fig. 4E, H). The number of CPCs in acf11/acf12 testes was also indistinguishable from wild-type controls (26.81 ± 0.78, n = 43 versus 27.60 ± 0.78, n = 33, p-value > 0.05) (Fig. 4F, H). Thus, acf1 is not required for stem cell maintenance in the Drosophila testis. Instead, our results indicate that stem cell maintenance is not a property of ISWI family remodeling complexes in general but can be ascribed specifically to the function of a single ISWI-containing chromatin remodeling complex: NURF.
Although NURF is the sole ISWI family member required in the testis niche, it was not known if members of the other families of chromatin remodelers, which contain different types of ATPase subunits, are also required for stem cell maintenance in this system. We found that a GFP protein trap inserted in the dMi-2 gene (Buszczak et al., 2007) is broadly expressed throughout the testis apex, indicating that dMi-2 is expressed in this tissue (data not shown). Since dMi-2 encodes the core ATPase of the Drosophila Mi-2/NuRD complex, which is involved in the repression of homeotic genes during embryogenesis (Kehle et al., 1998), analyzing the role of dMi-2 in the testis enabled us to determine the requirement for the Mi-2/CHD family of remodelers in our system. Although dMi-2 is essential for viability, 0.1% of dMi-2 null adults of the genotype dMi-25/Df(3L)BSC1 survive to adulthood (Yamasaki and Nishida, 2006) (Yamasaki and Nishida, 2006). Immunostaining testes of dMi-25/Df(3L)BSC1 adults as described above revealed that they contained a similar number of GSCs as heterozygous controls (6.44 ± 0.73, n = 9 versus 5.92 ± 0.30, n = 25, p-value > 0.05). Therefore, dMi-2 is not required for GSC maintenance in the Drosophila testis. Thus, the maintenance of Drosophila testis stem cells is not dependent on all chromatin remodelers but is a property unique to specific complexes including NURF.
NURF maintains testis stem cells by positively regulating the JAK-STAT pathway
Our data demonstrate that the NURF complex is required to maintain both GSCs and CPCs in the Drosophila testis. Since JAK-STAT signaling is also required autonomously to maintain both GSCs and CPCs (Issigonis et al., 2009; Kiger et al., 2001; Leatherman and Dinardo, 2008; Tulina and Matunis, 2001) we postulated that NURF could prevent stem cell differentiation in the testis by promoting the activity of the JAK-STAT pathway within stem cells. To test this hypothesis, we monitored JAK-STAT activity in negatively marked nurf301 GSC clones by immunostaining for STAT92E, since enrichment of STAT92E indicates pathway activity (Chen et al., 2002). In nurf301 heterozygous testes before clone induction, STAT92E is enriched in all GSCs surrounding the hub and reduced in gonialblast daughters, in a manner indistinguishable from wild-type (Fig. 5A, A'). At 4 days ACI, GSCs null for either nurf3012 (n = 24) or nurf3013 (n = 23) had significantly reduced levels of STAT92E staining relative to neighboring heterozygous GSCs (Fig. 5B, B', E). Instead, the level of STAT92E in GSCs lacking Nurf301 was less than or similar to that typically seen in heterozygous gonialblast daughters (Fig. 5B, B'). This decline in STAT92E enrichment upon loss of Nurf301 suggests that nurf301 positively regulates the JAK-STAT pathway in GSCs, thus promoting their maintenance in the niche.
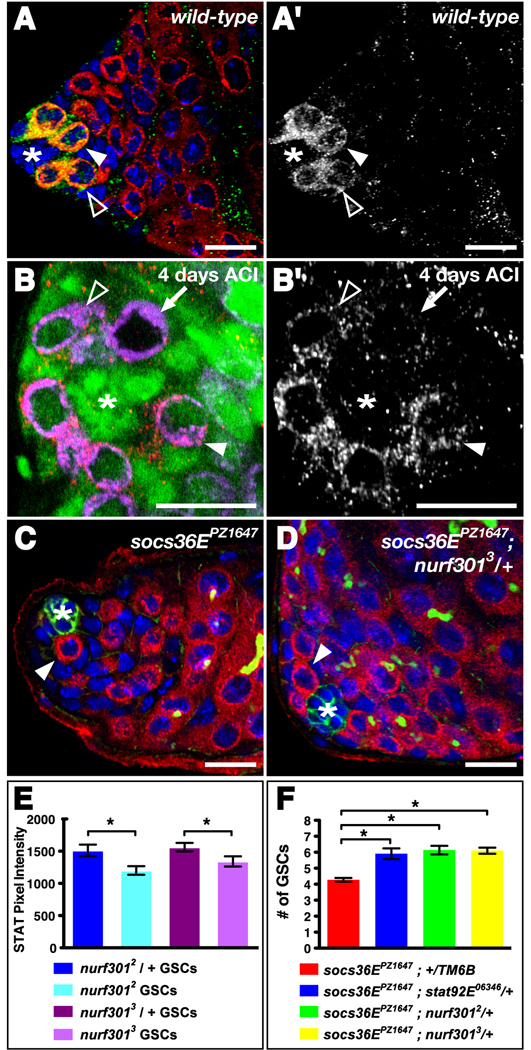
Figure 5. NURF regulates JAK-STAT signaling to maintain GSCs and CPCs in the Drosophila testis stem cell niche.
(A) nurf301 heterozygous testis immunostained with anti-Vasa (red), anti-dSTAT92E (green), and DAPI (blue). STAT92E is expressed primarily in GSCs (arrowhead) surrounding the hub (*), whereas gonialblasts have reduced STAT92E levels (open arrowhead). (A') STAT92E channel alone. (B) Testis immunostained with anti-Vasa (magenta), anti-GFP (green), anti-dSTAT92E (red), and DAPI (blue) containing a single negatively marked nurf3012 homozygous mutant GSC (arrow) indicated by the presence of Vasa and absence of GFP, 4 days ACI. The homozygous nurf301 GSC has reduced levels of STAT92E protein compared to the wild-type neighboring GSCs (one indicated, arrowhead) but similar levels to a neighboring wild-type gonialblast (open arrowhead). An asterisk denotes the hub. (B') STAT92E channel alone. (C–D) Testes immunostained with anti-Vasa (red), anti-Armadillo (green), anti-1B1 (green), and DAPI (blue). (C) socs36EPZ1647 homozygous mutant testes have a reduced number of GSCs (arrowhead) surrounding the hub (*). (D) Testes homozygous for socs36EPZ1647 and heterozygous for nurf3013 have a near wild-type number of GSCs (one indicated, arrowhead) surrounding the hub (*). (E) nurf301 null GSCs have significantly less dSTAT92E staining compared to neighboring wild-type GSCs (*, p-value < 0.05). (F) Removing one copy of stat92E or nurf301 partially rescues the socs36EPZ1647 GSC loss phenotype (*, p-value < 0.001). Data are represented as mean +/− SEM. Bars, 10 microns.
To confirm this hypothesis, we asked if nurf301 genetically interacts with the JAK-STAT pathway in the testis niche. Suppressor of cytokine signaling 36E (socs36E) is a highly conserved target of the JAK-STAT pathway and functions in a classical negative feedback loop by down-regulating pathway activity in CPCs (Callus and Mathey-Prevot, 2002; Issigonis et al., 2009; Karsten et al., 2002). In socs36EPZ1647 homozygous mutant testes, CPCs have aberrantly high JAK-STAT activity and consequently displace neighboring GSCs from the niche, resulting in GSC loss (Issigonis et al., 2009) (Fig. 5C, F). When Stat92E levels were genetically lowered in socs36EPZ1647 mutant flies, fewer GSCs were lost (Fig. 5F). Similarly, if Nurf301 levels were genetically reduced in socs36EPZ1647 mutant flies, fewer GSCs were lost (Fig. 5D, F). Thus, global reduction of either Stat92E or Nurf301 partially rescues the socs36EPZ1647 phenotype. Since nurf301 genetically interacts with the JAK-STAT pathway member socs36E in a manner consistent with that of a positive regulator, our data suggest that both GSCs and CPCs require NURF to effectively activate the JAK-STAT pathway, thus ensuring their maintenance within the testis niche. Considering its role as a chromatin remodeler, we hypothesized that NURF could promote transcription of JAK-STAT pathway activators. To test this hypothesis, we asked if boosting levels of STAT92E specifically within CPCs lacking Nurf301 could overcome the CPC loss phenotype. We found that restoration of STAT92E expression partially rescued nurf301 null CPC loss at 6 days ACI (Fig. 3E, Table 2). Although it is likely that Nurf301 regulates many genes, our data suggest that a major role of NURF in the maintenance of testis stem cells is to ensure sufficient STAT92E expression. Together these data support the hypothesis that NURF positively regulates JAK-STAT signaling in the testis niche.
DISCUSSION
This work reveals that the ATP-dependent chromatin remodeler NURF cooperates with local JAK-STAT signaling in the Drosophila testis niche to ensure stem cell maintenance. This may be a unique feature of NURF as three additional ATP-dependent chromatin remodelers are dispensable for stem cell maintenance in the testis.
The role of NURF in stem cell maintenance
We propose that NURF plays a critical role in maintaining a chromatin configuration that is essential for germline and somatic stem cell maintenance in the Drosophila testis. In the germline, NURF promotes expression of the stem cell maintenance factor STAT92E and prevents premature expression of the differentiation factor Bam. STAT92E expression is difficult to detect in CPCs due to inhibition of the JAK-STAT pathway by the suppressor Socs36E (Issigonis et al., 2009); however, expressing STAT92E in nurf301 null CPCs partially rescues their loss from the niche, suggesting that NURF also promotes JAK-STAT signaling in CPCs. Since both stem cell populations directly require JAK-STAT signaling for their maintenance, identifying targets of NURF in each lineage will be of interest. Interestingly, the JAK-STAT pathway is required for proper integrin expression in CPCs to maintain niche homeostasis (Issigonis et al., 2009), an intriguing possibility is that NURF may directly, or indirectly via regulation of JAK-STAT signaling, control expression of adhesion molecules in testis stem cells to ensure their maintenance in the niche.
Further insights into the epigenetic regulation of stem cells arise when comparing our work to studies of chromatin remodelers in the Drosophila ovary. ISWI prevents premature differentiation of testis GSCs, and as a component of the NURF complex, promotes JAK-STAT signaling. Similarly, ISWI prevents differentiation of ovarian GSCs by enabling them to respond to Dpp/Tgfß signals from their niche (Xi and Xie, 2005). This is not likely to involve JAK-STAT signaling, since female GSC maintenance does not require this pathway (Decotto and Spradling, 2005). However, the Dpp/Tgfß signaling pathway maintains GSCs in both the ovary (Xie and Spradling, 1998) and the testis (Kawase et al., 2004). Examining the interactions between Nurf301 and components of the Dpp/Tgfβ signaling pathway may reveal whether NURF regulates this signaling pathway in the testis niche. Interestingly, the ability of NURF to interact with the Dpp/Tgfß signaling pathway may be conserved; the mammalian orthologue of Drosophila Nurf301 (BPTF) may directly promote Dpp/Tgfß signaling via the NURF remodeling complex by recruiting Smad transcription factors to target sites in mouse ES cells and embryos (Landry et al., 2008). Thus, NURF may have a conserved role in stem cell maintenance.
NURF can positively or negatively regulate JAK-STAT signaling
Our finding that NURF promotes JAK-STAT signaling in the testis niche is surprising, given that it is thought to repress STAT targets during Drosophila hematopoiesis by interacting with the transcriptional repressor and JAK-STAT pathway inhibitor Ken and Barbie (Ken) (Arbouzova et al., 2006; Kwon et al., 2008). In contrast, STAT92E expression in GSCs requires NURF, reintroduction of STAT92E into nurf301 null CPCs partially rescues their loss from the testis niche, and NURF genetically interacts with the JAK-STAT inhibitor SOCS36E in a manner consistent with it being a positive regulator of this pathway in the testis. We propose that nurf301 likely regulates the JAK-STAT pathway in a tissue-specific manner and it will be important to identify factors that can interact directly with Nurf301 in the testis niche. Furthermore, determining whether Ken plays a role in the testis niche should be informative. Since Nurf301 can both activate and repress the transcription of several hundred genes in Drosophila larvae (Badenhorst et al., 2005) and binds to STAT92E binding sites in vivo (Kwon et al., 2008), identifying targets of both NURF and STAT92E in testis stem cells will reveal whether NURF promotes JAK-STAT signaling directly by activating transcription, or indirectly, by prohibiting the expression of JAK-STAT inhibitors.
NURF is the sole ISWI family member needed for testis stem cell maintenance
Although the Drosophila ISWI family of chromatin remodelers has three members (NURF, ACF, and CHRAC), NURF alone is required for GSC and CPC maintenance in the testis. Interestingly, germline and follicle stem cells in the ovary use distinct chromatin remodeling factors to control self-renewal; ISWI is required for maintenance of GSCs, but is dispensable in follicle stem cells, while the INO80 family ATPase Domino promotes follicle stem cell self-renewal but is not required in GSCs (Xi and Xie, 2005). Thus, within endogenous niches, each type of stem cell requires a unique constellation of genetic and epigenetic regulators. Since NURF is required for GSC maintenance and primary spermatocyte differentiation (Kwon et al., 2009), but is dispensable for spermatogonial differentiation, additional means of epigenetic regulation must exist to support the dramatic changes in chromatin structure that accompany spermatogenesis. The well-characterized polycomb-group (PcG) proteins, which epigenetically silence target genes via covalent histone modification and are essential for maintenance of mammalian hematopoietic and spermatogonial stem cells (Buaas et al., 2004; Lessard and Sauvageau, 2003; Park et al., 2003), are essential regulators of spermatogonial differentiation in Drosophila (Chen et al., 2005). It will be interesting to determine if PcG proteins also function in GSCs within the testis. Furthermore, since somatic stem cells also require NURF but their daughter cells do not, it will be interesting to learn whether differentiation in this lineage requires additional chromatin remodelers. Since chromatin immunoprecipitation (ChIP) has been used successfully in the testis (Chen et al., 2005), and we have devised a genetic means to greatly expand the pool of stem cells and identify genes with enriched expression in stem cells (Terry et al., 2006), identification of NURF and STAT92E targets in both testis stem cell lineages is now possible, and should greatly enhance our understanding of how genetic and epigenetic mechanisms coordinately regulate stem cells in an endogenous tissue.
EXPERIMENTAL PROCEDURES
Fly Stocks
w1118;; nurf3014/TM6B Hu, w;; nurf30112/TM3 Ser (Badenhorst et al., 2005), w;; nurf3012/TM6B Hu, and w;; nurf3013/TM3 Ser (Badenhorst et al., 2002) were from P. Badenhorst. w; P[w+ bamP::GFP] Sp/CyO (Chen and McKearin, 2003) was from D. McKearin. y w; acf11 and acf12 (Fyodorov et al., 2004) were from D. Fyodorov. w; al b cn ISWI2 sp/SM5, sp (Deuring et al., 2000) was from J. Birchler. UAS-ISWI-RNAi (24505) and UAS-WDS-RNAi (38926) (Dietzl et al., 2007) were from the Vienna Drosophila RNAi Center. P[c587-Gal4] (Kai and Spradling, 2003) and socs36EPZ1647 (Issigonis et al., 2009) were from A. Spradling. P[nanos-Gal4-VP16] (Van Doren et al., 1998) was from M. Van Doren. UAS-STAT92E was from D. Montell (Silver et al., 2005). y w was used as wild-type; additional fly stocks came from the Bloomington Stock Center and all flies were raised at 25°C on standard molasses/yeast medium unless stated otherwise.
Mosaic Analysis
Negatively marked clones: nurf3012, nurf3013, nurf38k16102, or iswi2 clones were induced using the FLP, FRT-mediated mitotic recombination technique (Xu and Rubin, 1993) in flies of the following genotypes: y w, P[hs-FLP]/Y;; P[Ubi-GFP] P[neoFRT]80B/nurf3012 or 3 P[neoFRT]80B ry506, +/Y; P[neoFRT]42D P[Ubi-GFP.nls]/P[neoFRT]42D nurf38 k16102; MKRS, P[hs-FLP]/+, +/Y; P[w+ FRT]42B P[Ubi-GFP.nls] P[Ubi-GFP.nls]/P[w+ FRT]42B iswi2; MKRS, P[hs-FLP]/+. Control clones were induced in P[hs-FLP]/Y;; P[Ubi-GFP] P[neoFRT]80B/P[neoFRT]80B ry506, +/Y; P[neoFRT]42D P[Ubi-GFP.nls]/P[neoFRT]42D; MKRS, P[hs-FLP]/+, or +/Y; P[w+ FRT]42B P[Ubi-GFP.nls] P[Ubi-GFP.nls]/P[w+FRT]42B; MKRS, P[hs-FLP]/+. To induce clones, 0–5 day old adult male flies were subjected to either two 1 hr. heat shocks at 37°C separated by 5 hrs. at 25°C (nurf301 and nurf38 clones) or three 30 min. heat shocks at 37°C separated by 30 min. intervals at 25°C (iswi clones). After the final heat shock, flies were kept at 25°C and dissected and stained at 2, 4, 6, 8, or 10 days after clone induction (ACI). GSC clones were identified as Vasa-positive, GFP-negative cells contacting the hub.
Positively marked clones: nurf3012 or nurf3013 were induced using the mosaic analysis with a repressible cell marker (MARCM) technique (Lee and Luo, 1999) in y w, P[hs-FLP], P[tub-Gal4] P[UAS-CD8-GFP];; P[tub-Gal80] P[w+ FRT]2A/nurf3012 or 3 P[w+ FRT]2A flies. Control clones were induced in y w, P[hs-FLP], P[tub-Gal4] P[UAS-CD8-GFP];; P[tub-Gal80] P[w+ FRT]2A/P[w+ FRT]2A (Wang and Struhl, 2004) (a gift from G. Struhl). For overexpression of STAT92E in nurf301 null CPCs y w, P[hs-FLP], P[tub-Gal4] P[UAS-CD8-GFP];UAS-STAT92E/+; P[tub-Gal80] P[w+ FRT]2A/nurf3012 or 3 P[w+ FRT]2A flies were used. Adult males were heat shocked three times for 30 min. at 37°C separated by 30 min. intervals at 25°C. After the final heat shock, flies were kept at 25°C and dissected and stained at 2, 4, 6, 8, 10, and 14 days ACI. CPC clones were identified as Vasa-negative, GFP-positive cells contacting the hub.
RNAi Knockdown
RNA interference (RNAi) knockdown of ISWI in CPCs was accomplished in P[c587-Gal4];; UAS-ISWI-RNAi 24505 flies. Control RNAi was performed with P[c587-Gal4]; P[UAS-AvicGFP.dsRNA] flies processed in parallel. 0–3 day old males raised at 18°C were shifted to 29°C for 7 days to induce robust expression of the RNAi construct. ISWI protein levels were monitored by staining with rabbit anti-ISWI and comparing the levels of ISWI in GSCs and CPCs of the same testis.
Genetic Interactions
To assay for genetic interactions between socs36E and stat92E or nurf301, socs36EPZ1647; nurf3012 or 3 /+ and socs36EPZ1647; stat92E06346 /+ flies were generated by crossing socs36EPZ1647; nurf3012 or 3/TM6B Hu or socs36EPZ1647; stat92E06346/TM6B Hu males to socs36EPZ1647 virgins. socs36EPZ1647; +/TM6B Hu siblings were used as controls. Testes from 0-3 day old males raised at 25°C were dissected and analyzed as described above.
Additional Loss-of-Function Experiments
To assay GSC number in nurf301 null larval testes, w;; nurf3012/nurf3013 third instar larvae were generated by crossing w;; nurf3012/TM3, P[Kr-GAL4], P[UAS-GFP], Sb males to w;; nurf3013/TM3, P[Kr-GAL4], P[UAS-GFP], Sb virgins; homozygous mutants were distinguished from heterozygous siblings by the absence of GFP. dMi-2 null flies (dMi-25/Df(3L)BSC1) were created by crossing w;; dMi-25/TM3 Sb males (Yamasaki and Nishida, 2006) (a gift from Y. Nishida) to w;; Df(3L)BSC1/TM3 Sb virgins. w;; dMi-25/TM3 Sb males were used as controls.
Immunostaining
Testes were dissected, fixed and stained as described previously (Matunis et al., 1997), except testes were incubated with anti-dSTAT92E for 48h at 4°C. The following antibodies were used: goat anti-Vasa (dC-13) (Santa Cruz Biotechnology; 1:400), rabbit anti-GFP (Torrey Pines Biolabs; 1:10,000), chicken anti-GFP (Abcam; 1:10,000), mouse anti-β-Galactosidase (Promega; 1:1000), mouse anti-1B1 (Developmental Studies Hybridoma Bank; 1:20), mouse anti-Armadillo (Developmental Studies Hybridoma Bank; 1:50), rabbit anti-ISWI (Deuring et al., 2000) (a gift from John Tamkun; 1:100), rabbit anti-Nurf301 (Kwon et al.) (a gift from P. Badenhorst; 1:1000), rabbit anti-dSTAT92E (Flaherty et al., 2010) (a gift from E. Bach; 1:400), rabbit anti-Zfh1 (a gift from R. Lehmann; 1:5000), and rabbit anti-phospho-Histone H3 (Upstate, 1:200). Alexa 488-, Alexa 555-, Alexa 568-, Alexa 633-conjugated (Molecular Probes/Invitrogen) and FITC-conjugated Donkey anti-Chicken (VWR International) secondary antibodies were used at 1:400. DNA was stained with 4,6-diamidino-2-phenylindole (DAPI; Sigma) at 1µg/ml.
Analysis of confocal images
Confocal images were obtained with a Zeiss LSM 5 Pascal or a Zeiss LSM 510 Meta microscope and were collected as serial confocal sections at similar detection settings. Figures were assembled using Adobe Photoshop 7.0 and Adobe Illustrator CS and graphs were created with Prism 5 (GraphPad Software, Inc.). The number of GSCs and CPCs were determined using serial confocal reconstructions of the testis niche. GSCs were scored as Vasa-positive cells (with a spherical fusome where specified) making contact with the hub. CPCs were scored as Zfh1-positive cells with medium to strong staining according to the rainbow indicator in the Zeiss Pascal software. dSTAT92E levels in nurf301 mutant GSCs were compared to those in nurf301 heterozygous neighboring GSCs. Pixel intensity was measured using the Zeiss Pascal software in a single confocal section through the middle of the nucleus of the nurf301 mutant GSC and the nearest heterozygous GSC in the same testis in the most proximal clockwise position. Statistical analysis of stem cell number was performed using Prism 5. Student’s T-test was used to compare two groups and ANOVA analysis was used to compare three or more groups.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues who have generously supplied us with reagents, Kristi Hohenstein for the initial characterization of nurf301 mutant larval gonads, and Dr. M. de Cuevas and M. Issigonis for comments on the manuscript. This work was supported by NIH grants HD052937 and HD040307-07 (EM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arbouzova NI, Bach EA, Zeidler MP. Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr Biol. 2006;16:80–88. doi: 10.1016/j.cub.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004;117:4355–4363. doi: 10.1242/jcs.01390. [DOI] [PubMed] [Google Scholar]
- Badenhorst P, Voas M, Rebay I, Wu C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002;16:3186–3198. doi: 10.1101/gad.1032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bouazoune K, Brehm A. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 2006;14:433–449. doi: 10.1007/s10577-006-1067-0. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–4821. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;13:1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- Chen HW, Chen X, Oh SW, Marinissen MJ, Gutkind JS, Hou SX. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–398. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev Cell. 2005;9:501–510. doi: 10.1016/j.devcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Elfring LK, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek SJ, Waldrip WR, Daubresse G, DePace A, et al. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics. 1998;148:251–265. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, Banerjee U, Bach EA. chinmo Is a Functional Effector of the JAK/STAT Pathway that Regulates Eye Development, Tumor Formation, and Stem Cell Self-Renewal in Drosophila. Dev Cell. 2010;18:556–568. doi: 10.1016/j.devcel.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory L, Came PJ, Brown S. Stem cell regulation by JAK/STAT signaling in Drosophila. Semin Cell Dev Biol. 2008;19:407–413. doi: 10.1016/j.semcdb.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, Matunis E. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science. 2009;326:153–156. doi: 10.1126/science.1176817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kai T, Spradling A. An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc Natl Acad Sci U S A. 2003;100:4633–4638. doi: 10.1073/pnas.0830856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten P, Hader S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117:343–346. doi: 10.1016/s0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- Kawase E, Wong MD, Ding BC, Xie T. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development. 2004;131:1365–1375. doi: 10.1242/dev.01025. [DOI] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- Kiger AA, White-Cooper H, Fuller MT. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature. 2000;407:750–754. doi: 10.1038/35037606. [DOI] [PubMed] [Google Scholar]
- Kwon SY, Xiao H, Glover BP, Tjian R, Wu C, Badenhorst P. The nucleosome remodeling factor (NURF) regulates genes involved in Drosophila innate immunity. Dev Biol. 2008;316:538–547. doi: 10.1016/j.ydbio.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Kwon SY, Xiao H, Wu C, Badenhorst P. Alternative splicing of NURF301 generates distinct NURF chromatin remodeling complexes with altered modified histone binding specificities. PLoS Genet. 2009;5:e1000574. doi: 10.1371/journal.pgen.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MS, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Loden M, van Steensel B. Whole-genome views of chromatin structure. Chromosome Res. 2005;13:289–298. doi: 10.1007/s10577-005-2166-z. [DOI] [PubMed] [Google Scholar]
- Matunis E, Tran J, Gonczy P, Caldwell K, DiNardo S. punt and schnurri regulate a somatically derived signal that restricts proliferation of committed progenitors in the germline. Development. 1997;124:4383–4391. doi: 10.1242/dev.124.21.4383. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Silver DL, Geisbrecht ER, Montell DJ. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development. 2005;132:3483–3492. doi: 10.1242/dev.01910. [DOI] [PubMed] [Google Scholar]
- Terry NA, Tulina N, Matunis E, DiNardo S. Novel regulators revealed by profiling Drosophila testis stem cells within their niche. Dev Biol. 2006;294:246–257. doi: 10.1016/j.ydbio.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Voog J, D'Alterio C, Jones DL. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature. 2008;454:1132–1136. doi: 10.1038/nature07173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–5380. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Xi R, Xie T. Stem cell self-renewal controlled by chromatin remodeling factors. Science. 2005;310:1487–1489. doi: 10.1126/science.1120140. [DOI] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yamasaki Y, Nishida Y. Mi-2 chromatin remodeling factor functions in sensory organ development through proneural gene repression in Drosophila. Dev Growth Differ. 2006;48:411–418. doi: 10.1111/j.1440-169X.2006.00880.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.