Abstract
New tetracycline and streptomycin resistance genes, tet(44) and ant(6)-Ib, were identified in Campylobacter fetus subsp. fetus within a transferable pathogenicity island that is typically unique to Campylobacter fetus subsp. venerealis. The 640-amino-acid tetracycline resistance determinant, Tet 44, belongs to a class of proteins that confers resistance to tetracycline and minocycline by ribosomal protection. The 286-amino-acid streptomycin resistance determinant, ANT(6)-Ib, belongs to a family of aminoglycoside nucleotidyltransferases. The resistance phenotypes were demonstrated by gene inactivation and expression.
Campylobacter fetus contains two closely related subspecies, Campylobacter fetus subsp. fetus and Campylobacter fetus subsp. venerealis, which exhibit distinct host and tissue specificities (19, 21). C. fetus subsp. venerealis is associated with genital tract infections in cattle and causes endemic abortion, embryonic death, and infertility. C. fetus subsp. fetus is associated with intestinal tract colonization in cattle and sheep and has been reported only in sporadic cases of abortion in these animals (12). C. fetus subsp. fetus is most frequently related to human infections causing several types of diseases (14, 22). The host specificity of C. fetus subsp. venerealis has been attributed to the presence of a unique pathogenicity island containing a type IV secretory pathway operon (6, 11). In our study, we describe the identification of new tetracycline and streptomycin resistance genes that were located within the C. fetus subsp. venerealis-specific pathogenicity island present in C. fetus subsp. fetus. The tetracycline- and streptomycin-resistant C. fetus subsp. fetus isolates were collected from veal calves in Switzerland (5) and were identified by PCR (1) and their tolerance to glycine (13, 21).
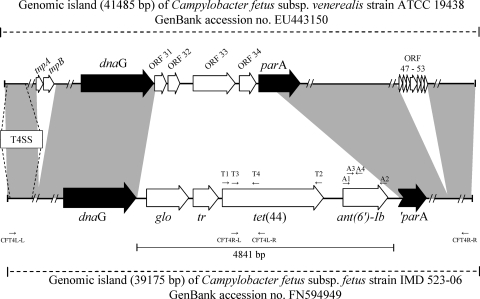
The nature of the tetracycline resistance mechanism was first investigated in the C. fetus subsp. fetus strain IMD523-06 by PCR using degenerate primers that amplify ribosome protection-type genes as previously described (2). Sequence comparison of the resulting 1,207-bp PCR product using the BLAST algorithm revealed homologies with tetracycline resistance determinants. The sequences of the 5′ and 3′ ends of the tetracycline resistance gene and the flanking regions (40,622 bp) were then obtained using the Illumina/Solexa sequencing technology (Fasteris SA, Geneva, Switzerland). Sequence analysis showed that the tetracycline resistance gene was preceded by two genes, a 471-bp putative regulatory gene (tr) and an 807-bp gene (glo) that was homologous to genes encoding glyoxalase, and followed by a novel streptomycin resistance gene (Fig. 1). This antibiotic resistance gene cluster was integrated within a pathogenicity island that was almost identical to the pathogenicity island described in C. fetus subsp. venerealis (6). The genomic regions 23,844 bp upstream of the glyoxalase gene and 12,286 bp downstream of the streptomycin resistance gene showed DNA alignment coverages of 86% and 87%, respectively, with the genomic island of C. fetus subsp. venerealis strain ATCC 19438 (Fig. 1). Differences were mainly due to an insertion sequence element, five additional small hypothetical proteins in C. fetus subsp. venerealis, and variable nucleotide polymorphisms (Fig. 1). Importantly, the genetic structures of the gene locus encoding the type IV secretion system were identical in the two subspecies, and the involved proteins VirB2 to VirB11, VirD4, and CagT (6) shared at least 95% identity.
FIG. 1.
Schematic organization of the antibiotic resistance genes tet(44) and ant(6)-Ib found in the genomic island of C. fetus subsp. fetus compared to the genomic island of C. fetus subsp. venerealis. Gray areas represent domains sharing more than 92% DNA identity. Open reading frames (ORFs) that are found in only one of the two genomic islands are represented by white arrows. tnpA and tnpB, transposases; ORF 31 to 34 and ORF 47 to 53, hypothetical open reading frames; dnaG, hypothetical DNA gyrase gene; glo, hypothetical glyoxalase gene; parA, ParA-like protein gene; ′parA, truncated parA gene. T4SS indicates the location of the type IV secretion system (T4SS) within the pathogenicity island and includes genes virB2 to virB11, virD4, and cagT (6). Primers are indicated by arrows: T1 [tet(44)-F] and T2 [tet(44)-R] and A1 (ant6Ib-F) and A2 (ant6Ib-R) indicate primer pairs used for the genetic cloning of tet(44) and ant(6)-Ib, respectively; T3 (CfetTetKO-L) and T4 (CfetTetKO-R) and A3 (CfetstreptKO-L) and A4 (CfetstreptKO-R) indicate primer pairs used for the disruption of tet(44) and ant(6)-Ib by homologous recombination; and CFT4L-L and CFT4L-R and CFT4R-L and CFT4R-R indicate primer pairs used for the PCR restriction analysis of the genomic island.
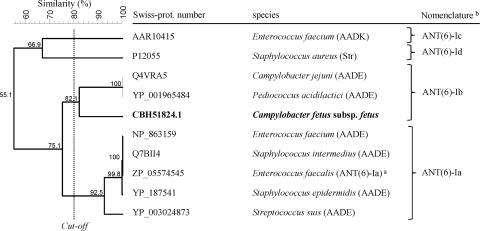
The tetracycline resistance gene encoded a 640-amino-acid (aa) protein, which shares less than 80% overall identity with amino acid sequences of other known tetracycline resistance determinants (Table 1) and was assigned the new name of tet(44) using the nomenclature of the tetracycline resistance determinants (10). Based on its amino acid similarity and resistance to tetracycline and minocycline, tet(44) is likely to confer resistance by ribosomal protection. The streptomycin resistance gene encoded a 286-aa protein that shares homology with aminoglycoside nucleotidyltransferase ANT(6)-I (Fig. 2). Based on its amino acid homology to ANT(6)-I and the resistance to streptomycin (16) (Fig. 2), the new aminoglycoside resistance gene was named ant(6)-Ib using the nomenclature proposed for genes encoding aminoglycoside-modifying enzymes (16, 20). GenBank sequence comparisons showed that tet(44) and ant(6)-Ib were also present in the Clostridium perfringens strain JGS1495, which is currently being completely sequenced (contig GenBank accession number NZ_ABDU01000081).
TABLE 1.
Nucleotide and amino acid identities of the new Tet 44 determinant compared with closely related Tet determinants conferring resistance to tetracycline and minocycline by ribosomal protection
| Determinant | GenBank accession no. | % of amino acid (aa) and nucleotide (nt) identity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tet 44 |
Tet W |
Tet O |
Tet S |
Tet M |
|||||||
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | ||
| Tet 44 | FN594949 | 100 | 100 | 63.6 | 67.0 | 69.1 | 67.5 | 69.0 | 64.6 | 69.5 | 66.0 |
| Tet W | AJ222769 | 100 | 100 | 62.7 | 67.0 | 61.0 | 66.0 | 64.1 | 68.4 | ||
| Tet O | M18896 | 100 | 100 | 70.3 | 70.9 | 75.5 | 76.8 | ||||
| Tet S | L09756 | 100 | 100 | 77.7 | 77.9 | ||||||
| Tet M | X04388 | 100 | 100 | ||||||||
FIG. 2.
Alignment of ANT(6)-Ib from C. fetus subsp fetus (in bold) with other characterized aminoglycoside 6-nucleotidyltransferase enzymes conferring streptomycin resistance. The alignment of the protein sequences was performed by UPGMA (unweighted-pair group method using average linkages) (pairwise, open gap penalty, 100%; unit gap penalty, 0%; FAST, 2.10; Gapcost, 0%) using Bionumerics 5.10 (Applied Maths, Kortrijk, Belgium). Original nomenclature is indicated in parentheses. a, nomenclature proposed by Shaw et al. (16); b, nomenclature proposed in this study based on clusters with similarity levels higher than 80% and as a continuation of the proposed nomenclature (16, 20).
The phenotypes of tet(44) and ant(6)-Ib were demonstrated by gene inactivation and expression. For expression, tet(44) was amplified by PCR using primers tet(44)-F (5′-aatagatctAAAATAATCAACATTGGTATTCTTGCTCA) and tet(44)-R (5′-aatactagtTAGTAACTTAATTTTCTTTTTTATTAAACATATGGCG), and ant(6)-Ib was amplified using primers ant6Ib-F (5′-aatggatccAAAATGAGAACAGAGAAACAAATATATGATACT) and ant6Ib-R (5′-aatactagtTTATCTTTGATATTTTTCTTTTTGTCTTATAACA) (sequences in lowercase indicate synthetic linkers that contain restriction sites [underlined]). The tet(44) primers contained BglII and SpeI restriction sites and the ant(6)-Ib primers contained BamHI and SpeI restriction sites to allow site-directed cloning into the Escherichia coli-Campylobacter shuttle vector pRYSK3, which contains a Campylobacter surface array protein A (SAP) promoter (9). The resulting plasmids, pCA73 and pCA75 (Table 2), were first transformed into E. coli S17-1 λpir (4) by heat shock and subsequently transferred from E. coli into the ciprofloxacin-resistant C. fetus subsp. fetus strain 1516477 by filter mating, as described previously (9). Transconjugants were obtained on heart infusion agar plates (Becton Dickinson, Franklin Lakes, NJ) containing 8 μg/ml of ciprofloxacin and either 8 μg/ml of tetracycline or 10 μg/ml of streptomycin after 48 h of incubation at 37°C under microaerophilic conditions (80% N2, 10% CO2, and 10% H2). The tet(44) and ant(6)-Ib genes were disrupted in C. fetus subsp. fetus strain IMD523-06 by single crossover homologous recombinations (15) using the suicide kanamycin resistance plasmid pCA48, which is a pRYSK3 derivative lacking a 90-bp AflII fragment in the Campylobacter replication region. Internal fragments of tet(44) were amplified by PCR using primers CfetTetKO-L (5′-tgcgcggccgcATGTTTTTAGAACGTCAGCG) and CfetTetKO-R (5′-atcgagctcAGCACTTCCATGGTATATAGG), while internal fragments of ant(6)-Ib were amplified using primers CfetstreptKO-L (5′-tgcgcggccgcGGTTACTTTAGAAGGTTCAAGAACA) and CfetstreptKO-R (5′-atcgagctcCCTTTCGACATAATCCTTTCAC). The PCR products were then cloned into the NotI and SacI restriction sites of pCA48. Single crossover recombinants were obtained by filter mating on heart infusion agar plates containing 25 μg/ml of kanamycin and 8 μg/ml of nalidixic acid, as described above.
TABLE 2.
Characteristics and MICs for tetracyclines and streptomycin for the C. fetus subsp. fetus strains used in this studya
| Strain | Characteristic(s) | MIC (μg/ml) |
|||
|---|---|---|---|---|---|
| TET | DOX | MIN | STR | ||
| IMD523-06 | tet(44) ant(6)-Ib; Nalr; bovine isolate | 16 | 8 | 16 | >64 |
| 1516477 | Recipient strain; Cipr; human isolate | 1 | 0.5 | 1 | ≤1 |
| CA73 | 1516477 containing tet(44) on pCA73 | >32 | 32 | 32 | ≤1 |
| CA74 | IMD523-06 with Δtet(44)::pCA64; Kanr | 1 | 0.5 | 2 | >64 |
| CA75 | 1516477 containing ant(6)-Ib on pCA75 | 1 | 0.5 | 1 | 64 |
| CA76 | IMD523-06 with Δant(6)-Ib::pCA76; Kanr | 16 | 8 | 16 | ≤1 |
| CA80 | 1516477 containing IMD523-06 pathogenicity island with tet(44) and ant(6)-Ib; Cipr | 16 | 4 | 16 | 64 |
TET, tetracycline; DOX, doxycycline; MIN, minocycline; STR, streptomycin. Resistance markers used for transformation: Nalr, intrinsic resistance to nalidixic acid; Cipr, resistance to ciprofloxacin; Kanr, resistance to kanamycin.
Resistance phenotypes were determined by MIC measurements in Mueller-Hinton broth supplemented with 5% laked horse blood (3). When expressed from pRYSK3 in C. fetus subsp. fetus, tet(44) mediated at least a 5-fold increase in tetracycline, doxycycline, and minocycline resistance (Table 2). Similarly, ant(6)-Ib expressed from pRYSK3 in C. fetus subsp. fetus also mediated at least a 5-fold increase in streptomycin resistance (Table 2). On the other hand, a 3- to 5-fold decrease in resistance to these drugs was observed when the genes were disrupted (Table 2). MICs of ciprofloxacin, nalidixic acid, meropenem, ampicillin, amoxicillin-clavulanic acid, chloramphenicol, florfenicol, gentamicin, neomycin, and erythromycin remained unchanged (data not shown).
The pathogenicity and resistance island was localized on the chromosome by DNA hybridization using a digoxigenin (DIG)-labeled probe for tet(44). It was transferred from C. fetus subsp. fetus strain IMD523-06 into C. fetus subsp. fetus strain 1516477 by filter mating. Transconjugants were selected on plates containing 10 μg/ml of streptomycin and 8 μg/ml of ciprofloxacin. Transconjugants were identified by the detection of tet(44) and ant(6)-Ib and by multilocus sequence typing (19) (http://pubmlst.org/cfetus/) with the donor strain belonging to ST2 and the recipient strain and the transconjugants belonging to ST35. ClaI restriction digests of two large PCR fragments (21,752 and 14,078 bp) covering the regions situated between tet(44) and both ends of the pathogenicity island (Fig. 1) generated identical restriction profiles in both the donor and transconjugants, demonstrating that the complete genomic island was transferred by conjugation. The fragments were amplified using the Expand Long Template PCR system (Roche Diagnostics AG, Rotkreuz, Switzerland) (extension time, 20 min; annealing temperature, 58°C) with primers specific to the 5′ end of the genomic island and to tet(44) (CFT4L-L, 5′-TGTTAGTCAAAAAAGATGATATGGCTTTTAGGC, and CFT4L-R, 5′-CATCTTTAGCAGAAATTACTAAAATTGCTCCATC) and primers specific to tet(44) and to the 3′ end of the genomic island (CFT4R-L, 5′-ATAATCAACATTGGTATTCTTGCTCATGTAGATG, and CFT4R-R, 5′-GAAGTGGGGTAATGTTGTTTTCATAGGAATT) (Fig. 1).
An identical PCR restriction profile was also obtained with 10 additional tetracycline- and streptomycin-resistant C. fetus subsp. fetus strains (data not shown), indicating that this genomic island has already disseminated into the C. fetus subsp. fetus population. Given the location of tet(44) and ant(6)-Ib within this pathogenicity island, it is likely that the use of antibiotics has been contributing to the spread of this element. Resistance to tetracycline has been reported in C. fetus subsp. fetus from both human and animal origins (8, 17, 18). Whether tet(44) and the pathogenicity island are present in these isolates remains to be determined.
The use of antibiotics poses the risk of selecting a transferable element in C. fetus subsp. fetus, which has so far been associated only with the virulence and host specificity of C. fetus subsp. venerealis. The presence of the C. fetus subsp. venerealis island in C. fetus subsp. fetus may also lead to the false identification of C. fetus subspecies because some PCR methods have been based on genes present in the genomic island to discriminate between the C. fetus subspecies (7, 11). This discovery gives a new insight into the host specificity and pathogenicity potential of C. fetus subsp. fetus and also into the diagnosis of diseases caused by C. fetus infections.
Nucleotide sequence accession number.
The sequences of tet(44), ant(6)-Ib, and the flanking genomic island of C. fetus subsp. fetus IMD523-06 have been deposited in the EMBL/GenBank/DDBJ database under accession number FN594949.
Footnotes
Published ahead of print on 17 May 2010.
REFERENCES
- 1.Abril, C., E. M. Vilei, I. Brodard, A. Burnens, J. Frey, and R. Miserez. 2007. Discovery of insertion element ISCfe1: a new tool for Campylobacter fetus subspecies differentiation. Clin. Microbiol. Infect. 13:993-1000. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa, T. M., K. P. Scott, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1:53-64. [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 8th ed., vol. 29, no. 2. Approved standard M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 5.Di Labio, E., G. Regula, A. Steiner, R. Miserez, A. Thomann, and U. Ledergerber. 2007. Antimicrobial resistance in bacteria from Swiss veal calves at slaughter. Zoonoses Public Health 54:344-352. [DOI] [PubMed] [Google Scholar]
- 6.Gorkiewicz, G., S. Kienesberger, C. Schober, S. R. Scheicher, C. Gully, R. Zechner, and E. L. Zechner. 2010. A genomic island defines subspecies-specific virulence features of the host-adapted pathogen Campylobacter fetus subsp. venerealis. J. Bacteriol. 192:502-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hum, S., K. Quinn, J. Brunner, and S. W. On. 1997. Evaluation of a PCR assay for identification and differentiation of Campylobacter fetus subspecies. Aust. Vet. J. 75:827-831. [DOI] [PubMed] [Google Scholar]
- 8.Inglis, G. D., D. W. Morck, T. A. McAllister, T. Entz, M. E. Olson, L. J. Yanke, and R. R. Read. 2006. Temporal prevalence of antimicrobial resistance in Campylobacter spp. from beef cattle in Alberta feedlots. Appl. Environ. Microbiol. 72:4088-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kienesberger, S., G. Gorkiewicz, M. M. Joainig, S. R. Scheicher, E. Leitner, and E. L. Zechner. 2007. Development of experimental genetic tools for Campylobacter fetus. Appl. Environ. Microbiol. 73:4619-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moolhuijzen, P. M., A. E. Lew-Tabor, B. M. Wlodek, F. G. Aguero, D. J. Comerci, R. A. Ugalde, D. O. Sanchez, R. Appels, and M. Bellgard. 2009. Genomic analysis of Campylobacter fetus subspecies: identification of candidate virulence determinants and diagnostic assay targets. BMC Microbiol. 9:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Office International des Epizooties. 2008. Bovine genital campylobacteriosis, p. 661-670. In Manual of diagnostic tests and vaccines for terrestrial animals. Office International des Epizooties, Paris, France.
- 13.On, S. L., and C. S. Harrington. 2001. Evaluation of numerical analysis of PFGE-DNA profiles for differentiating Campylobacter fetus subspecies by comparison with phenotypic, PCR and 16S rDNA sequencing methods. J. Appl. Microbiol. 90:285-293. [DOI] [PubMed] [Google Scholar]
- 14.Pacanowski, J., V. Lalande, K. Lacombe, C. Boudraa, P. Lesprit, P. Legrand, D. Trystram, N. Kassis, G. Arlet, J. L. Mainardi, F. Doucet-Populaire, P. M. Girard, and J. L. Meynard. 2008. Campylobacter bacteremia: clinical features and factors associated with fatal outcome. Clin. Infect. Dis. 47:790-796. [DOI] [PubMed] [Google Scholar]
- 15.Pei, Y., V. Parreira, V. M. Nicholson, and J. F. Prescott. 2007. Mutation and virulence assessment of chromosomal genes of Rhodococcus equi 103. Can. J. Vet. Res. 71:1-7. [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tremblay, C., and C. Gaudreau. 1998. Antimicrobial susceptibility testing of 59 strains of Campylobacter fetus subsp. fetus. Antimicrob. Agents Chemother. 42:1847-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tremblay, C., C. Gaudreau, and M. Lorange. 2003. Epidemiology and antimicrobial susceptibilities of 111 Campylobacter fetus subsp. fetus strains isolated in Quebec, Canada, from 1983 to 2000. J. Clin. Microbiol. 41:463-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Bergen, M. A., K. E. Dingle, M. C. Maiden, D. G. Newell, L. van der Graaf-van Bloois, J. P. van Putten, and J. A. Wagenaar. 2005. Clonal nature of Campylobacter fetus as defined by multilocus sequence typing. J. Clin. Microbiol. 43:5888-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanhoof, R., E. Hannecart-Pokorni, and J. Content. 1998. Nomenclature of genes encoding aminoglycoside-modifying enzymes. Antimicrob. Agents Chemother. 42:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Véron, M., and R. Chatelain. 1973. Taxonomic study of the genus Campylobacter Sebald and Véron and designation of the neotype strain for the type species, Campylobacter fetus (Smith and Taylor) Sebald and Véron. Int. J. Syst. Bacteriol. 23:122-134. [Google Scholar]
- 22.Zonios, D. I., G. D. Panayiotakopoulos, E. O. Kabletsas, E. L. Tzima, I. Stefanou, and A. J. Archimandritis. 2005. Campylobacter fetus bacteraemia in a healthy individual: clinical and therapeutical implications. J. Infect. 51:329-332. [DOI] [PubMed] [Google Scholar]