Abstract
CMX157 is a lipid (1-0-hexadecyloxypropyl) conjugate of the acyclic nucleotide analog tenofovir (TFV) with activity against both wild-type and antiretroviral drug-resistant HIV strains, including multidrug nucleoside/nucleotide analog-resistant viruses. CMX157 was consistently >300-fold more active than tenofovir against multiple viruses in several different cell systems. CMX157 was active against all major subtypes of HIV-1 and HIV-2 in fresh human peripheral blood mononuclear cells (PBMCs) and against all HIV-1 strains evaluated in monocyte-derived macrophages, with 50% effective concentrations (EC50s) ranging between 0.20 and 7.2 nM. The lower CMX157 EC50s can be attributed to better cellular uptake of CMX157, resulting in higher intracellular levels of the active antiviral anabolite, TFV-diphosphate (TFV-PP), inside target cells. CMX157 produced >30-fold higher levels of TFV-PP in human PBMCs exposed to physiologically relevant concentrations of the compounds than did TFV. Unlike conventional prodrugs, including TFV disoproxil fumarate (Viread), CMX157 remains intact in plasma, facilitating uptake by target cells and decreasing relative systemic exposure to TFV. There was no detectable antagonism with CMX157 in combination with any marketed antiretroviral drug, and it possessed an excellent in vitro cytotoxicity profile. CMX157 is a promising clinical candidate to treat wild-type and antiretroviral drug-resistant HIV, including strains that fail to respond to all currently available nucleoside/nucleotide reverse transcriptase inhibitors.
Nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs) remain the backbone for HIV combination therapy, despite the availability of multiple alternative drug classes targeting HIV replication (2009 U.S. Department of Health and Human Services [DHHS] guidelines). Although treatment regimens for HIV have improved dramatically since the advent of monotherapy, antiretroviral drug toxicities, difficulties with regimen adherence, and HIV resistance remain significant challenges for many patients (11, 17, 27, 45). Further complicating the issue of resistance are recent data demonstrating that mutations present at levels below the limit of detection by standard genotyping assays are relatively prevalent and may negatively impact antiretroviral efficacy (19, 23). For these reasons, there is an ongoing need for new NRTIs that diminish or eliminate these obstacles to optimum clinical antiviral efficacy.
Tenofovir (TFV) disoproxil fumarate (TDF; Viread) (16), a prodrug of TFV, is one of the most widely used NRTIs for treatment of HIV. TDF was initially developed for therapy-experienced patients, and two key studies of the development of TDF, GS-902 and GS-907, enrolled NRTI-therapy-experienced patients. Retrospective analysis of these studies identified patient populations that responded poorly to TDF, based on HIV reverse transcriptase genotype and the corresponding phenotype. Many of the patients who were unresponsive to TDF in these studies were infected with an NRTI-resistant virus that would have been unlikely to respond to any NRTI. Notably, specific patterns of thymidine analog mutations (TAMs) were strongly associated with poor response, and patients with the K65R mutation, although uncommon in the cohort, failed to respond virologically (28). Small (<4-fold) changes in phenotypic resistance to TFV were associated with loss of clinical antiviral effect (16, 28).
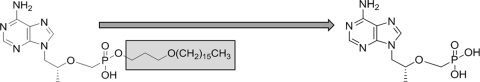
CMX157 {3-(hexadecyloxy)propyl hydrogen [(R)-1-(6-amino-9H-purin 9-yl)propan-2-yloxy]methylphosphonate; hexadecyloxypropyl TFV [HDP-TFV]}, a lipid conjugate of TFV, was designed to mimic lysophosphatidylcholine to take advantage of natural lipid uptake pathways and to achieve high intracellular concentrations of the active antiviral, with the aim of increasing the effectiveness of TFV against wild-type and mutant HIV (18, 33, 34). The structure of CMX157 is shown in Fig. 1. In vivo, TDF is rapidly converted by plasma esterases to the TFV dianion, which is not readily taken up by target cells for HIV therapy but is a substrate for organic anion transporters that are highly expressed on renal proximal tubule epithelial cells (RPTECs). This leads to relatively high 50% effective concentrations (EC50s) for TFV against HIV and, infrequently, to renal toxicity (28, 48). We sought to address these limitations through the development of a lipid conjugate of TFV designed for efficient uptake by generalized mechanisms rather than transporters preferentially expressed on RPTECs. Here, we report the results from our analysis of the in vitro activity and cytotoxicity profile of CMX157. CMX157 demonstrated potential to effectively suppress replication of multi-NRTI-resistant (MNR) HIV that cannot be treated with any currently available NRTIs, including TDF.
FIG. 1.
Structures of CMX157 (1) and TFV (2). The hexadecyloxypropyl lipid moiety is highlighted in gray and is cleaved inside cells to liberate TFV.
MATERIALS AND METHODS
Materials.
The synthesis of CMX157 has been previously described (33). TFV-monophosphate (TFV-MP) and TFV-diphosphate (TFV-PP) were obtained from Moravek Biochemicals and Radiochemicals (Brea, CA). The NRTIs lamivudine (3TC), abacavir (ABC), zidovudine (ZDV; AZT), stavudine (d4T), zalcitabine (ddC), didanosine (ddI), emtricitabine (FTC), TFV (PMPA), and TDF; the non-NRTIs (NNRTIs) efavirenz (EFV), etravirine (ETV; Intelence) (TMC125 from Tibotec, Inc.), and nevirapine (NVP); the protease inhibitors (PIs) amprenavir (APV), atazanavir (ATV; sulfate form of compound), darunavir (DRV; Tibotec, Inc.), indinavir (IDV; sulfate form of compound), lopinavir (LPV), nelfinavir (NFV), ritonavir (RTV), saquinavir (SQV), and tipranavir (TPV); the entry inhibitors maraviroc (MVC) and enfuvirtide (T-20; Roche); and the integrase inhibitor raltegravir (RAL; Merck & Company, Inc.) were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. The NNRTI delavirdine (DLV) was purchased from Biomol International, LP (Plymouth Meeting, PA). Ribavirin (RBV) was purchased from Sigma (St. Louis, MO).
Viruses and cells.
Virus isolates and cell lines were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, as follows: HIV-1 isolates 92RW009, 92UG001, 92UG024, 92UG029, 92UG037, 92UG046, 92BR014, 92BR025, 93BR019, 93BR020, 93BR029, 92TH014, 92TH026, and 93TH073 from the UNAIDS Network for HIV Isolation and Characterization (10); HIV-1 isolates 93IN101 and 93MW959 from Robert Bollinger and the UNAIDS Network for HIV Isolation and Characterization (10); HIV-1 isolates CMU06 and CMU08 from Kenrad Nelson and the UNAIDS Network for HIV Isolation and Characterization (10); HIV-1 isolates JV1083 and G3 from Alash'le Abimiku (1); HIV-1 isolates BCF01, BCF02, and BCF03 from Sentob Saragosti, Françoise Brun-Vézinet, and François Simon (26); HIV-1IIIB from Robert C. Gallo (38, 39, 42); HIV-1Ba-L from Suzanne Gartner, Mikulas Popovic, and Robert Gallo (12, 37); HIV-1Ada-M from Howard Gendelman (13-15, 50); HIV-196USHIPS7 from D. Ellenberger, P. Sullivan, and R. B. Lal (5, 8, 44, 47); HIV-1JR-CSF from Irvin Chen (4, 22); HIV-1RU132 from A. Bobkov and Jonathon Weber; HIV-1 molecular clone pNL4-3 from Malcolm Martin (2); the multidrug-resistant HIV-1 reverse transcriptase panel (entire panel of 14 viruses, including HIV-1 clones 7324-1, 7324-4, 10076-4, 7295-1, 4755-5, 6463-13, 7303-3, 1617-1, 35764-2, 29129-2, 52534-2, 56252-1, 71361-1, and 8415-2) from Robert W. Shafer (7); HIV-2CDC310319 from Stefan Wiktor and Mark Rayfield (32); HIV-2CDC310342 from Mark Rayfield and Stephen Wiktor; HIV-2CBL-20/H9 from Robin Weiss (46); CEM-SS cells from Peter L. Nara (9, 30, 31); and MAGI-CCR5 cells from Julie Overbaugh (6). Each of these viruses was propagated in fresh human cells as described in the product profile provided by the NIAID AIDS Research and Reference Reagent Program.
Anti-HIV assays in human PBMCs.
HIV human peripheral blood mononuclear cell (PBMC) assays were performed as described previously (41). Briefly, fresh PBMCs, seronegative for HIV and hepatitis B virus (HBV), were isolated from blood samples of the screened donors (Biological Specialty Corporation, Colmar, PA) by using lymphocyte separation medium (LSM; density, 1.078 ± 0.002 g/ml; Cellgro; Mediatech, Inc.) by following the manufacturer's instructions. Cells were stimulated by incubation in 4 μg/ml phytohemagglutinin (PHA; Sigma) for 48 to 72 h. Mitogenic stimulation was maintained by the addition of 20 U/ml recombinant human interleukin-2 (rhIL-2; R&D Systems, Inc.) to the culture medium. PHA-stimulated PBMCs from at least two donors were pooled, diluted in fresh medium, and added to 96-well plates at 5 × 104 cells/well. Cells were infected (final multiplicity of infection [MOI] of ∼0.1) in the presence of nine different concentrations of test compounds (triplicate wells/concentration) and incubated for 7 days. To determine the level of virus inhibition, cell-free supernatant samples were collected for analysis of reverse transcriptase activity (3). Following removal of supernatant samples, compound cytotoxicity was measured by the addition of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS; CellTiter 96 reagent; Promega) by following the manufacturer's instructions.
In vitro evaluation of the effect of human serum.
The effect of human serum on CMX157 antiviral activity was determined using a modification of the HIV PBMC assay described above. In this experiment, the standard assay was modified slightly to include a preincubation of virus (HIV-1 JR-CSF) with the PBMCs prior to the addition of drug and human serum in order to obtain adequate and more-consistent levels of virus infection in the various cultures that contained different levels of human serum. In the modified assay, 8 × 106 cells from pooled donors were incubated with 1.0 ml of stock virus for 1 h at 37°C and 5% CO2 followed by centrifugation for 1 h at 900 rpm (200 × g). Cells were then gently resuspended and incubated for an additional 2 h. During this second incubation period, 2.5 × 104 uninfected PBMCs from pooled donors were added to the interior 60 wells of a round-bottom 96-well plate in a volume of 50 μl of medium, followed by the addition of 100 μl of diluted compounds in medium at 2× concentration to the appropriate wells. At the end of the second incubation period, infected cells were diluted in medium to a concentration of 5 × 105 cells/ml (without washout of virus), and 50 μl (2.5 × 104 cells) were added to each well of the plate. The remaining steps of the assay were performed in the same manner as for the standard assay. For this study, the 0% human serum control included 15% fetal bovine serum (FBS; i.e., standard assay conditions). For the 10%, 15%, 20%, and 30% human serum evaluations, the appropriate amount of human serum was added in addition to the 15% FBS that is in the standard medium.
Anti-HIV-1 assays in human macrophages.
To isolate monocyte-derived macrophages (MDMs), PBMCs were isolated from blood by using LSM as described above and subsequently washed two times with phosphate-buffered saline (PBS) by low-speed centrifugation. The cells were diluted to 4 × 106 cells/ml in Dulbecco's modified Eagle's medium (DMEM) with 10% heat-inactivated human pooled AB serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. MDMs were allowed to adhere to the interior 60 wells (100 μl/well) of a 96-well flat-bottom plate for 2 to 18 h at 37°C and 5% CO2. Following adherence, the cultures were washed with sterile Dulbecco's PBS (DPBS) to remove nonadherent cells (lymphocytes and contaminating red blood cells [RBCs]). Two hundred microliters of RPMI 1640 medium supplemented with 15% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin was subsequently added to the wells. Culture medium was replaced once per week until the cells were used in the antiviral assay. Following 6 to 14 days in culture, the MDM cultures were washed three times to remove any nonadherent cells, and serially diluted test compounds (nine concentrations; triplicate wells/concentration) were added, followed by the addition of a predetermined titer of HIV-1 (final MOI ≅ 0.1). To reduce p24 background signal in the assay, cultures were washed a final time by medium removal 24 h postinfection, fresh compound was added, and the cultures were continued for an additional 6 days. At assay termination, virus replication was measured by collecting cell-free supernatant samples, which were analyzed for HIV-1 p24 antigen content by using a commercially available p24 enzyme-linked immunosorbent assay (ELISA; PerkinElmer) by following the manufacturer's instructions. Following removal of supernatant samples, compound cytotoxicity was measured by addition of MTS to the plates for determination of cell viability.
Cross-resistance analysis using the PhenoSense assay.
The susceptibility of the HIV-1 to marketed NRTIs was assessed using a recombinant virus assay, PhenoSense HIV (Monogram Biosciences). Briefly, PhenoSense HIV uses nucleic acid amplification to derive viral sequences from a patient's plasma. A resistance test vector is constructed by incorporating the patient-derived segment into a viral vector with an indicator gene (luciferase) inserted within the deleted HIV-1 envelope (env) gene. The recombinant virus is tested in the lab against increasing concentrations of the antiretroviral drugs, and the resulting data are plotted as an inhibition curve. The inhibition curve of the patient virus is compared to that of a drug-sensitive reference virus for each drug. The results from this assay are expressed as an EC50, which equals the concentration of drug that inhibits the virus by 50% (36). Reduced drug susceptibility is indicated by a shift in the patient inhibition curve toward higher drug concentrations, which may be expressed as a ratio of the test recombinant EC50 to the wild-type control EC50. This ratio is referred to here as the fold change in resistance. CMX157 activity against wild-type and NRTI-resistant HIV was determined for a panel of 30 NRTI isolates with major NRTI mutations, including K65R with or without M184V, multiple TAM combinations with or without M184V, and K70E in various combinations, and multinucleoside-resistant complexes, including T69SXX and Q151M isolates.
Antiretroviral combination studies.
Two-drug combination experiments were performed in CEM-SS cells infected with HIV-1IIIB by using a virus-induced cytopathic effect (CPE) inhibition assay (all combinations except those with MVC) or in MAGI-CCR5 cells infected with HIV-1Ba-L by using a β-galactosidase reporter gene induction assay (MVC only) as previously described (24, 41). Each two-drug combination was tested with three independent experiments. The concentrations of CMX157 used in these evaluations were selected in order to test a broad range of concentrations and to provide as complete a dose response curve as possible under the limitations of eight total concentrations. Similarly, the concentrations of the FDA-approved antiretroviral drugs were selected to provide as complete of a dose response curve as possible under the limitations of five total concentrations. The positive antagonism control of d4T in combination with RBV was tested in parallel with each of the assays. Analysis of drug interactions for each of the two-drug combinations was performed using the Prichard and Shipman MacSynergy II three-dimensional model for statistical evaluation of combination assays (40). For these studies, synergy is defined as drug combinations yielding synergy volumes of >50. Slightly synergistic activity and highly synergistic activity have been operationally defined as yielding synergy volumes of 50 to 100 and >100, respectively. Additive drug interactions have synergy volumes in the range of −50 to 50, while synergy volumes between −50 and −100 are considered slightly antagonistic and those of <−100 are highly antagonistic.
In vitro anabolism in PBMCs.
Fresh human PBMCs were isolated and stimulated with PHA/IL-2 as described above for HIV PBMC assays. Unstimulated and stimulated PBMCs were resuspended at 2 × 106 cells/ml in RPMI 1640 medium supplemented with 15% FBS, 2 mM l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and nonessential amino acids. The PBMCs were then split into 25-ml aliquots (5 × 107 cells) in T75 tissue culture flasks. CMX157 and TFV were diluted in culture medium and added to separate cultures of both unstimulated and stimulated PBMCs to give the following final incubation concentrations: CMX157, 10 μM, 1.0 μM, 0.1 μM, and 0.01 μM; TFV, 10 μM and 1.0 μM. Control flasks in which the cultures were treated with dimethyl sulfoxide (DMSO; single concentration equivalent to the amount of DMSO present in the 10 μM TFV-treated cultures) or MeOH:distilled water (dH2O):NH4OH (50:50:0.2; single concentration equivalent to the concentration present in the 10 μM CMX157-treated culture) were also prepared. Following the addition of test compounds or controls, the cultures were incubated for 24 h in 5% CO2 at 37°C. Upon completion of the 24-h incubation period, the PBMCs from each culture were washed and processed as follows. Cells were resuspended and transferred to 50-ml conical centrifuge tubes. The T75 flasks were rinsed twice with 10 ml of 0.9% NaCl (+4°C), and the resulting solutions were added to the 50-ml conical centrifuge tubes. Samples were kept on ice from this point forward. Samples were immediately centrifuged for 10 min at ∼800 × g at +4°C (Sorvall H1000B rotor at 2,000 rpm). The supernatants were carefully removed and discarded by draining them out of the tubes. Forty milliliters of 0.9% NaCl solution (+4°C) was added to each tube, followed by gentle inversion of the tube three to four times to resuspend the cell pellet. Samples were immediately centrifuged again for 10 min close to 800 × g at +4°C. The supernatants were carefully removed and discarded by draining. Four milliliters of 0.9% NaCl solution (+4°C) was added to the cell pellets, and the cells were resuspended by gentle pipetting. NaCl (0.9% +4°C) solution was added to a final volume of 40 ml, followed by gentle inversion of the tube three to four times. Samples were immediately centrifuged again for 10 min at ∼800 × g at +4°C. The supernatants were carefully removed and discarded by draining. Using a Pipetman pipettor, 2 ml of 0.9% NaCl solution (+4°C) was added to the cell pellets and used to resuspend the cells by gentle pipetting. The cells were counted by trypan blue dye exclusion by the use of a hemacytometer and then adjusted to a concentration of 1 × 107 cells/ml by adding 0.9% NaCl (+4°C). For each sample, 1-ml aliquots of the cells were distributed to each of three 1.5-ml Eppendorf microcentrifuge tubes (screw cap with O-ring), which were centrifuged at ∼1,800 × g for 10 min at +4°C (Sorvall Legend RT refrigerated table top centrifuge; Micro Liter rotor 7500 3332). The supernatants were carefully removed and discarded by draining. The cell pellets were resuspended in 100 μl of ice-cold 70:30 MeOH:dH2O and quickly frozen in a dry-ice/ethanol bath and immediately stored at −80°C.
Analyses of the PBMC samples to determine concentrations of the nucleotide analog TFV and anabolites TFV-MP and TFV-PP were performed by Optimized Analytical Solutions, LLC (OpAns, Durham, NC), and conducted utilizing high-performance liquid chromatography (HPLC) with tandem mass spectrometric detection (MS/MS). TFV, TFV-MP, and TFV-PP were separated by gradient, reverse-phase, ion-pairing chromatography and detected by positive-ion electrospray.
Cytotoxicity in primary and transformed human cells.
CMX157, TDF, and TFV were evaluated in parallel with Triton X-100 as a control by using a six-concentration dose response (10, 3.2, 1, 0.32, 0.1, and 0.032 μM for CMX157 and TDF; 100, 32, 10, 3.2, 10 and 0.32 μM for TFV) for the inhibition of cell growth of the following cell lines: CEM-SS (human T4-lymphoid), HeLa (human cervical epithelial carcinoma), ME180 (CD4-negative human cervical epithelial carcinoma), Huh-7 (human liver cell carcinoma), and HepG2 (human hepatocyte). Cytotoxicity was also evaluated in primary human cells, including PHA-stimulated and unstimulated PBMCs, monocyte/macrophages, dendritic cells, and hepatocytes. Cell viability was quantified by XTT tetrazolium dye uptake.
Established cell preparation.
Established cell lines CEM-SS, HeLa, ME180, Huh-7, and HepG2 were passaged in T75 flasks prior to use in the assay in cell culture medium recommended by the ATCC. On the day preceding the assay, the cells were split 1:2 to ensure they were in an exponential-growth phase at the time of compound treatment. Total cell and viability quantification was performed using a hemacytometer and trypan blue dye exclusion. Cell viability was greater than 95% for the cells to be utilized in the assay. The cells were resuspended at the appropriate number of cells per ml (5 × 103 cells per well to 2 × 104 cells per well) in tissue culture medium and added to the microtiter plates in a volume of 100 μl. Adherent cell lines were incubated overnight at 37°C and 5% CO2 to allow for cell adherence to the plates.
Fresh PBMC preparation.
PBMCs were prepared for use in the toxicity evaluations as described above.
Monocyte/macrophage cell preparation.
Monocyte/macrophage cultures for use in the toxicity evaluations were prepared as described above.
Monocyte-derived dendritic cell preparation.
Freshly separated PBMCs (from one donor) were suspended in DPBS at 4 × 106 cells/ml, and 15 ml was transferred into a 75-cm2 cell culture flask. The flask was incubated at 37°C and 5% CO2 for 90 min and washed five to seven times with DPBS to remove the nonadherent cells. Fifteen milliliters of RPMI 1640 medium supplemented with 10% FBS, 2 mM l-glutamine, 25 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, 50 ng/ml recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF), and 50 ng/ml rhIL-4 was added to the flask. The flask was incubated at 37°C and 5% CO2 for 7 days. The cells were cultured in the same cytokine cocktails with the addition of lipopolysaccharide (LPS; 10 ng/ml) for 2 days, and then the cell monolayer was washed. Visual examination for contamination and cell morphology change was performed prior to cytotoxicity evaluations.
Fresh human hepatocyte preparation.
Primary human hepatocytes with a Matrigel (0.2 mg/ml) overlay were obtained from Celsis (catalog number F91565; female lot number FHU-L-022708). Upon receipt, the medium was replaced with fresh hepatocyte culture medium (In Vitro Technologies catalog number Z990012) prewarmed to 37°C, and the plate was placed in the 37°C/5% CO2 incubator overnight. Following the incubation, the medium was removed and replaced with 100 μl of fresh medium. Diluted test material was added in triplicate in a 100-μl volume. Plates were returned to the incubator at 37°C and 5% CO2 for 24 h and then stained for cell viability with XTT.
GM-CFU cell preparation.
Fresh human bone marrow mononuclear cells were obtained from Lonza (Walkersville, MD; catalog number IM-125C, lot number 080815C) on the day of assay initiation. Total cell and viability quantification was performed using a hemacytometer and trypan blue dye exclusion. The cells were resuspended in cell culture medium (Iscove modified Dulbecco medium; Lonza) containing 15% heat-inactivated FBS (Gibco), 10% giant cell tumor conditioned medium (Bone Marrow Plus; Sigma), 10 ng/ml rhIL-6, 10 ng/ml rhIL-3, and 25 ng/ml rhGM-CFU and a final concentration of 1% methylcellulose (Sigma) and were added to the drug-containing six-well plates in a volume of 1 ml at 105 cells per well.
BFU-E cell preparation.
Fresh human bone marrow mononuclear cells were obtained from Lonza on the day of assay initiation. Total cell and viability quantification was performed using a hemacytometer and trypan blue dye exclusion. The cells were resuspended in cell culture medium (Iscove modified Dulbecco medium; Lonza) containing 20% heat-inactivated FBS (Gibco), 10% T-cell conditioned medium (RPMI 1640 medium, 5% FBS, 2 mM l-glutamine, 5 × 10−5 M 2-mercaptoethanol), 1 U/ml human erythropoietin (R&D Systems), and a final concentration of 1% methylcellulose (Sigma) and were added to the drug-containing six-well plates in a volume of 1 ml at 105 cells per well.
Cellular toxicity evaluations.
Each test material was evaluated in triplicate at 5, 0.5, and 0.05 μM. AZT was evaluated in parallel as a negative (i.e., cytotoxic) control. Following 14 days of incubation at 37°C and 5% CO2, colonies of greater than 30 cells in the GM-CFU and erythroid colony-forming (BFU-E) assay plates were counted by microscopic observation. Colonies in the BFU-E assay were confirmed to be of erythroid nature by using benzidene. Briefly, the plates were stained with 12% glacial acetic acid containing 0.4% benzidene (Sigma) and 0.3% hydrogen peroxide solution for 10 min at room temperature. The plates were then washed with 12% glacial acetic acid and fixed with methanol. Blue precipitation was an indication of hemoglobin in the erythroid cells.
RESULTS
Antiretroviral activity of CMX157 against wild-type HIV in PBMCs and macrophages.
CMX157 was active against all major subtypes of HIV-1 in PBMCs, with EC50s ranging from 0.20 to 7.2 nM for the 27 different viruses tested (mean EC50 ± standard deviation [SD] = 2.6 ± 2.0 nM). No cytotoxicity was observed up to the high test concentration of 1,000 nM CMX157. By comparison, TFV EC50s against HIV-1 group M subtypes A to G and group O have been reported to range from 1,600 to 4,900 nM (16). Table 1 summarizes the mean CMX157 EC50 results for HIV-1 group M subtypes A to G and HIV-1 group O. Six isolates were tested for subtype B, and three isolates were tested for all other subtypes/groups. Similar results were obtained for CMX157 against three HIV-2 isolates in PBMCs (Table 1) (mean EC50 ± SD = 3.5 ± 1.5; EC50 range, 1.8 to 4.5 nM) and against six HIV-1 isolates in MDMs (Table 2) (mean EC50 ± SD = 2.5 ± 1.7 nM; EC50 range, 0.56 to 4.6 nM). AZT was included in these studies as a positive-control inhibitor of HIV replication (Tables 1 and 2). The EC50 of CMX157 against the HIV-1 strain JR-CSF in PBMCs was affected by the addition of human serum (EC50s of 1.1 nM, 4.6 nM, 6.4 nM, 11 nM, and 16 nM obtained for 0%, 10%, 15%, 20%, and 30% human serum, respectively). Extrapolating from the EC50s obtained in 0% to 30% human serum resulted in an estimated EC50 of 53 nM for CMX157 in 100% human serum. AZT activity was unaffected by human serum in these assays (data not shown).
TABLE 1.
Median activity of CMX157 and AZT against HIV subtype isolates in PBMCs
| Subtype (no. of isolates)a | Mean EC50 ± SD (nM) for: |
|
|---|---|---|
| CMX157 | AZT | |
| A (3) | 3.3 ± 2.2 | 14 ± 5.4 |
| B (6) | 1.5 ± 1.7 | 2.0 ± 1.2 |
| C (3) | 2.8 ± 2.1 | 4.9 ± 1.5 |
| D (3) | 2.1 ± 2.6 | 6.3 ± 7.1 |
| E (3) | 2.3 ± 1.1 | 5.5 ± 4.4 |
| F (3) | 2.7 ± 3.2 | 9.6 ± 13 |
| G (3) | 2.2 ± 0.53 | 4.7 ± 0.90 |
| Group O (3) | 5.0 ± 2.4 | 1.7 ± 0.85 |
| HIV-2 (3) | 3.5 ± 1.5 | 9.3 ± 8.1 |
The following HIV-1 isolates were used for this study: subtype A isolates 92RW009, 92UG029, 92UG037; subtype B isolates Ba-L, ADA, 92BR014, 96USHIPS7, JR-CSF, 92TH026; subtype C isolates 92BR025, 93IN101, 93MW959; subtype D isolates 92UG001, 92UG024, 92UG046; subtype E isolates 93TH073, CMU06, CMU08; subtype F isolates 93BR019, 93BR020, 93BR029; subtype G isolates JV1083, RU132, G3; group O isolates BCF01, BCF02, BCF03; HIV-2 isolates CDC310319, CDC310342, CBL-20. No toxicity was observed up to the highest concentration tested (1,000 nM).
TABLE 2.
Activity of CMX157 and AZT against HIV-1 isolates in MDMsa
| Subtype (no. of isolates)a | Mean EC50 ± SD (nM) for: |
|
|---|---|---|
| CMX157 | AZT | |
| A (2) | 1.3 ± 0.92 | 1.2 ± 1.4 |
| B (4) | 3.1 ± 1.8 | 2.7 ± 1.1 |
The following HIV-1 isolates were used for this study: subtype A isolates 92UG029, 92UG037; subtype B isolates Ba-L, ADA, JR-CSF, 92TH014. No toxicity was observed up to the highest concentration tested (1,000 nM).
Antiretroviral activity of CMX157 against NRTI-resistant HIV-1 mutants in PBMCs.
The activity of CMX157 against HIV-1 with clinically important mutations engendering resistance to NRTIs was assessed in a PBMC assay by using a panel of 14 multidrug-resistant HIV-1 reverse transcriptase mutants. This virus panel includes clones in an HIV-1NL4-3 backbone with each of the published nucleoside analog reverse transcriptase mutations in the combinations that occur most frequently in HIV-infected individuals (7). As summarized in Table 3, HIV-1 genotypes associated with pan-NRTI resistance were sensitive to CMX157. The 41L/44D/67N/69D/118I/210W/215Y mutant had a mean EC50 of 19 nM for CMX157 versus >8,500 nM for TFV, while the 41L/44D/67N/69D/118I/184V/210W/215Y mutant had a mean EC50 of 3.1 nM for CMX157 versus 5,400 nM for TFV. Similarly, the 65R mutant had a mean EC50 of 13 nM for CMX157 versus >8,000 nM for TFV, and the 65R/184V mutant had a mean EC50 of 1.8 nM for CMX157 versus 1,000 nM for TFV. These results demonstrate the ameliorative effect of M184V on resistance to these agents conferred by TAMs. The seven additional MNR HIV-1 clones from the panel that are not listed in Table 3 and that were examined in this assay all had low nanomolar CMX157 EC50s.
TABLE 3.
Activity of CMX157, TFV, and AZT against NRTI-resistant HIV-1 mutants in PBMCs
| HIV reverse transcriptase mutant genotype (clone) | Value(s) for: |
|||||
|---|---|---|---|---|---|---|
| CMX157 |
TFV |
AZT |
||||
| Mean EC50 ± SD (n)a | FCb | Mean EC50± SD (n)a | FCb | Mean EC50 ± SD (n)a | FCb | |
| Wild type (HIV-1NL4-3) | 1.5 ± 0.95 (3) | 300 ± N/Ac (1) | 0.53 ± 0.55 (3) | |||
| 41L/67N/69N/70R/215F/219E (7324-1) | 3.8 ± 1.3 (3) | 2.5 | 6,500 ± N/Ac (1) | 22 | 330 ± 270 (3) | 620 |
| 41L/44D/67N/69D/118I/210W/215Y/184V (4755-5) | 3.1 ± 0.62 (2) | 2.1 | 5,400 ± N/Ac (1) | 18 | 100 ± 16 (2) | 190 |
| 41L/44D/67N/69D/118I/210W/215Y (7303-3) | 19 ± 15 (3) | 13 | >8,500 ± >2,100d (2) | >28 | 3,000 ± 2,800 (3) | 5,700 |
| 69K/70G/75I/77L/116Y/151M/184V (1617-1) | 5.0 ± 3.6 (3) | 3.3 | >6,500 ± >5,000d (2) | >22 | >340 ± >570d (3) | >640 |
| 41L//74V/210W/215Y/184V/69SSS (52534-2) | 9.0 ± 6.6 (3) | 6.0 | >6,500 ± >5,000d (2) | >22 | 350 ± 290 (3) | 660 |
| 65R (71361-1) | 13 ± 1.3 (2) | 8.7 | >8,000 ± >2,800d (2) | >27 | 2.7 ± 3.6 (2) | 5.1 |
| 65R/184V (8415-2) | 1.8 ± 1.2 (3) | 1.2 | 1,000 ± N/Ac (1) | 3.3 | 1.8 ± 1.5 (3) | 3.4 |
EC50s are shown in nM as mean ± SD of one to three (n) independent experiments.
FC, fold change in mean EC50 compared to mean EC50 for wild-type HIV-1NL4-3.
N/A, SD not applicable due to single available EC50.
Values indicate that one of the EC50s was greater than the high test concentration used in the experiment.
Activity of CMX157 and approved NRTIs with the PhenoSense assay.
Cross-resistance to CMX157 was assessed in a panel of 30 HIV mutants with resistance to NRTIs and two wild-type viruses by using the PhenoSense assay. Historical median coefficients of variance for this assay have ranged from 12% for TFV to 32% for AZT (35). CMX157 showed fold changes in EC50s similar to those shown by TFV (Table 4), but the absolute EC50s (Table 5) for CMX157 (mean EC50 = 5.33 nM) were on average 334-fold lower than the EC50s for TFV (mean EC50 = 1,780 nM). Wild-type HIV from patients yielded CMX157 EC50s of 1.72 and 1.61 nM (isolate numbers 056 and 057) versus EC50s of 613 and 652 nM for TFV. EC50s for CMX157 ranged from 0.66 nM for the L74V/M184V mutant to 57 nM for the A62V/T69SVG/V75I/T215I mutant; corresponding EC50s for TFV were 230 nM and 17,000 nM.
TABLE 4.
Fold changes in EC50 of mutants versus wild type for CMX157 and comparator nucleosides
| MBSa no. | NRTI-associated mutation(s) | Phenotype fold change from wild-type control |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CMX157 | TFV | ZDV | d4T | ddI | ABC | 3TC | FTC | ||
| 056 | Wild type | 0.93 | 0.79 | 0.72 | 0.86 | 1.0 | 0.86 | 0.86 | 0.78 |
| 057 | Wild type | 0.87 | 0.85 | 0.61 | 0.81 | 1.0 | 0.94 | 1.2 | 1.4 |
| 058 | M41L/D67N/K70R/L210W/T215F/K219Q | 4.6 | 3.5 | 350 | 3.7 | 1.6 | 4.4 | 3.7 | 5.0 |
| 060 | M41L/D67N/K70R/L210W/T215Y/K219E | 3.3 | 4.0 | 400 | 3.1 | 1.5 | 4.7 | 5.1 | 6.9 |
| 061 | M41L/D67N/K70R/M184V/L210W/T215Y/K219E | 2.1 | 2.2 | 58 | 2.6 | 2.1 | 9.1 | >71 | >63 |
| 063 | M41L/D67N/K70R/M184V/L210W/T215Y/K219E | 2.8 | 2.2 | 59 | 2.3 | 2.1 | 7.9 | >80 | >88 |
| 066 | 184V | 0.73 | 0.69 | 0.63 | 0.77 | 1.4 | 2.7 | >80 | >88 |
| 067 | A62V/D67G/T69SVG/V75I/T215I | 31 | 22 | >920 | 11 | 4.0 | 18 | 12 | 26 |
| 069 | D67E/T69SSG/V75M/M184V/L210W/T215Y | 6.1 | 4.7 | 320 | 11 | 4.4 | 26 | >80 | >88 |
| 070 | 65R/S68G | 1.9 | 1.7 | 0.44 | 1.3 | 1.7 | 3.2 | 19 | 17 |
| 071 | 65R/S68N/184V | 1.5 | 1.2 | 0.41 | 0.99 | 2.6 | 6.6 | >80 | >88 |
| 153 | L74V | 0.66 | 0.62 | 0.41 | 1.0 | 1.6 | 1.9 | 1.9 | 1.9 |
| 157 | L74V/M184V | 0.36 | 0.29 | 0.22 | 0.71 | 2.0 | 5.2 | >80 | >88 |
| 160 | S68G/V75(I/T)/F77L/Y115F/F116Y/Q151M | 2.0 | 2.3 | 93 | 9.3 | 12 | 19 | 24 | 30 |
| 162 | A62V/V75I/F77L/Y115F/F116Y/Q151M/M184V | 1.6 | 1.4 | 150 | 7.1 | 11 | >29 | >80 | >88 |
| 166 | L74V in SDM | 0.80 | 0.69 | 0.50 | 1.1 | 1.5 | 1.9 | 1.6 | 1.5 |
| 174 | M184V in SDM | 0.62 | 0.54 | 0.34 | 0.68 | 1.2 | 2.6 | >80 | >88 |
| 180 | K65R in SDM | 2.6 | 2.0 | 0.51 | 1.4 | 2.0 | 2.6 | 12 | 11 |
| 185 | T215Y | 2.7 | 2.4 | 38 | 1.4 | 1.1 | 2.1 | 1.6 | 2.3 |
| 188 | T215Y/M184V | 0.90 | 0.81 | 2.6 | 1.1 | 1.4 | 4.3 | >80 | >88 |
| 192 | M41L/T215Y | 2.1 | 1.8 | 20 | 1.4 | 1.1 | 1.9 | 2.0 | 2.7 |
| 194 | M41L/T215Y/M184V | 1.6 | 1.3 | 6.5 | 1.5 | 1.8 | 5.4 | >80 | >88 |
| 200 | M41L/L210W/T215Y | 3.2 | 2.8 | 100 | 2.1 | 1.4 | 2.8 | 2.7 | 3.7 |
| 201 | M41L/L210W/T215Y | 3.5 | 3.0 | 98 | 1.8 | 1.1 | 2.4 | 1.6 | 1.9 |
| 203 | M41L/L210W/T215Y/M184V | 1.0 | 0.97 | 3.2 | 1.5 | 1.5 | 4.6 | >80 | >88 |
| 204 | M41L/L210W/T215Y/M184V | 1.4 | 1.0 | 3.9 | 1.8 | 1.6 | 5.0 | >80 | >88 |
| 206 | D67N/K70R | 3.0 | 2.6 | 28 | 1.3 | 1.2 | 1.4 | 2.1 | 2.9 |
| 211 | D67N/K70R/T215F/K219E/M184V | 1.0 | 0.91 | 7.0 | 1.3 | 1.6 | 4.4 | >80 | >88 |
| 212 | L210W/T215Y | 1.5 | 1.3 | 7.6 | 1.6 | 1.0 | 2.1 | 1.9 | 2.3 |
| 215 | D67N/K70E | 1.2 | 0.96 | 0.21 | 0.94 | 1.4 | 1.4 | 2.9 | 3.0 |
| 217 | D67N/K70E/M184V | 0.78 | 0.64 | 0.15 | 0.70 | 1.3 | 2.9 | >80 | >88 |
| 220 | K70E/M184V | 0.71 | 0.61 | 0.13 | 0.65 | 1.8 | 5.9 | >80 | >88 |
MBS, Monogram Biosciences.
TABLE 5.
CMX157 and comparator nucleoside activity against NRTI-resistant mutants by the use of PhenoSense
| MBSd no. | NRTI-associated mutation(s) | EC50 (μM)a,b |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CMX157 | TFV | ZDV | d4T | ddI | ABC | 3TC | FTC | ||
| 056 | Wild type | 0.0017 | 0.61 | 0.020 | 0.51 | 5.5 | 1.5 | 3.2 | 0.89 |
| 057 | Wild type | 0.0016 | 0.65 | 0.017 | 0.48 | 5.6 | 1.7 | 4.4 | 1.6 |
| 058 | M41L/D67N/K70R/L210W/T215F/K219Q | 0.0085 | 2.7 | 9.6 | 2.2 | 8.8 | 7.9 | 14 | 5.7 |
| 060 | M41L/D67N/K70R/L210W/T215Y/K219E | 0.0096 | 3.6 | 16 | 2.8 | 8.9 | 11 | 22 | 11 |
| 061 | M41L/D67N/K70R/M184V/L210W/T215Y/K219E | 0.0061 | 2.0 | 2.2 | 2.4 | 12 | 22 | >300 | >100 |
| 063 | M41L/D67N/K70R/M184V/L210W/T215Y/K219E | 0.0052 | 1.7 | 1.6 | 1.4 | 11 | 14 | >300 | >100 |
| 066 | 184V | 0.0014 | 0.53 | 0.017 | 0.46 | 7.6 | 4.8 | >300 | >100 |
| 067 | A62V/D67G/T69SVG/V75I/T215I | 0.057 | 17 | >25 | 6.4 | 22 | 33 | 46 | 30 |
| 069 | D67E/T69SSG/V75M/M184V/L210W/T215Y | 0.011 | 3.6 | 8.7 | 6.8 | 24 | 47 | >300 | >100 |
| 070 | 65R/S68G | 0.0036 | 1.3 | 0.012 | 0.77 | 9.2 | 5.7 | 71 | 19 |
| 071 | 65R/S68N/184V | 0.0027 | 0.94 | 0.011 | 0.59 | 14 | 12 | >300 | >100 |
| 153 | L74V | 0.0012 | 0.48 | 0.011 | 0.60 | 8.9 | 3.4 | 7.0 | 2.1 |
| 157 | L74V/M184V | 0.00066 | 0.23 | 0.006 | 0.42 | 11 | 9.2 | >300 | >100 |
| 160 | S68G/V75(I/T)/F77L/Y115F/F116Y /Q151M | 0.0037 | 1.7 | 2.5 | 5.5 | 63 | 34 | 90 | 35 |
| 162 | A62V/V75I/F77L/Y115F/F116Y/Q151M/M184V | 0.0031 | 1.1 | 4.2 | 4.2 | 58 | >52 | >300 | >100 |
| 166 | L74V in SDMc | 0.0015 | 0.54 | 0.014 | 0.63 | 8.1 | 3.3 | 6.1 | 1.7 |
| 174 | M184V in SDMc | 0.0012 | 0.41 | 0.009 | 0.40 | 6.8 | 4.7 | >300 | >100 |
| 180 | K65R in SDMc | 0.0047 | 1.6 | 0.014 | 0.83 | 11 | 4.6 | 43 | 13 |
| 185 | T215Y | 0.0050 | 1.8 | 1.0 | 0.83 | 6.3 | 3.7 | 6.1 | 2.6 |
| 188 | T215Y/M184V | 0.0017 | 0.62 | 0.071 | 0.65 | 7.9 | 7.6 | >300 | >100 |
| 192 | M41L/T215Y | 0.0039 | 1.4 | 0.56 | 0.84 | 6.3 | 3.4 | 7.6 | 3.1 |
| 194 | M41L/T215Y/M184V | 0.0029 | 1.0 | 0.18 | 0.86 | 10 | 9.6 | >300 | >100 |
| 200 | M41L/L210W/T215Y | 0.0060 | 2.2 | 2.7 | 1.2 | 7.5 | 5.0 | 10.3 | 4.2 |
| 201 | M41L/L210W/T215Y | 0.0066 | 2.3 | 2.7 | 1.1 | 5.9 | 4.2 | 5.9 | 2.2 |
| 203 | M41L/L210W/T215Y/M184V | 0.0019 | 0.75 | 0.087 | 0.88 | 8.2 | 8.2 | >300 | >100 |
| 204 | M41L/L210W/T215Y/M184V | 0.0025 | 0.79 | 0.11 | 1.1 | 8.6 | 8.9 | >300 | >100 |
| 206 | D67N/K70R | 0.0056 | 2.0 | 0.77 | 0.78 | 6.6 | 2.5 | 8.1 | 3.3 |
| 211 | D67N/K70R/T215F/K219E/M184V | 0.0019 | 0.70 | 0.19 | 0.79 | 8.7 | 7.7 | >300 | >100 |
| 212 | L210W/T215Y | 0.0028 | 1.0 | 0.21 | 0.94 | 5.5 | 3.6 | 7.3 | 2.6 |
| 215 | D67N/K70E | 0.0021 | 0.74 | 0.0060 | 0.56 | 7.7 | 2.4 | 11 | 3.4 |
| 217 | D67N/K70E/M184V | 0.0015 | 0.50 | 0.0040 | 0.42 | 7.3 | 5.1 | >300 | >100 |
| 220 | K70E/M184V | 0.0013 | 0.47 | 0.0040 | 0.39 | 9.9 | 10 | >300 | >100 |
Represent values from a single PhenoSense assay.
Historical median coefficients of variance for nucleosides of 12 to 32%.
SDM, site-directed mutant of pNL4-3.
MBS, Monogram Biosciences.
One of the most clinically important TFV-resistant virus types includes M41L/L210W/T215Y mutations. Two clinically derived recombinants with these mutations are represented by viruses 200 and 201, which exhibited CMX157 EC50s of 6.0 and 6.6 nM, respectively, versus 2,200 and 2,300 nM for TFV, respectively. Addition of M184V to this mutation pattern reduced these EC50s ∼3-fold, as shown with viruses 203 and 204 (EC50s of 1.9 and 2.5 nM for CMX157, and 750 and 790 nM for TFV, respectively). The K65R mutation resulted in CMX157 EC50s of 3.60 nM in a clinically derived recombinant also containing S68G (isolate number 070) and 4.73 nM by itself in the site-directed mutant of the laboratory strain pNL4-3 (isolate number 180); EC50s for TFV in these viruses were 1,300 and 1,600 nM, respectively. With the exception of viruses with the insertion at position 69 (isolate numbers 67 and 69), all mutants had low nanomolar EC50s for CMX157.
Antiretroviral activity of CMX157 in combination with other antiretroviral drugs.
The anti-HIV-1 activity of CMX157 was evaluated in two-drug combination studies with each FDA-approved antiretroviral drug. Additivity, synergy, and antagonism were defined as detailed in Materials and Methods. No antagonistic interactions were observed within the concentration ranges examined for antiviral efficacy between CMX157 and any FDA-approved drug, as summarized in Table 6. Additive to synergistic interactions were observed between CMX157 and all approved antiretroviral drugs. In contrast, the positive antagonism control of d4T in combination with RBV exhibited a highly antagonistic interaction in all experiments. There was no evidence of synergistic cytotoxicity for any combination at the concentrations examined (10 μM = highest test concentration for CMX157).
TABLE 6.
Antiviral efficacy of CMX157 in combination with approved antiretroviral drugs in CEM-SS or MAGI-CCR5 cellsa
| Compound | Mean synergy vol (nM2%, μM2%, or nM μM%)b | Mean antagonism vol (nM2%, μM2%, or nM μM%)b | Interpretation of resultsc |
|---|---|---|---|
| NRTIs | |||
| 3TC | 73 ± 19 | −14 ± 25 | Slightly synergistic |
| ABC | 15 ± 13 | −17 ± 17 | Additive |
| ZDV | 13 ± 18 | −23 ± 39 | Additive |
| d4T | 15 ± 22 | −0.61 ± 1.1 | Additive |
| ddC | 120 ± 38 | −5.3 ± 9.2 | Highly synergistic |
| ddI | 12 ± 15 | −0.34 ± 0.58 | Additive |
| FTC | 73 ± 11 | 0.00 ± 0.00 | Slightly synergistic |
| TFV | 28 ± 9.9 | −0.18 ± 0.31 | Additive |
| NNRTIs | |||
| DLV | 74 ± 76 | −0.86 ± 1.5 | Slightly synergistic |
| EFV | 70 ± 23 | 0.00 ± 0.00 | Slightly synergistic |
| ETV | 69 ± 36 | −0.54 ± 0.47 | Slightly synergistic |
| NVP | 39 ± 37 | 0.00 ± 0.00 | Additive |
| PIs | |||
| APV | 51 ± 49 | 0.00 ± 0.00 | Slightly synergistic |
| ATV | 61 ± 46 | −0.24 ± 0.42 | Slightly synergistic |
| DRV | 43 ± 57 | −0.77 ± 1.3 | Additive |
| IDV | 53 ± 32 | −0.19 ± 0.32 | Slightly synergistic |
| LPV | 56 ± 26 | −0.14 ± 0.16 | Slightly synergistic |
| NFV | 81 ± 47 | 0.00 ± 0.00 | Slightly synergistic |
| RTV | 67 ± 43 | −0.87 ± 1.5 | Slightly synergistic |
| SQV | 45 ± 68 | −1.1 ± 1.3 | Additive |
| TPV | 52 ± 51 | −2.9 ± 5.0 | Slightly synergistic |
| Entry inhibitors | |||
| MVC | 85 ± 66 | −2.3 ± 2.0 | Slightly synergistic |
| T-20 | 1.0 ± 1.8 | 0.00 ± 0.00 | Additive |
| Integrase inhibitor | |||
| RAL | 34 ± 43 | 0.00 ± 0.00 | Additive |
| Positive antagonism control (CEM-SS cells) | |||
| d4T/RBV | 3.2 ± 8.5d | −300 ± 110d | Highly antagonistic |
| Positive antagonism control (MAGI-CCR5 cells) | |||
| d4T/RBV | 190 ± 320 | −110 ± 47 | Highly antagonistic |
The antiviral efficacy results of CMX157 in combination with MVC were performed in MAGI-CCR5 cells. All other evaluations were performed in CEM-SS cells.
Synergy and antagonism volumes were calculated at the 95% confidence level; values are means ± SD from the results for three independent determinations. For an explanation of the units, see the MacSynergy analysis program, which was used to analyze the combination data as described in reference 40.
See definitions in Materials and Methods.
Values represent means ± SD from 24 independent experiments conducted in parallel with the CMX157 combinations with FDA-approved antiretroviral drugs.
Intracellular levels of TFV-PP following incubation of human PBMCs with CMX157 or TFV.
To examine the relative levels of TFV-PP produced by CMX157 and TFV in relevant HIV target cells, human PBMCs were incubated with concentrations of CMX157 or TFV that bracketed the maximum concentration of TFV in serum (Cmax) in humans following standard dosing of TDF (16). PHA/IL-2-activated human PBMCs exposed to 1.0 μM CMX157 for 24 h produced approximately 34 times more TFV-PP than that produced following the same exposure to TFV (Table 7). One-hundred-fold less CMX157 (10 nM) produced levels of TFV-PP similar to those produced by 1,000 nM TFV in resting and activated PBMCs. The fold increase in TFV-PP in CMX157- versus TFV-exposed cells appeared lower, as the concentration of CMX157 was increased compared to that of TFV in resting cells.
TABLE 7.
Levels of active anabolite in unstimulated and stimulated human PBMCs incubated with TFV or CMX157 for 24 h
| Cells | Drug and concn | No. of pmol/1 × 106 cells (% relative SD)a |
||
|---|---|---|---|---|
| TFV | TFV-P | TFV-PP | ||
| Unstimulated PBMCs | TFV, 1.0 μM | ND | BLQ | 0.05 (2.4) |
| TFV, 10 μM | 0.11 (13.3) | 0.05 (5.1) | 0.15 (9.4) | |
| CMX157, 0.01 μM | ND | BLQ | 0.07 (8.9) | |
| CMX157, 0.1 μM | 0.05 (2.3) | BLQ | 0.07 (3.6) | |
| CMX157, 1.0 μM | 0.07 (7.4) | 0.05 (6.8) | 0.22 (3.2) | |
| CMX157, 10 μM | 0.19 (5.9) | 0.12 (7.7) | 1.1 (9.8) | |
| PHA/IL-2-stimulated PBMCs | TFV, 1.0 μM | BLQ | BLQ | 0.05 (4.9) |
| TFV, 10 μM | 0.09 (41.5) | BLQ | 0.22 (8.0) | |
| CMX157, 0.01 μM | BLQ | BLQ | 0.07 (12.4) | |
| CMX157, 0.1 μM | BLQ | BLQ | 0.25 (9.5) | |
| CMX157, 1.0 μM | 0.56 (14.7) | 0.30 (6.9) | 1.7 (6.3) | |
| CMX157, 10 μM | 1.9 (5.9) | 0.90 (6.2) | 5.5 (14.2) | |
ND, not done; BLQ, below limit of quantification.
Cell toxicity in primary and transformed human cells.
CMX157, TDF, and TFV were evaluated for toxicity to primary human cells (PBMCs ± activation, dendritic cells, monocytes/macrophages, and hepatocytes) and transformed human cell lines (CEM-SS, HeLa, ME180, Huh-7, and HepG2). No toxicity was detected for CMX157 or TFV, even at concentrations well above those found in patients taking TDF or concentrations targeted as a Cmax with CMX157.
CMX157 was less toxic than AZT and similarly toxic to TFV in a GM-CFU assay and a BFU-E assay comparing AZT, TFV, TDF, and CMX157 exposures to human bone marrow. In the GM-CFU assay, TFV, CMX157, TDF, and AZT gave CC50 values of 4.7, 2.5, 1.9, and 0.89 μM, respectively. In the BFU-E assay, TFV, CMX157, TDF, and AZT gave 50% cytotoxicity concentration (CC50) values of 3.5, 4.6, 0.9, and 0.50 μM, respectively.
DISCUSSION
Viread (TDF) is part of the preferred NRTI backbone in current HIV treatment guidelines but has limitations due primarily to concerns about renal toxicity and resistance. In vivo, TDF is rapidly converted to the TFV dianion by plasma esterases. High levels of circulating TFV are required for efficacy because TFV is not readily absorbed by the CD4+ cells that are targets for HIV infection. TFV in plasma is actively taken up by organic anion transporters (hOATs) on RPTECs and effluxed by MRP4 (43). CMX157 has the potential to increase efficacy and decrease toxicity of TFV because it circulates as the lipid-TFV conjugate, which is not a substrate for human organic anion transporters (hOATs 1 to 3) (data not shown) but is readily taken up and anabolized to the active antivirus in human PBMCs.
HIV that is unresponsive to TFV is typically unresponsive to most, and frequently all, other NRTIs as well. TFV-PP inhibits HIV DNA synthesis by competing with the natural substrate, dATP, for binding to HIV reverse transcriptase; following incorporation, TFV acts as a chain terminator since, like all current NRTIs, it lacks a 3′-hydroxyl group, which is required for addition of the next base. The mechanisms of resistance to TFV and the diminution of antiviral activity in vitro and in vivo are relatively well understood (28, 51). Mutations in HIV reverse transcriptase associated with resistance to TFV may decrease incorporation of TFV into the nascent HIV DNA or increase excision of incorporated TFV from the nascent chain. In general, TAMs and insertion at position 69 increase NRTI excision, while K65R and the 151 complex decrease incorporation, although some mutations affect both mechanisms. An important aspect for development of CMX157 is that the fold change in EC50 for all of these mutants is relatively small, suggesting that it is reasonable to expect that resistance can be overcome by raising the level of TFV-PP in target cells without causing unacceptable toxicity.
In this study, we examined the phenotypic resistance of CMX157 with a large panel of NRTI-resistant viruses derived from clinical specimens in both primary and transformed cells. Mutants associated with decreased incorporation and/or increased excision of TFV were well represented. In vitro efficacy was examined against HIV with TAMs (ZDV/d4T) M41L, D67N, K70R, L210W, T215Y/F, K219Q/E, the ddI/ABC/TDF-associated mutation K65R, the ddI/ABC-associated mutation L74V, the ABC/ddI/3TC/FTC-associated mutation M184V, and the MNR complexes marked by Q151M or T69SXX (20). The highest EC50 obtained for CMX157 in the PhenoSense assay was 57 nM (A62V/D67G/T69SVG/V75I/T215I); the corresponding EC50 for TFV was 17,000 nM. The ∼300-fold improvement in EC50 for CMX157 versus TFV for this mutant was typical of the other 29 NRTI-resistant isolates tested in this assay. Notably, this mutant and many others tested were resistant to all NRTIs examined (TFV, ZDV, d4T, ddI, ABC, 3TC, FTC), as shown in Tables 4 and 5. Similar data showing much lower EC50s for CMX157 versus TFV were obtained in primary cultures, as summarized in Table 3. A notable discrepancy between the PhenoSense data and the PBMC data is in the fold changes observed for CMX157 versus TFV. Given an identical active antiviral anabolite, it would be expected that the fold change in EC50 should be very similar, as seen across the mutants tested in the PhenoSense assay (Table 4). The apparent lack of correlation in the PBMC data between fold changes observed for CMX157 and TFV shown in Table 3, particularly for mutants 7324-1, 4755-5, 1617-1, and 52534-2, was surprising. This may be due to differences in the cells (e.g., uptake, anabolism) or in the viruses utilized (e.g., whole virus versus PhenoSense chimeras) but is more likely due to variability in the PBMC assays. Among the four viruses noted, two had large SDs for TFV, and two had only a single assay for TFV. The concordant fold changes for CMX157 and TFV across the 32 isolates tested in the PhenoSense assay suggest that these drugs are merely delivering the same agent (TFV-PP) with greater and lower efficiencies, respectively.
Overall, the in vitro data suggest CMX157 will be effective against MNR mutants, including those that are unresponsive to all currently available NRTIs. Notably, the average EC50 in PBMCs for CMX157 against a panel of 27 wild-type HIV-1 isolates representing group M subtypes A to G and group O was 2.6 nM (range, 0.20 to 7.2 nM). Similarly, single-digit nanomolar EC50s were obtained for HIV-2 in PBMCs and HIV-1 in primary macrophages, suggesting that relatively low exposures to CMX157 may be adequate to treat infection by wild-type and most NRTI-resistant strains.
The mechanisms behind the increased in vitro potency of CMX157 are believed to relate to high cellular and viral uptake promoted by the lipid moiety. Data presented here demonstrate that higher intracellular concentrations of the active anabolite, TFV-PP, are seen following incubation of CMX157 with primary human PBMCs than equimolar, physiologically relevant, concentrations of TFV. For example, 24-h treatment of activated human PBMCs with a level of TFV that approximates the human Cmax (1 μM) results in 50 fmol of TFV-PP per 1 million cells; the same amount of CMX157 produced 1,700 fmol/million cells (34-fold more TFV-PP). Notably, the median level of TFV-PP in humans taking Viread is 76 fmol/million PBMCs (21).
However, higher intracellular levels of TFV-PP could also increase mitochondrial toxicity. Recent data suggest that CMX157, like TFV, has a low potential to induce mitochondrial toxicity, showing no effect on biogenesis of a mitochondrially encoded and expressed protein or a nuclear-encoded, cytoplasmically translated mitochondrial protein at concentrations more than 10-fold higher than the clinical target. In these studies CMX157 and TFV were identical in their toxicity to mitochondria (29, 49). Studies designed to evaluate the preclinical safety of CMX157 were encouraging. CMX157 showed no cytotoxicity in a panel of 10 primary and transformed human cell types up to 10,000 nM. In colony-forming assays using human bone marrow progenitor cells, CMX157 was similar to TFV and less toxic than AZT.
Unlike TFV, CMX157 also appears to reduce infectivity of HIV when purified virus is preincubated with CMX157 prior to infection of untreated human cell lines, presumably by fusion of the CMX157 lipid side chain with the lipid membrane of HIV such that HIV carries CMX157 into the target cell (25). This mechanism of virus carrying the antiviral agent into target cells may be a significant advantage for CMX157, potentially permitting treatment of HIV in privileged compartments; however, further studies are required to fully explore this putative mechanism of antiviral distribution/activity.
As demonstrated previously, the lipid conjugate appears stable in vivo, resulting in high levels of CMX157 in plasma after oral administration in animals (25, 33). In addition, no antagonistic effects were observed for antiviral activity evaluating CMX157 and FDA-approved antiretroviral drugs in two-drug combination studies.
The data presented here suggest CMX157 has potential to treat both wild-type and antiretroviral drug-resistant HIV, including strains that are resistant to all approved NRTIs. Dose optimization in the clinic may be based on plasma PK and intracellular PBMC TFV-PP, in addition to safety, prior to starting human efficacy trials. It is encouraging that 100 nM CMX157 yielded five times the levels of TFV-PP observed with TFV at approximately the human Cmax (1 μM) in relevant human cells (PBMCs). This suggests that CMX157 may simultaneously reduce the level of circulating TFV and raise the level of intracellular TFV-PP in target cells for HIV, effectively shifting the preferential accumulation of drug from the site of toxicity to the site of efficacy. Taken together, these data demonstrate the potential for CMX157 to reduce renal toxicity associated with TDF and to address an unmet medical need in the treatment of patients infected with MNR HIV.
Acknowledgments
We thank Julie Russell and Katherine Marotte for support in performing two-drug combination studies.
Footnotes
Published ahead of print on 3 May 2010.
REFERENCES
- 1.Abimiku, A. G., T. L. Stern, A. Zwandor, P. D. Markham, C. Calef, S. Kyari, W. C. Saxinger, R. C. Gallo, M. Robert-Guroff, and M. S. Reitz. 1994. Subgroup G HIV type 1 isolates from Nigeria. AIDS Res. Hum. Retroviruses 10:1581-1583. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckheit, R. W., Jr., and R. Swanstrom. 1991. Characterization of an HIV-1 isolate displaying an apparent absence of virion-associated reverse transcriptase activity. AIDS Res. Hum. Retroviruses 7:295-302. [DOI] [PubMed] [Google Scholar]
- 4.Cann, A. J., J. A. Zack, A. S. Go, S. J. Arrigo, Y. Koyanagi, P. L. Green, Y. Koyanagi, S. Pang, and I. S. Chen. 1990. Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J. Virol. 64:4735-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. 1996. Persistent lack of detectable HIV-1 anitbody in a person with HIV infection. MMWR Morb. Mortal. Wkly. Rep. 45:181-185. [PubMed] [Google Scholar]
- 6.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupnik, K., M. Gonzales, and R. Shafer. 2001. Most multidrug-resistant HIV-1 reverse transcriptase clones in plasma encode functional reverse transcriptase enzymes. Antivir. Ther. 6(Suppl.):42. [Google Scholar]
- 8.Ellenberger, D. L., P. S. Sullivan, J. Dorn, C. Schable, T. J. Spira, T. M. Folks, and R. B. Lal. 1999. Viral and immunologic examination of human immunodeficiency virus type 1-infected, persistently seronegative persons. J. Infect. Dis. 180:1033-1042. [DOI] [PubMed] [Google Scholar]
- 9.Foley, G. E., H. Lazarus, S. Farber, B. G. Uzman, B. A. Boone, and R. E. McCarthy. 1965. Continuous culture of human lymphoblasts from peripheral blood of a child with acute leukemia. Cancer 18:522-529. [DOI] [PubMed] [Google Scholar]
- 10.Gao, F., L. Yue, S. Craig, C. L. Thornton, D. L. Robertson, F. E. McCutchan, J. A. Bradac, P. M. Sharp, and B. H. Hahn. 1994. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. WHO Network for HIV Isolation and Characterization. AIDS Res. Hum. Retroviruses 10:1359-1368. [DOI] [PubMed] [Google Scholar]
- 11.Gardner, E. M., S. Sharma, G. Peng, K. H. Hullsiek, W. J. Burman, R. D. Macarthur, M. Chesney, E. E. Telzak, G. Friedland, and S. B. Mannheimer. 2008. Differential adherence to combination antiretroviral therapy is associated with virological failure with resistance. AIDS 22:75-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gartner, S., P. Markovits, D. M. Markovitz, M. H. Kaplan, R. C. Gallo, and M. Popovic. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215-219. [DOI] [PubMed] [Google Scholar]
- 13.Gendelman, H. E., L. M. Baca, C. A. Kubrak, P. Genis, S. Burrous, R. M. Friedman, D. Jacobs, and M. S. Meltzer. 1992. Induction of IFN-alpha in peripheral blood mononuclear cells by HIV-infected monocytes. Restricted antiviral activity of the HIV-induced IFN. J. Immunol. 148:422-429. [PubMed] [Google Scholar]
- 14.Gendelman, H. E., J. M. Orenstein, L. M. Baca, B. Weiser, H. Burger, D. C. Kalter, and M. S. Meltzer. 1989. The macrophage in the persistence and pathogenesis of HIV infection. AIDS 3:475-495. [DOI] [PubMed] [Google Scholar]
- 15.Gendelman, H. E., J. M. Orenstein, M. A. Martin, C. Ferrua, R. Mitra, T. Phipps, L. A. Wahl, H. C. Lane, A. S. Fauci, and D. S. Burke. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilead Sciences, Inc. 2008. Viread (tenofovir disoproxyl fumurate) tablets prescribing information. http://www.gilead.com/pdf/viread_pi.pdf.
- 17.Harrigan, P. R., R. S. Hogg, W. W. Dong, B. Yip, B. Wynhoven, J. Woodward, C. J. Brumme, Z. L. Brumme, T. Mo, C. S. Alexander, and J. S. Montaner. 2005. Predictors of HIV drug-resistance mutations in a large antiretroviral-naive cohort initiating triple antiretroviral therapy. J. Infect. Dis. 191:339-347. [DOI] [PubMed] [Google Scholar]
- 18.Hostetler, K. Y., J. R. Beadle, G. D. Kini, M. F. Gardner, K. N. Wright, T. H. Wu, and B. A. Korba. 1997. Enhanced oral absorption and antiviral activity of 1-O-octadecyl-sn-glycero-3-phospho-acyclovir and related compounds in hepatitis B virus infection, in vitro. Biochem. Pharmacol. 53:1815-1822. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. A., J.-F. Li, X. Wei, J. Lipscomb, D. Irlbeck, C. Craig, A. Smith, D. E. Bennett, M. Monsour, P. Sandstrom, E. R. Lanier, and W. Heneine. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment—naïve populations and associate with reduced treatment efficacy. PLoS Med. 5:e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2009. Update of the drug resistance mutations in HIV-1: December 2009. Top. HIV Med. 17:138-145. [PubMed] [Google Scholar]
- 21.Kiser, J. J., C. L. Aquilante, P. L. Anderson, T. M. King, M. L. Carten, and C. V. Fletcher. 2008. Clinical and genetic determinants of intracellular tenofovir diphosphate concentrations in HIV-infected patients. J. Acquir. Immune. Defic. Syndr. 47:298-303. [DOI] [PubMed] [Google Scholar]
- 22.Koyanagi, Y., S. Miles, R. T. Mitsuyasu, J. E. Merrill, H. V. Vinters, and I. S. Chen. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 23.Kuritzkes, D. R., C. M. Lalama, H. J. Ribaudo, M. Marcial, W. A. Meyer III, C. Shikuma, V. A. Johnson, S. A. Fiscus, R. T. D'Aquila, B. R. Schackman, E. P. Acosta, and R. M. Gulick. 2008. Preexisting resistance to nonnucleoside reverse-transcriptase inhibitors predicts virologic failure of an efavirenz-based regimen in treatment-naive HIV-1-infected subjects. J. Infect. Dis. 197:867-870. [DOI] [PubMed] [Google Scholar]
- 24.Lai, M. T., V. Munshi, S. Touch, R. M. Tynebor, T. J. Tucker, P. M. McKenna, T. M. Williams, D. J. DiStefano, D. J. Hazuda, and M. D. Miller. 2009. Antiviral activity of MK-4965, a novel nonnucleoside reverse transcriptase inhibitor. Antimicrob. Agents Chemother. 53:2424-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanier, R. 2009. Proceedings of the 16th Conference on Retrovirus and Opportunistic Infections, Montreal, Canada.
- 26.Loussert-Ajaka, I., M. L. Chaix, B. Korber, F. Letourneur, E. Gomas, E. Allen, T. D. Ly, F. Brun-Vezinet, F. Simon, and S. Saragosti. 1995. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J. Virol. 69:5640-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marks, K., and R. Gulick. 2005. Progress in HIV treatment. In S. Butera (ed.), HIV chemotherapy. Caister Academic Press, Norwich, United Kingdom.
- 28.Miller, M. D., N. Margot, B. Lu, L. Zhong, S. S. Chen, A. Cheng, and M. Wulfsohn. 2004. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate treatment in antiretroviral-experienced patients. J. Infect. Dis. 189:837-846. [DOI] [PubMed] [Google Scholar]
- 29.Nadanaciva, S., J. H. Willis, M. L. Barker, D. Gharaibeh, R. A. Capaldi, M. F. Marusich, and Y. Will. 2009. Lateral-flow immunoassay for detecting drug-induced inhibition of mitochondrial DNA replication and mtDNA-encoded protein synthesis. J. Immunol. Methods 343:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Nara, P. L., and P. J. Fischinger. 1988. Quantitative infectivity assay for HIV-1 and -2. Nature 332:469-470. [DOI] [PubMed] [Google Scholar]
- 31.Nara, P. L., W. C. Hatch, N. M. Dunlop, W. G. Robey, L. O. Arthur, M. A. Gonda, and P. J. Fischinger. 1987. Simple, rapid, quantitative, syncytium-forming microassay for the detection of human immunodeficiency virus neutralizing antibody. AIDS Res. Hum. Retroviruses 3:283-302. [DOI] [PubMed] [Google Scholar]
- 32.Owen, S. M., D. Ellenberger, M. Rayfield, S. Wiktor, P. Michel, M. H. Grieco, F. Gao, B. H. Hahn, and R. B. Lal. 1998. Genetically divergent strains of human immunodeficiency virus type 2 use multiple coreceptors for viral entry. J. Virol. 72:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Painter, G. R., M. R. Almond, L. C. Trost, B. M. Lampert, J. Neyts, E. De Clercq, B. E. Korba, K. A. Aldern, J. R. Beadle, and K. Y. Hostetler. 2007. Evaluation of hexadecyloxypropyl-9-R-[2-(phosphonomethoxy)propyl]-adenine, CMX157, as a potential treatment for human immunodeficiency virus type 1 and hepatitis B virus infections. Antimicrob. Agents Chemother. 51:3505-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Painter, G. R., and K. Y. Hostetler. 2004. Design and development of oral drugs for the prophylaxis and treatment of smallpox infection. Trends Biotechnol. 22:423-427. [DOI] [PubMed] [Google Scholar]
- 35.Parkin, N. T., N. S. Hellmann, J. M. Whitcomb, L. Kiss, C. Chappey, and C. J. Petropoulos. 2004. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:437-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popovic, M., S. Gartner, E. Read-Connole, B. Beaver, and M. Reitz. 1988. Cell tropism and expression of HIV-1 isolates in natural targets. In G. A. Valette (ed.), Retroviruses of human AIDS and related animal diseases. Colloque des Cent Gardes, Marnes-la-Coquette, Paris, France.
- 38.Popovic, M., E. Read-Connole, and R. C. Gallo. 1984. T4 positive human neoplastic cell lines susceptible to and permissive for HTLV-III. Lancet 2:1472-1473. [DOI] [PubMed] [Google Scholar]
- 39.Popovic, M., M. G. Sarngadharan, E. Read, and R. C. Gallo. 1984. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science 224:497-500. [DOI] [PubMed] [Google Scholar]
- 40.Prichard, M. N., and C. Shipman, Jr. 1990. A three-dimensional model to analyze drug-drug interactions. Antiviral Res. 14:181-205. [DOI] [PubMed] [Google Scholar]
- 41.Ptak, R. G., P. A. Gallay, D. Jochmans, A. P. Halestrap, U. T. Ruegg, L. A. Pallansch, M. D. Bobardt, M. P. de Bethune, J. Neyts, E. De Clercq, J. M. Dumont, P. Scalfaro, K. Besseghir, R. M. Wenger, and B. Rosenwirth. 2008. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob. Agents Chemother. 52:1302-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, et al. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 43.Ray, A. S., T. Cihlar, K. L. Robinson, L. Tong, J. E. Vela, M. D. Fuller, L. M. Wieman, E. J. Eisenberg, and G. R. Rhodes. 2006. Mechanism of active renal tubular efflux of tenofovir. Antimicrob. Agents Chemother. 50:3297-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reimer, L., S. Mottice, C. Schable, P. Sullivan, A. Nakashima, M. Rayfield, R. Den, and C. Brokopp. 1997. Absence of detectable antibody in a patient infected with human immunodeficiency virus. Clin. Infect. Dis. 25:98-100. [DOI] [PubMed] [Google Scholar]
- 45.Robbins, G. K., B. Daniels, H. Zheng, H. Chueh, J. B. Meigs, and K. A. Freedberg. 2007. Predictors of antiretroviral treatment failure in an urban HIV clinic. J. Acquir. Immune. Defic. Syndr. 44:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulz, T. F., D. Whitby, J. G. Hoad, T. Corrah, H. Whittle, and R. A. Weiss. 1990. Biological and molecular variability of human immunodeficiency virus type 2 isolates from The Gambia. J. Virol. 64:5177-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan, P. S., C. Schable, W. Koch, A. N. Do, T. Spira, A. Lansky, D. Ellenberger, R. B. Lal, C. Hyer, R. Davis, M. Marx, S. Paul, J. Kent, R. Armor, J. McFarland, J. Lafontaine, S. Mottice, S. A. Cassol, N. Michael, and Seronegative AIDS Clinical Study Group. 1999. Persistently negative HIV-1 antibody enzyme immunoassay screening results for patients with HIV-1 infection and AIDS: serologic, clinical, and virologic results. AIDS 13:89-96. [DOI] [PubMed] [Google Scholar]
- 48.Szczech, L. A. 2008. Renal dysfunction and tenofovir toxicity in HIV-infected patients. Top. HIV Med. 16:122-126. [PubMed] [Google Scholar]
- 49.Trost, L. C., E. R. Lanier, B. M. Lampert, and G. R. Painter. 2010. Preclinical safety evaluation of CMX157: a lipid-conjugated nucleotide analog for treatment of HIV. Abstr. 49th SOT Meeting, Salt Lake City, UT.
- 50.Westervelt, P., H. E. Gendelman, and L. Ratner. 1991. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc. Natl. Acad. Sci. U. S. A. 88:3097-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White, K. L., J. M. Chen, J. Y. Feng, N. A. Margot, J. K. Ly, A. S. Ray, H. L. MacArthur, M. J. McDermott, S. Swaminathan, and M. D. Miller. 2006. The K65R reverse transcriptase mutation in HIV-1 reverses the excision phenotype of zidovudine resistance mutations. Antivir. Ther. 11:155-163. [DOI] [PubMed] [Google Scholar]