Abstract
cis-Acting RNA elements in the leaders of bacterial mRNA often regulate gene transcription, especially in the context of amino acid metabolism. We determined that the transcription of the auxiliary, antibiotic-resistant tryptophanyl-tRNA synthetase gene (trpRS1) in Streptomyces coelicolor is regulated by a ribosome-mediated attenuator in the 5′ leader of its mRNA region. This regulatory element controls gene transcription in response to the physiological effects of indolmycin and chuangxinmycin, two antibiotics that inhibit bacterial tryptophanyl-tRNA synthetases. By mining streptomycete genome sequences, we found several orthologs of trpRS1 that share this regulatory element; we predict that they are regulated in a similar fashion. The validity of this prediction was established through the analysis of a trpRS1 ortholog (SAV4725) in Streptomyces avermitilis. We conclude that the trpRS1 locus is a widely distributed and self-regulating antibiotic resistance cassette. This study provides insights into how auxiliary aminoacyl-tRNA synthetase genes are regulated in bacteria.
Streptomyces is a large genus of Gram-positive, soil-dwelling bacteria. Interest in these organisms is based largely on the fact that they produce half of the 10,000 known antibiotics and two-thirds of the antibiotics used in clinical medicine (2). Since antibiotic resistance determinants accompany genes for antibiotic biosynthesis (7), it is a widely held notion that these bacteria represent a reservoir of antibiotic resistance genes (8, 28, 38). The mining of streptomycete genomes has led to the discovery of novel antibiotic resistance genes (40). The high frequency of antibiotic resistance observed in many Streptomyces species indicates that much lateral transfer of resistance genes has taken place (3, 8, 38, 42). In many cases, a Streptomyces species will harbor multiple genes conferring resistance to antibiotics that it does not produce (8). As many resistance genes are needed conditionally, the expression of antibiotic resistance genes often is induced upon exposure to antibiotics (9).
Bioinformatic analysis of the genome sequence of Streptomyces coelicolor (the model organism of the genus) revealed several putative genes that confer resistance to antibiotics that the organism does not produce, including erythromycin, vancomycin, chloramphenicol, the pristinamycins, and the lincosamides (2). Many of the bioinformatic predictions have been validated by antibiotic susceptibility assays and by genetic and biochemical analyses (11, 17, 20, 24, 40). The transcription of some resistance genes is tightly regulated. For example, in the presence of vancomycin and the pristinamycins, S. coelicolor activates the transcription of the corresponding resistance genes (11, 17, 20).
The focus of this study is an auxiliary tryptophanyl-tRNA synthetase gene (trpRS1) in S. coelicolor that confers resistance to indolmycin and chuangxinmycin (25, 39), antibiotics that competitively inhibit bacterial tryptophanyl-tRNA synthetases (34). The primary tryptophanyl-tRNA synthetase encoded by the trpRS2 gene is sensitive to these antibiotics (25). Indolmycin is an antibiotic produced by Streptomyces griseus ATCC 12648 (35). Although indolmycin is not used clinically, its selective inhibition of bacterial tryptophanyl-tRNA synthetases and potent activity against methicillin-resistant Staphylococcus aureus (MRSA) and Helicobacter pylori recently has renewed interest in its pharmacological potential (19). Chuangxinmycin, produced by Actinoplanes tsinanensis, also has demonstrable activity against multidrug-resistant pathogens (6). We previously reported that these antibiotics did not affect the transcription of trpRS2, whereas the open reading frame (ORF) of the trpRS1 transcript could be detected only in cultures that were treated with indolmycin or chuangxinmycin (39). These observations indicated that the gene was subject to regulation at the level of transcription (39). Further, our finding that trpRS1 is transcribed with a 157-nucleotide leader raised the possibility that the gene is transcriptionally regulated via the formation of distinct secondary structures in the leader that either cause or repress the premature termination of transcription. The regulation of gene transcription by cis-acting RNA regulatory structures such as T-box riboswitches and ribosome-mediated transcriptional attenuators is a recurring theme in bacteriology, especially in the context of amino acid metabolism (15, 26, 29). Computational analyses often enable the facile classification of RNA sequences into one of these two categories of RNA regulatory elements (13, 29, 37, 41). On this basis, we analyzed the trpRS1 leader sequence in search of any canonical RNA regulatory elements. Sequence elements resembling those of ribosome-mediated transcriptional attenuators were identified in the trpRS1 leader.
In this study, we used molecular genetic approaches to determine if the apparent attenuator regulates trpRS1 transcription. Herein, we report the mechanism by which trpRS1 transcription is induced by antibiotics that inhibit TrpRS2.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All of the strains that were utilized in this study are listed in Table 1. S. coelicolor strains were grown at 30°C on mannitol soya flour medium (SFM) or Difco nutrient agar medium (DNA) (24). For RNA isolation, S. avermitilis ATCC 31267 was grown in minimal liquid medium (NMMP) as previously described (40). Escherichia coli strains DH5α and ET12567/pUZ8002 were grown on Luria-Bertani medium at 37°C (36). The MICs for Streptomyces strains were assessed on DNA solid medium after a 48-h incubation period. For selecting Escherichia coli, ampicillin, apramycin, chloramphenicol, hygromycin, and kanamycin were employed at 100, 50, 25, 80, and 50 μg/ml, respectively. Nalidixic acid was used at 20 μg/ml to counterselect E. coli in conjugations with S. coelicolor. Apramycin and hygromycin were used at 50 μg/ml for selecting S. coelicolor. Indolmycin was chemically synthesized by the authors as described by Kamiyama et al. (14) and used at 10 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype/description | Reference/source |
|---|---|---|
| S. avermitilis ATCC 31267 | 21, 32 | |
| S. coelicolor | ||
| M600 | Prototroph; SCP1− SCP2 | 24 |
| B725 | M600 trpRS1::apr | 39 |
| B734 | B725 TrpRS2(H48N) | 40 |
| B735 | B725 TrpRS2(H48Q) | 40 |
| B771 | B734 pJS340 | This study |
| B772 | B735 pJS340 | This study |
| B774 | M600 pJS340 | This study |
| B775 | M600 pJS341 | This study |
| B776 | M600 pJS342 | This study |
| B779 | M600 pJS345 | This study |
| B780 | M600 pJS346 | This study |
| B782 | M600 pJS348 | This study |
| B784 | M600 pJS350 | This study |
| B785 | M600 pJS351 | This study |
| B786 | M600 pJS352 | This study |
| B788 | M600 pJS354 | This study |
| B790 | M600 pJS356 | This study |
| B791 | M600 pJS357 | This study |
| E. coli | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK− mK−) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| ET12567 | dam dcm hsdS cat tet | 39 |
Plasmids and primers.
All plasmids used in this study are listed in Table 2. The plasmids used in this study were constructed using standard cloning procedures (36). DNA sequencing was performed by Davis Sequencing (Davis, CA). The site-specific integrating vector pMS81 (12) was used to introduce the various reporter constructs (various trpRS1 leader fragments fused to the neo open reading frame) into S. coelicolor. The primers used in this work, all of which were synthesized by Invitrogen, are listed in Table 3. PCR was performed with either Taq (Invitrogen) or Pfu (Stratagene, Agilent Technologies) under standard conditions for G-C-rich DNA templates (24).
TABLE 2.
Plasmids used in this studya
| Plasmid | Description | Vector backbone | Reference or source |
|---|---|---|---|
| pBluescript KS+ | pUC ori Ampr | Agilent Technologies (Stratagene) | |
| pGemT | pUC-derived, lacZ′, Ampr | Promega | |
| pMS81 | oriT φattB-int Hygr | 12 | |
| pJS340 | trpRS1 wild-type leader-neo | pMS81 | This study |
| pJS341 | trpRS1 leader lacking 3:4 stem-loop-neo | pMS81 | This study |
| pJS342 | trpRS1 leader lacking part of A+U-rich tract-neo | pMS81 | This study |
| pJS345 | trpRS1 leader G(−126)A-neo | pMS81 | This study |
| pJS346 | trpRS1 leader C(−127)A-neo | pMS81 | This study |
| pJS348 | trpRS1 leader G(−129)C-neo | pMS81 | This study |
| pJS350 | trpRS1 leader C(−131)A-neo | pMS81 | This study |
| pJS351 | trpRS1 leader T(−132)C-neo | pMS81 | This study |
| pJS354 | SAV4725 wild-type leader-neo | pMS81 | This study |
| pJS356 | SAV4725 leader G(−128)C-neo | pMS81 | This study |
| pJS357 | SAV4725 leader G(−125)C-neo | pMS81 | This study |
| pJS358 | trpRS1 wild-type leader | pBluescript KS+ | This study |
| pJS359 | trpRS1 wild-type leader-neo | pBluescript KS+ | This study |
| pJS360 | SAV4725 wild-type leader | pBluescript KS+ | This study |
Numbers in parentheses refer to positions relative to the predicted translation start site. Antibiotic resistance markers: Ampr, ampicillin resistance; Hygr, hygromycin resistance; neo, kanamycin resistance.
TABLE 3.
Primers used in this studya
| Entry | Primer name | Application/function | Sequence |
|---|---|---|---|
| 1 | SCO3334 (−380) For | Cloning of the trpRS1 leader | 5′-GATATCTTCTTTCGAACGTGGATTCCTC-3′b |
| 2 | SCO3334 (−1) Rev | Cloning of the trpRS1 leader | 5′-CATATGCTCTCCACCTCCTGGTCAG-3′c |
| 3 | SCO3334 (−59)A Rev | Cloning of the trpRS1 leader lacking part of the A+U-rich tract | 5′-CATATGCTCTCCACCTCCTTACGAGAACGGCCGCC-3′c,e |
| 4 | SCO3334 (−100)A Rev | Cloning of the trpRS1 leader lacking 3:4 hairpin | 5′-CATATGCTCTCCACCTCCTTGTGTACGGCCGCCGTC-3′c,e |
| 5 | TrpRS1 G(−126)A F | Site-directed mutagenesis of the trpRS1 leader, creation of G(−126)A mutation | 5′-GTACCCAGCTCTGGCAGGCCGCCTG-3′ |
| 6 | TrpRS1 G(−126)A R | Site-directed mutagenesis of the trpRS1 leader, creation of G(−126)A mutation | 5′-CGCCGTCAGGCGGCCTGCCAGAGCTG-3′ |
| 7 | TrpRS1 C(−127)A F | Site-directed mutagenesis of the trpRS1 leader, creation of C(−127)A mutation. | 5′-GTGTACCCAGCTCTGGAGGGCCGCCTG-3′ |
| 8 | TrpRS1 C(−127)A R | Site-directed mutagenesis of the trpRS1 leader, creation of C(−127)A mutation. | 5′-GCCGTCAGGCGGCCCTCCAGAGCTG-3′ |
| 9 | TrpRS1 G(−129)C F | Site-directed mutagenesis of the trpRS1 leader, creation of G(−129)C mutation | 5′-CCAGCTCTCGCGGGCCG-3′ |
| 10 | TrpRS1 G(−129)C R | Site-directed mutagenesis of the trpRS1 leader, creation of G(−129)C mutation. | 5′-CGGCCCGCGAGAGCTG-3′ |
| 11 | TrpRS1 C(−131)A F | Site-directed mutagenesis of the trpRS1 leader, creation of C(−131)A mutation | 5′-GTACGTGTACCCAGCTATGGCGGGCCGCCTG-3′ |
| 12 | TrpRS1 C(−131)A R | Site-directed mutagenesis of the trpRS1 leader, creation of C(−131)A mutation | 5′-GTCAGGCGGCCCGCCATAGCTGGGTAC-3′ |
| 13 | TrpRS1 T(−132)C F | Site-directed mutagenesis of the trpRS1 leader, creation of T(−132)C mutation | 5′-GTACGTGTACCCAGCCCTGGCGGGCCGCCTG-3′ |
| 14 | TrpRS1 T(−132)C R | Site-directed mutagenesis of the trpRS1 leader, creation of T(−132)C mutation | 5′-CAGGCGGCCCGCCAGGGCTGGGTAC-3′ |
| 15 | SAV4725 UF | Cloning of the SAV4725 leader | 5′- ACTAGTCGGAATCCTTGCTCCC-3′d |
| 16 | SAV4725 UR | Cloning of the SAV4725 leader | 5′-CATATGCTCTCCACCTCCTGGTCG-3′c |
| 17 | SAV4725 G(−125)C F1 | Site-directed mutagenesis of the SAV4725 leader, creation of G(−125)C mutation | 5′-GTACCCAGCAGTGGTCGGCCGCCTGAC-3′ |
| 18 | SAV4725 G(−125)C R1 | Site-directed mutagenesis of the SAV4725 leader, creation of G(−125)C mutation | 5′-CGCCGTCAGGCGGCCGACCACTGCTG-3′ |
| 19 | SAV4725 G(−128)C F1 | Site-directed mutagenesis of the SAV4725 leader, creation of G(−128)C mutation | 5′-CGTGTACCCAGCAGTCGTGGGCCGCC-3′ |
| 20 | SAV4725 G(−128)C R1 | Site-directed mutagenesis of the SAV4725 leader, creation of G(−128)C mutation | 5′-CGTCAGGCGGCCCACGACTGCTGGGTAC-3′ |
| 21 | SAV4725 RC A | SAV4725 transcript, 5′ RACE | 5′-CTGAAGATCCGCTTCATCTCTC-3′ |
| 22 | SAV4725 RC C | SAV4725 transcript, 5′ RACE | 5′-CCGTGCTGATCGACGTC-3′ |
| 23 | SAV4725 RC D | SAV4725 transcript, 5′ RACE | 5′-GTCGACGACGCAGAACAG-3′ |
Numbers in parentheses refer to positions relative to the predicted translation start site.
The engineered EcoRV site is underlined.
The engineered NdeI site is underlined.
The engineered SpeI site is underlined.
The predicted trpRS1 ribosome-binding site is in boldface.
Creation of reporter constructs.
Using wild-type strain S. coelicolor M600 genomic DNA as the template, a 380-bp region spanning the trpRS1 leader and promoter was amplified by PCR using primers 1 and 2 (Table 3). The PCR product, with an engineered NdeI site (introduced from primer 2) at its 3′ end to enable fusion to the neo (kanamycin resistance) gene, was ligated into pBluescript KS+ to give pJS358 (Table 2). The trpRS1 leader-neo fusion then was subcloned into the integrative vector pMS81, yielding pJS340. pJS340 was introduced into E. coli strain ET12567/pUZ8002 and then into S. coelicolor strains B734 and B735 (yielding strains B771 and B772 [Table 1]), and M600 (yielding strain B774; Table 1) by way of conjugation (23).
A 280-bp segment spanning the trpRS1 promoter and the proposed 1:2 hairpin of the trpRS1 leader was PCR amplified using pJS358 as the template with primers 1 and 4. The native trpRS1 ribosome-binding site was engineered into primer 4 to enable fusion to the truncated leader (Table 3, boldface segment of primer 4 sequence). The PCR product derived from primers 1 and 4 was fused to the neo gene as described above. The truncated trpRS1 leader-neo fusion was subcloned into pMS81 to yield pJS341, which was introduced into wild-type strain S. coelicolor M600 by way of conjugation to yield strain B775 (Table 1).
A 322-bp segment spanning the trpRS1 promoter and leader, with the exception of the four terminal residues of the A+U-rich tract, was PCR amplified using pJS358 as the template with primers 1 and 3. The native trpRS1 ribosome-binding site was engineered into primer 3 to enable fusion to the truncated leader (boldface segment of primer 3 sequence in Table 3). The PCR product was ligated to the neo gene, subcloned into pMS81 to give pJS342, and introduced into wild-type strain S. coelicolor M600 by way of conjugation to yield strain B776 (Table 1).
Using wild-type S. avermitilis ATCC 31267 genomic DNA as the template, a 400-bp region spanning the SAV4725 leader and promoter was amplified by PCR using primers 15 and 16 (Table 3). The PCR product was cloned into pBluescript KS+ (yielding pJS360; Table 2). It then was subcloned into pJS341 for fusion to the neo gene, yielding pJS354 (Table 2). pJS354 was introduced into wild-type S. coelicolor M600 by way of conjugation, yielding strain B788 (Table 1).
Site-directed mutagenesis.
Specific point mutations in the S. coelicolor trpRS1 leader were generated using the Stratagene QuikChange site-directed mutagenesis procedure. The primer pairs used to generate all point mutations in the trpRS1 leader are provided in Table 3 (primers 5 to 14). Plasmid pJS358 was used as the template in these reactions. All mutant trpRS1 leaders were fused to the neo gene and subcloned into pMS81, yielding plasmids pJS345 to pJS351 (Table 2). Plasmids pJS345 to pJS351 were independently introduced into wild-type S. coelicolor strain M600 via conjugation, yielding strains B779 to B785 (Table 1).
The primer pairs used for the site-directed mutagenesis of the SAV4725 leader are provided in Table 3 (primers 17 to 20). Plasmid pJS360 was used as the template in these reactions. The mutant SAV4725 leaders were fused to the neo gene and subcloned into pMS81, yielding plasmids pJS356 and pJS357 (Table 2). Plasmids pJS356 and pJS357 were introduced into wild-type S. coelicolor strain M600 by conjugation, yielding strains B790 and B791 (Table 1).
Mapping of the SAV4725 transcription start site.
The SAV4725 transcription start site was mapped using the 5′ rapid amplification of cDNA ends (5′-RACE) system, version 2.0 (Invitrogen), as previously described (39). Primers used in this analysis (21-23) are described in Table 3. The final PCR product was ligated into vector pGemT (Table 2) and identified by sequencing. The location of the transcription start site was corroborated through independent trials.
Nucleotide sequence accession numbers.
The nucleotide sequences of all trpRS1 orthologs are available at the NCBI database under the following accession numbers: Streptomyces albus (ABYC01000233), Streptomyces avermitilis (NC_003155), Streptomyces coelicolor (NC_003888), Streptomyces ghanaensis (ABYA01000261), Streptomyces griseoflavus (ACFA01000404), Streptomyces hygroscopicus (ACEX01000367), Streptomyces lividans (ACEY01000177), Streptomyces pristinaespiralis (ABJI01000428), Streptomyces sviceus (ABJJ01000233), and Streptomyces viridochromogenes (ACEZ01000149).
RESULTS
Bioinformatic analysis of the trpRS1 leader revealed features that are reminiscent of a ribosome-mediated transcriptional attenuator.
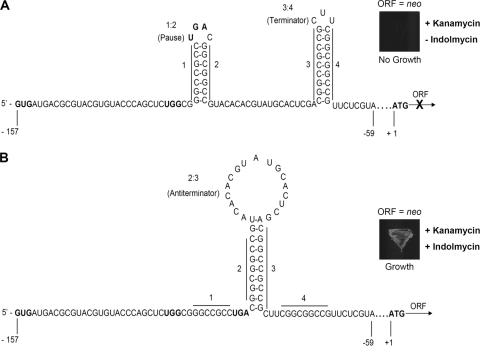
Analysis of the trpRS1 leader revealed features found in ribosome-mediated transcriptional attenuators with rho-independent terminators, especially those in tryptophan metabolism operons (10, 43, 44). Specifically, we identified two distinct dyads of symmetry within the leader sequence that were capable of forming mutually exclusive RNA secondary structures (Fig. 1). In one of the presumptive RNA secondary structures, each dyad of symmetry forms a hairpin. The two sequences of the upstream dyad of symmetry (sequences 1 and 2) form the 1:2 hairpin, while the two sequences of the downstream dyad (sequences 3 and 4) form the 3:4 hairpin (Fig. 1A). As it is G+C rich and immediately upstream of an A+U-rich tract, the 3:4 hairpin resembles a rho-independent transcription terminator (10, 27). By analogy to well-characterized transcription attenuators, the 3:4 hairpin is likely to mediate premature transcription. In the other presumptive RNA secondary structure, sequence 2 of the upstream dyad base pairs with sequence 3 of the downstream dyad to form the 2:3 hairpin (Fig. 1B). As this structure precludes the formation of the rho-independent terminator, it is likely that the 2:3 hairpin is an antiterminator structure permissive to the transcription of the trpRS1 ORF.
FIG. 1.
Schematic representation of the mutually exclusive secondary structures formed by the 157-nucleotide leader of the trpRS1 transcript. The start codon (GUG), tryptophan codon (UGG), and stop codon (UGA) of the trpRS1 leader peptide ORF are in boldface. (A) The proposed terminator structure of the wild-type trpRS1 leader peptide attenuator. An S. coelicolor strain harboring the reporter construct with the wild-type promoter/leader fails to grow on medium containing only kanamycin (growth is indolmycin dependent). (B) The proposed antiterminator structure of the trpRS1 leader peptide attenuator. An S. coelicolor strain harboring the wild-type reporter construct grows on medium containing kanamycin and indolmycin.
The presumptive switch between the transcriptional terminator and antiterminator structures is mediated by the translation of a small ORF positioned adjacent to sequence 1 of the upstream dyad (Fig. 1). The position of the coding region relative to the dyads is indicative of ribosome-mediated transcriptional attenuation (33). The apparent 39-nucleotide ORF encodes a 13-amino-acid peptide with the sequence VMTRTCTQLWRAA. The translation start site of the small ORF coincides with the transcription start site of trpRS1. The small peptide encoded by the ORF is distinct from those found in other bacterial tryptophan metabolism operons in that it only has a single tryptophan residue (37). The relative proximity of the tryptophan codon to sequence 1 of the upstream dyad suggests that the stalling of the ribosome at this codon would physically sequester sequence 1 and thus preclude the formation of the 3:4 hairpin (the apparent rho-independent terminator). Therefore, the ribosome-mediated transcriptional attenuation model can explain the transcriptional response of trpRS1 to indolmycin and chuangxinmycin (inhibitors of tryptophanyl-tRNA synthetase). Specifically, the inhibition of TrpRS2 by these antibiotics would decrease intracellular levels of charged tryptophanyl-tRNA. Under these conditions, the ribosome would stall at the trp codon and trigger antitermination, leading to the transcription of the trpRS1 ORF.
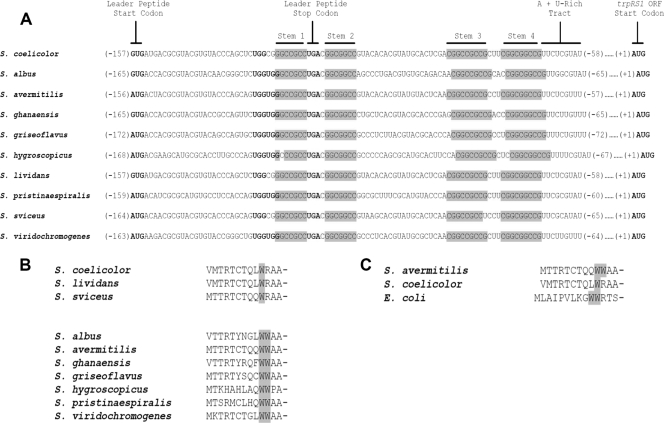
trpRS1 orthologs are found in the genomes of many species of Streptomyces bacteria.
A comparative analysis of the available streptomycete genome sequences revealed that several species possess auxiliary tryptophanyl-tRNA synthetases that are homologous to trpRS1 (data not shown). The nucleotide sequences immediately upstream of the orthologs were homologous to the trpRS1 leader (Fig. 2 A). The highest degrees of homology corresponded to the units of dyad symmetry and to the small ORF in the trpRS1 leader. Further, the position of the small ORF relative to the dyads was absolutely conserved (Fig. 2A). All of the apparent leader peptides were 13 amino acids in length and had a tryptophan residue at position 10 (Fig. 2B). Interestingly, some leader peptides had tandem tryptophan residues at positions 10 and 11 (Fig. 2B). On this basis, we organized the leader peptides into two groups (Fig. 2B). The latter group is typical of the canonical attenuators of trp operons in bacteria, in that the leader peptide has tandem tryptophan residues (29, 30). We predict that trpRS1 and these orthologs are regulated by ribosome-mediated transcriptional attenuation.
FIG. 2.
Sequence alignments of the regions upstream of trpRS1 and its orthologs in various Streptomyces species and of the cognate leader peptides. (A) The regions upstream of the trpRS1 orthologs are highly homologous to the attenuator elements of the trpRS1 leader. The sequences of the dyads of symmetry are shaded gray. The start codon (GUG), tryptophan codon (UGG), and stop codon (UGA) of the leader peptide ORFs are in boldface. (B) Alignment of the predicted peptide encoded by the small ORF in the trpRS1 leader with those of its orthologs. The peptides are grouped based on their tryptophan content. The tryptophan residues are shaded gray. (C) Comparison of the leader peptide sequences of S. avermitilis (SAV4725), S. coelicolor (trpRS1), and E. coli (trpL).
Targeted deletions and missense point mutations within the S. coelicolor trpRS1 leader deregulate transcription.
The sequence and predicted RNA secondary structures of the trpRS1 leader motivated a series of experiments to examine their significance in the regulation of trpRS1. The key to these experiments was the construction of a reporter system in which the promoter and the leader of the trpRS1 gene were fused to the neo gene ORF, which encodes an aminoglycoside phosphotransferase that is a kanamycin resistance determinant. An integrative plasmid harboring the reporter system was introduced into wild-type strain S. coelicolor M600, which is kanamycin sensitive. As expected, the resulting strain (S. coelicolor B774) exhibited kanamycin resistance only when grown on media supplemented with indolmycin (Fig. 1).
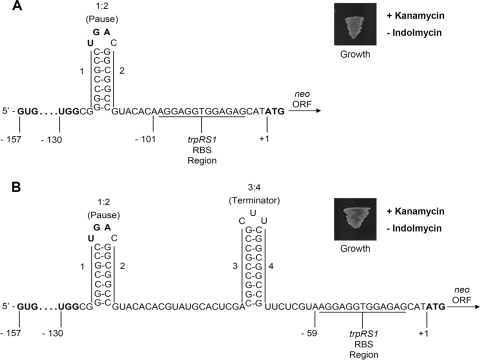
Initially, we tested the hypothesis that the 3:4 hairpin is a rho-independent transcriptional terminator. We deleted the sequence comprising the 3:4 hairpin from the aforementioned reporter construct and introduced the resulting plasmid into wild-type S. coelicolor M600. The resulting strain (S. coelicolor B775) did not require indolmycin for growth on media supplemented with kanamycin (Fig. 3 A). The observed deregulation of the neo reporter gene was consistent with the proposed function of the 3:4 hairpin as a rho-independent transcriptional terminator. This proposed role for the 3:4 hairpin also was based on the fact that it was accompanied by an A+U-rich sequence. These sequences often flank rho-independent terminators and are required for the efficient termination of transcription. The truncation of the A+U-rich tract of rho-independent terminators has been shown to compromise transcriptional termination (4). To evaluate the significance of the trpRS1 leader's A+U-rich tract, we prepared a reporter construct in which the four terminal nucleotides of the tract were replaced with the native trpRS1 ribosome-binding site. S. coelicolor strain B776, harboring this truncated reporter construct, exhibited kanamycin resistance in the absence of indolmycin. The deregulated expression of the neo gene from this reporter construct indicated that the A+U-rich tract is required for efficient transcriptional termination. Taken together, these observations support our proposal that the 3:4 hairpin functions as a rho-independent terminator.
FIG. 3.
Schematic representation of the engineered trpRS1 leaders defective with respect to transcription termination. (A) Schematic representation of the trpRS1 reporter construct in which the 3:4 hairpin is replaced by the native trpRS1 ribosome-binding site. The strain harboring this construct grows in an indolmycin-independent fashion on media supplemented with kanamycin. (B) Schematic representation of the trpRS1 reporter construct in which the terminal four nucleotides of the A+U-rich tract are replaced by the native trpRS1 ribosome-binding site. The strain harboring this construct grows in an indolmycin-independent fashion on media supplemented with kanamycin. The start codon (GUG), tryptophan codon (UGG), and stop codon (UGA) of the leader peptide ORF are in boldface. The start codon (ATG) of the neo open reading frame at position +1 also is in boldface.
The presence of a small ORF in the trpRS1 leader is central to our prediction that the trpRS1 gene is subject to ribosome-mediated transcriptional attenuation. Our model for antitermination is based on the antibiotic-induced stalling of the ribosome at the tryptophan codon of the leader peptide ORF. On this basis, we hypothesized that missense mutations affecting this codon would deregulate the transcription of trpRS1. The wild-type reporter system was used to test this hypothesis. Using site-directed mutagenesis, we introduced a missense mutation (UGG to UCG) that changed the tryptophan codon to a serine codon. This construct was introduced into wild-type S. coelicolor M600, yielding strain S. coelicolor B782. This strain failed to grow on media supplemented with both kanamycin and indolmycin or with kanamycin alone; apparently, the transcription of the reporter gene no longer was induced by indolmycin. In contrast, missense mutations affecting other codons of the small open reading frame did not perturb the indolmycin dependence of reporter gene transcription (as evident in the phenotype of S. coelicolor strains B779 to B780 and B784 to B785; data not shown). Based on these observations, it is clear that antibiotic-induced antitermination is dependent on the tryptophan codon within the small ORF of the trpRS1 leader.
Tryptophanyl-tRNA synthetase activity is inversely correlated with antitermination of trpRS1 transcription.
S. coelicolor transcribes the trpRS1 ORF only in the presence of indolmycin and chuangxinmycin. This phenomenon is not a response to the antibiotics but to their inhibition of TrpRS2 activity (39). We proposed that the reduced availability of charged tryptophanyl-tRNA resulting from TrpRS2 inhibition is directly sensed via the trpRS1 leader. Accordingly, we suspected that S. coelicolor strains with a compromised ability to charge tryptophanyl-tRNA would transcribe the trpRS1 ORF in the absence of indolmycin. To test this hypothesis, we introduced the wild-type neo reporter construct into S. coelicolor trpRS1 null strains with point mutations in the trpRS2 ORF, encoding TrpRS2(H48N) or TrpRS2(H48Q) (40). In homologous tryptophanyl-tRNA synthetases, this histidine residue directly participates in catalysis, and the replacement of this residue with asparagine is known to compromise enzymatic activity (19, 25). The resulting strains, S. coelicolor B771 and S. coelicolor B772, exhibited indolmycin-independent growth on media supplemented with kanamycin. Presumably, strains with genetic mutations that affect tryptophanyl-tRNA charging have insufficient charged tryptophanyl-tRNA for the efficient synthesis of the trpRS1 leader peptide; the ensuing ribosomal stalling leads to antitermination. Similar observations have been made in E. coli, where it has been reported that strains harboring mutations in tryptophanyl-tRNA synthetase exhibit a high degree of antitermination at the trp operon via ribosome-mediated transcriptional attenuation (6, 43, 44).
Analysis of a trpRS1 ortholog in S. avermitilis.
We predict that the trpRS1 orthologs identified in streptomycete genome sequences also are regulated by ribosome-mediated transcriptional attenuation. Some of these orthologs have two tryptophan codons in their apparent leader peptide ORF rather than one, as in the trpRS1 leader peptide ORF. To test predictions about the regulation of trpRS1 orthologs and to investigate the significance of tryptophan content in the leader peptide, we analyzed the trpRS1 ortholog (SAV4725) in S. avermitilis (21, 31).
A series of experiments was performed to characterize the transcription of SAV4725. Using reverse transcription-PCR (RT-PCR), we found that the full-length transcript was detected only in S. avermitilis cultures that had been treated with indolmycin (40). 5′-RACE analysis revealed that the SAV4725 transcript has a leader that is 156 nucleotides in length and highly homologous to the trpRS1 leader (90% identity). The sequences comprising the dyads of symmetry are identical in the trpRS1 and SAV4725 leaders (Fig. 2A). Furthermore, a positionally conserved small ORF was observed in the SAV4725 leader. As was the case for trpRS1, the start codon of the apparent SAV4725 leader peptide was coincident with the transcription start site. There were few differences between the trpRS1 and SAV4725 leader peptide sequences; most notably, the SAV4725 peptide had two tryptophan residues (Fig. 2C). Both leader peptides resemble the one encoded in the leader of the E. coli trp operon (Fig. 2C), which has been extensively studied (29, 30).
Site-directed mutagenesis experiments, analogous to those described in our analysis of the trpRS1 leader, were performed to characterize the SAV4725 leader. The experiments were based on the construction of a reporter construct in which the SAV4725 promoter region and leader were fused to the neo gene ORF. This construct was introduced into wild-type S. coelicolor M600. As expected, the resulting strain (S. coelicolor B788) exhibited indolmycin-dependent growth on media supplemented with kanamycin. Independent missense mutations were introduced at each of the tryptophan codons of the leader peptide ORF; the codons were changed to serine codons. S. coelicolor strains harboring reporter constructs with mutations in either of the tryptophan codons of the leader peptide ORF (i.e., S. coelicolor B790 and S. coelicolor B791) exhibited indolmycin-dependent growth on media supplemented with kanamycin. These observations indicate that trp codons at either position 10 or 11 within the leader peptide ORF enable transcriptional attenuation; the stalling of the ribosome at either of these tryptophan codons will lead to antitermination.
It is noteworthy that the strains harboring reporter constructs with single missense mutations in either of the trp codons of the SAV4725 leader peptide ORF (S. coelicolor 790 and 791) required 2.5 times more indolmycin (10 versus 25 μg/ml) for growth on media supplemented with kanamycin than did the strain harboring the wild-type reporter construct (S. coelicolor B788). These findings are consistent with observations of a direct correlation between the number of consecutive regulatory codons in the leader peptide and the sensitivity of ribosome-mediated transcriptional attenuators to changes in levels of charged tRNA (44). Thus, we predict that trpRS1 orthologs with two consecutive tryptophan residues in the leader peptide attenuator are more sensitive to antibiotic-induced perturbations in levels of charged tryptophanyl-tRNA than those with a single tryptophan residue.
DISCUSSION
Auxiliary aminoacyl-tRNA synthetase genes in bacteria often are associated with resistance to antibiotics that inhibit aminoacyl-tRNA synthetases (25, 39, 40, 45). Given that the auxiliary enzyme isoforms are needed only in the presence of antibiotics, one might anticipate that the corresponding genes are regulated in a fundamentally different way than those encoding the primary isoforms. Streptomyces coelicolor provided an interesting opportunity to test this premise, since it has two tryptophanyl-tRNA synthetase genes (2). Apparently, the transcription of trpRS1 (the auxiliary, antibiotic-resistant tryptophanyl-tRNA synthetase) is an adaptive response to indolmycin and chuangxinmycin exposure. The transcription of other tryptophan metabolism genes in S. coelicolor is regulated in a growth phase- and growth rate-dependent fashion (16, 18).
The regulated expression of genes encoding aminoacyl-tRNA synthetases and other enzymes involved in amino acid metabolism is a well-known phenomenon in many bacterial genera (29, 30). Often, the messenger RNAs have regulatory structures in their 5′ leaders that are affected by regulatory proteins, uncharged tRNA, cofactors, or rates of leader peptide synthesis (1, 29, 30). We determined that the trpRS1 leader controls transcription via ribosome-mediated attenuation. trpRS1 orthologs were identified in a number of Streptomyces species, and their proposed leader sequences were found to be highly homologous to that of the trpRS1 leader. We demonstrated that the trpRS1 ortholog in S. avermitilis (SAV4725) is regulated in the same fashion as trpRS1. Thus, it appears that the trpRS1 locus is a widely distributed and self-regulating antibiotic resistance cassette.
The closest analogy to the mode of regulation for trpRS1 and SAV4725 is that of the trp operon in E. coli (29, 30). Its transcription is controlled by a 162-nucleotide leader region designated trpL (43, 44). In trpL, attenuation is governed by the formation of competing terminator and antiterminator RNA secondary structures that either arrest or permit, respectively, the transcription of genes for tryptophan biosynthesis (29, 30, 43). When charged tryptophanyl-tRNA levels are low, the stalling of the ribosome at either of two tandem tryptophan codons in the trpL leader peptide ORF precludes the formation of a rho-independent terminator and allows the transcription of the tryptophan biosynthesis genes. The results of all of our experiments were consistent with the trpL model of transcriptional attenuation (30, 44).
It is interesting that the SAV4725 attenuator, but not the trpRS1 attenuator, was identified in a bioinformatic search for RNA regulatory elements in actinobacterial genomes (37). These attenuators differ only in the number of trp codons in the leader peptide ORF. Our findings suggest that future bioinformatic analyses should account for leader peptide attenuators with a single regulatory codon. Further, we predict that the tryptophan content of the trpRS1 leader peptide is physiologically meaningful. Generally, leader peptide attenuators play a critical role in cellular responses to amino acid starvation. The number of regulatory codons in the leader peptide ORF determines the sensitivity of the attenuator to amino acid availability (22). Attenuators with higher numbers of regulatory codons are more sensitive to changes in the availability of charged tRNA than are those with fewer numbers of regulatory codons (44). Since the trpRS1 leader has a single trp codon, we suspect that the trpRS1 gene is not transcribed under typical starvation conditions. Indeed, the trpRS1 ORF was not detected in response to tryptophan starvation (25). It appears that the trpRS1 gene is transcribed only under the adverse physiological conditions brought about by antibiotics that inhibit tryptophanyl-tRNA synthetases. Only under these conditions is the level of charged tryptophanyl-tRNA low enough to cause the ribosome to stall at the single trp codon in the trpRS1 leader. The stringent requirements for trpRS1 transcription likely minimize the fitness cost of maintaining the gene on the S. coelicolor chromosome.
The expression of antibiotic resistance genes is often an adaptive response to the physiological consequence of antibiotic action (9). The molecular logic of trpRS1 regulation is an ingenious strategy for controlling the expression of an antibiotic-resistant tryptophanyl-tRNA synthetase gene. One can envision that adaptive responses to antibiotics that inhibit aminoacyl-tRNA synthetases are based on the detection of either substrate accumulation (i.e., amino acids or uncharged tRNA) or product diminution (i.e., charged tRNA). The peptide attenuator mechanism for trpRS1 regulation enables a response to the diminution of charged tRNA levels triggered by indolmycin and chuangxinmycin. In contrast, the presence of a T-box motif upstream of an auxiliary tryptophanyl-tRNA synthetase (homologous to TrpRS1) in Bacillus anthracis suggests that its transcription is a response to the uncharged tRNA that would accumulate in the presence of these antibiotics (13, 40). In any case, this study provides insights into how auxiliary aminoacyl-tRNA synthetase genes are regulated in bacteria.
Acknowledgments
Brown University is gratefully acknowledged for financial support. J.J.V. was supported by an NSF EPSCoR graduate fellowship.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Babitzke, P., J. Yealy, and D. Campanelli. 1996. Interaction of the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis with RNA: effects of the number of GAG repeats, the nucleotides separating adjacent repeats, and RNA secondary structure. J. Bacteriol. 178:5159-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor. Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Benveniste, R., and J. Davies. 1973. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. U. S. A. 70:2276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertrand, K., L. J. Korn, F. Lee, and C. Yanofsky. 1977. The attenuator of the tryptophan operon of Escherichia coli: heterogeneous 3′-OH termini in vivo and deletion mapping of functions. J. Mol. Biol. 117:227-247. [DOI] [PubMed] [Google Scholar]
- 5.Bogosian, G., P. V. Haydock, and R. L. Somerville. 1983. Indolmycin-mediated inhibition and stimulation of transcription at the trp promoter of Escherichia coli. J. Bacteriol. 153:1120-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, M. J., P. S. Carter, A. E. Fenwick, A. P. Fosberry, D. W. Hamprecht, M. J. Hibbs, R. L. Jarvest, et al. 2002. The antimicrobial natural product chuangxinmycin and some synthetic analogues are potent and selective inhibitors of bacterial tryptophanyl tRNA synthetase. Bioorg. Med. Chem. Lett. 12:3171-3174. [DOI] [PubMed] [Google Scholar]
- 7.Cundliffe, E. 1989. How antibiotic producing organisms avoid suicide. Annu. Rev. Microbiol. 43:207-233. [DOI] [PubMed] [Google Scholar]
- 8.D'Costa, V. M., K. M. McGrann, D. W. Hughes, and G. D. Wright. 2006. Sampling the antibiotic resistome. Science 311:374-377. [DOI] [PubMed] [Google Scholar]
- 9.Depardieu, F., I. Podglajen, R. Leclercq, E. Collatz, and P. Courvalin. 2007. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20:79-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farnham, P., and T. Platt. 1981. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 9:563-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folcher, M., R. P. Morris, G. Dale, K. Salah-Bey-Hocini, P. H. Viollier, and C. J. Thompson. 2001. A transcriptional regulator of a pristinamycin resistance gene in Streptomyces coelicolor. J. Biol. Chem. 276:1479-1485. [DOI] [PubMed] [Google Scholar]
- 12.Gregory, M. A., R. Till, and M. C. M. Smith. 2003. Integration site for Streptomyces phage ΦBT1 and development of site-specific integrating vectors. J. Bacteriol. 185:5320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutiérrez-Preciado, A., T. M. Henkin, F. J. Grundy, C. Yanofsky, and E. Merino. 2009. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol. Mol. Biol. Rev. 73:36-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasuoka, A., Y. Nakayama, M. Adachi, H. Kamiguchi, and K. Kamiyama. 2001. Development of a stereoselective practical synthetic route to indolmycin, a candidate anti-H. pylori agent. Chem. Pharm. Bull. 49:1604-1608. [DOI] [PubMed] [Google Scholar]
- 15.Henkin, T. M., and C. Yanofsky. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24:700-707. [DOI] [PubMed] [Google Scholar]
- 16.Hodgson, D. A. 2000. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 42:47-238. [DOI] [PubMed] [Google Scholar]
- 17.Hong, H.-J., M. I. Hutchings, J. M. Neu, M. S. Paget, and M. J. Buttner. 2004. Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (VanK) required for drug resistance. Mol. Microbiol. 59:1107-1121. [DOI] [PubMed] [Google Scholar]
- 18.Hu, D. S., D. W. Hood, R. Heidstra, and D. A. Hodgson. 1999. The expression of the trpD, trpC and trpBA genes of Streptomyces coelicolor A3(2) is regulated by growth rate and growth phase but not by feedback repression. Mol. Microbiol. 32:869-880. [DOI] [PubMed] [Google Scholar]
- 19.Hurdle, J. G., A. J. O' Neill, and I. Chopra. 2004. Anti-staphylococcal activity of indolmycin, a potential topical agent for control of staphylococcal infections. J. Antimicrob. Chemother. 54:549-552. [DOI] [PubMed] [Google Scholar]
- 20.Hutchings, M. I., H.-J. Hong, and M. J. Buttner. 2006. The vancomycin resistance VanRS two-component signal transduction system of Streptomyces coelicolor. Mol. Microbiol. 59:923-935. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, et al. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 22.Johnston, H. M., W. M. Barnes, F. G. Chumley, L. Bossi, and J. R. Roth. 1980. Model for regulation of the histidine operon of Salmonella. Proc. Natl. Acad. Sci. U. S. A. 77:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, G. H., M. S. B. Paget, L. Chamberlin, and M. J. Buttner. 1997. Sigma-E is required for the production of the antibiotic actinomycin in Streptomyces antibioticus. Mol. Microbiol. 23:169-178. [DOI] [PubMed] [Google Scholar]
- 24.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, CT.
- 25.Kitabatake, M., K. Ali, A. Demain, K. Sakamoto, S. Yokoyama, and D. Söll. 2002. Indolmycin resistance of Streptomyces coelicolor A3(2) by induced expression of one of its two tryptophanyl-tRNA synthetases. J. Biol. Chem. 277:23882-23887. [DOI] [PubMed] [Google Scholar]
- 26.Kolter, R., and C. Yanofsky. 1982. Attenuation in amino acid biosynthetic operons. Annu. Rev. Genet. 16:113-134. [DOI] [PubMed] [Google Scholar]
- 27.Lesnik, E. A., R. Sampath, H. B. Levene, T. J. Henderson, J. A. McNeil, and D. J. Ecker. 2001. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 29:3583-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez, J. L. 2009. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. R Soc. B 276:2521-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merino, E., and C. Yanofsky. 2005. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 21:260-264. [DOI] [PubMed] [Google Scholar]
- 30.Merino, E., R. A. Jensen, and C. Yanofsky. 2008. Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 11:78-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnishi, Y., J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, et al. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, et al. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. U. S. A. 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oxender, D. L., G. Zurawski, and C. Yanofsky. 1979. Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region. Proc. Natl. Acad. Sci. U. S. A. 76:5524-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pohlmann, J., and H. Brötz-Oesterhelt. 2004. New aminoacyl-tRNA synthetase inhibitors as antibacterial agents. Curr. Drug Targets Infect. Disord. 4:261-272. [DOI] [PubMed] [Google Scholar]
- 35.Routien, J. B. 1966. Identity of streptomycete producing antibiotic PA 155. J. Bacteriol. 91:1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Seliverstov, A. V., H. Putzer, M. S. Gelfand, and V. A. Lyubetsky. 2005. Comparative analysis of RNA regulatory elements of amino acid metabolism genes in Actinobacteria. BMC Microbiol. 5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, C. J., J. M. Ward, and D. A. Hopwood. 1980. DNA cloning in Streptomyces: resistance genes from antibiotic-producing species. Nature 286:525-527. [DOI] [PubMed] [Google Scholar]
- 39.Vecchione, J. J., and J. K. Sello. 2008. Characterization of an inducible, antibiotic-resistant aminoacyl-tRNA synthetase gene in Streptomyces coelicolor. J. Bacteriol. 190:6253-6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vecchione, J. J., and J. K. Sello. 2009. A novel tryptophanyl-tRNA synthetase gene confers high-level resistance to indolmycin. Antimicrob. Agents Chemother. 53:3972-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vitreschak, A. G., A. A. Mironov, V. A. Lyubetsky, and M. S. Gelfand. 2008. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14:717-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiener, P., S. Egan, and E. M. H. Wellington. 1998. Evidence for transfer of antibiotic-resistance genes in soil populations of streptomycetes. Mol. Ecol. 7:1205-1216. [DOI] [PubMed] [Google Scholar]
- 43.Yanofsky, C. 1981. Attenuation in the control of expression of bacterial operons. Nature 289:751-758. [DOI] [PubMed] [Google Scholar]
- 44.Yanofsky, C. 2003. Reflections: using studies on tryptophan metabolism to answer basic biological questions. J. Biol. Chem. 278:10859-10878. [DOI] [PubMed] [Google Scholar]
- 45.Zeng, Y., H. Roy, P. B. Patil, M. Ibba, and S. Chen. 2009. Characterization of two seryl-tRNA synthetases in albomycin-producing Streptomyces sp. strain ATCC 700974. Antimicrob. Agents Chemother. 53:4619-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]