Abstract
We report here the isolation and recombinational cloning of a large plasmid, pZL12, from endophytic Streptomyces sp. 9R-2. pZL12 comprises 90,435 bp, encoding 112 genes, 30 of which are organized in a large operon resembling bacteriophage genes. A replication locus (repA) and a conjugal transfer locus (traA-traC) were identified in pZL12. Surprisingly, the supernatant of a 9R-2 liquid culture containing partially purified phage particles infected 9R-2 cured of pZL12 (9R-2X) to form plaques, and a phage particle (φZL12) was observed by transmission electron microscopy. Major structural proteins (capsid, portal, and tail) of φZL12 virions were encoded by pZL12 genes. Like bacteriophage P1, linear φZL12 DNA contained ends from a largely random pZL12 sequence. There was also a hot end sequence in linear φZL12. φZL12 virions efficiently infected only one host, 9R-2X, but failed to infect and form plaques in 18 other Streptomyces strains. Some 9R-2X spores rescued from lysis by infection of φZL12 virions contained a circular pZL12 plasmid, completing a cycle comprising autonomous plasmid pZL12 and lytic phage φZL12. These results confirm pZL12 as the first example of a plasmid-phage in Streptomyces.
Streptomyces species, a major source of antibiotics and pharmacologically active metabolites, are Gram-positive, mycelial bacteria with high G+C content in their DNA (15). They usually harbor conjugative circular and/or linear plasmids, propagating in autonomous and/or chromosomally integrated forms (14). Most Streptomyces circular plasmids reported are small (8 to 14 kb), including rolling-circle-replication (RCR) plasmids (pIJ101, pJV1, pSG5, pSN22, pSVH1, pSB24.2, pSY10, pSNA1, pSLG33, pEN2701, etc.) (12, 14) and chromosomally integrating/autonomous plasmids (SLP1 and pSAM2) (4, 27, 28). Some theta replication plasmids are of intermediate size (31 to 39 kb), such as SCP2, pFP1, and pFP11 (13, 40). These theta replication loci comprise a rep gene and an adjacent noncoding or iteron sequence, to which Rep protein binds specifically in vitro (10, 40). The occurrence of an ∼163-kb large plasmid, pSV1, in Streptomyces violaceoruber SANK95570 was confirmed (1, 37), but this plasmid could not be physically isolated by standard procedures for plasmid preparation (17). In contrast to more than 30 genes for conjugal transfer on the Escherichia coli F plasmid (20), Streptomyces plasmids usually need a single tra gene (encoding a DNA translocase containing a cell division FtsK/SpoIIIE domain) (15, 29). The transfer of Streptomyces circular plasmids involves binding of the nonnicked double-stranded DNA (dsDNA) by multimers of Tra proteins at a noncoding sequence and ATP hydrolysis-dependent translocation of this DNA through the hyphal tips of the Streptomyces mycelium (15, 32).
Numerous Streptomyces phages have been described, including φC31 (22), SAt1 (26), TG1 (11), FP43 (24), φSPK1 (19), φSC623 (34), DAH2/DAH4/DAH5/DAH6 (6), and mu1/6 (9). They range in size from 36 kb (19) to 121 kb (6), with 50 to 71.2% GC content (9, 23, 35). Streptomyces phages often have a wide host range; for example, 16 of 27 Streptomyces strains are susceptible to infection by φSPK1 (19), and phage FP43 transduces species of Streptoverticillium, Chainia, and Sacchropolyspora (24). φC31 is the most-studied Streptomyces phage and cloning vector (8). The sequences of the φC31 head proteins (e.g., portal, capsid, and head protease) resemble those of other bacterial dsDNA phages, suggesting evolutionary relationships to other viruses (35).
We report here the isolation and recombinational cloning of a 90,435-bp plasmid, pZL12, from endophytic Streptomyces sp. 9R-2 and the characterization of its replication and transfer. Surprisingly, the supernatant of 9R-2 liquid culture infected 9R-2 cured of pZL12 to form plaques. A cycle comprising autonomous plasmid pZL12 and lytic phage φZL12 is described.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general methods.
The strains and plasmids used in this work are listed in Table 1. The isolation of endophytic actinomycetes from Chinese medicinal herbs was done in accordance with the method of Cao et al. (7). The 16S rRNA genes were amplified by PCR with primers (5′-AGACTTTGATCCTGGCTCAG-3′ and 5′-CGGCTACCTTGTTACGACTTC-3′). Plasmid isolation, transformation of Escherichia coli, and Southern hybridization were done in accordance with the method of Sambrook et al. (33). Plasmids pSP72, pQC578, and pQC156 were used as cloning vectors (30). E. coli DH5α was used as a cloning host. Streptomyces lividans ZX7 (41) was the host for propagating plasmids. Streptomyces culture, plaque formation, phage isolation, pulsed-field gel electrophoresis, plasmid isolation, preparation of protoplasts, transformation, and conjugation were done in accordance with the method of Kieser et al. (18). Shotgun cloning and sequencing of pZL12 were performed with the FLX 454 genome sequencer system (Roche) at the Chinese Human Genome Center in Shanghai. Analysis of Streptomyces protein coding regions was performed with FramePlot 3.0 beta (http://watson.nih.go.jp/∼jun/cgi-bin/frameplot-3.0b.pl). Sequence comparisons were done with software from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Blast.cgi).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| S. coelicolor M145 | SCP1− SCP2− | 18 |
| S. lividans ZX7 | pro-2 str-6 rec-46 dnd SLP2− SLP3− | 41 |
| S. venezuelae ISP5230 | Jadomycin B producer | 38 |
| S. glaucescens GLA 4-26 | Tetracenomycin C producer | 25 |
| S. violaceoruber SANK95570 | Contains pSV1 | 1 |
| S. avermitilis MMR630 | Avermectin producer | 36 |
| S. hygroscopicus 5008 | Validoxylamine A producer | 2 |
| Endophytic Streptomyces | ||
| 13 strains (9R-2, 14R-10, 14R-3-2, 13R-3A, 21R-9-1A, 37L-3, 9L-2A, 9R-4C, 7R-9-2, 41S-1, H2GS-3B, 56LL-1, and 75R-4B) | Isolated from Chinese medicinal herbs in the Shanghai Botanical Garden, harboring pZL1-pZL14 | This work |
| 9R-2X | Strain 9R-2 cured of pZL12 | This work |
| Escherichia coli | ||
| DH5α | F−deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | Life Technologies |
| ET12567 (pUZ8002) | dam dcm hsdM cm kan | 18 |
| Plasmids | ||
| pSP72 | amp colEI-ori | Life Technologies |
| pBluescript II SK | amp colEI-ori lacZ | Stratagene |
| pIJ702 | melC tsr pIJ101 origin | 16 |
| pQC156 | 2.6-kb BclI-fragment of melC tsr cloned in pSP72 | 30 |
| pQC578 | 6-kb MluI fragment of pSLA2 rep-rlrA-rorA cloned in pQC156 | 30 |
| pHAQ31 | amp colEI-ori cos melC tsr | 36 |
| pSET152 | Streptomyces phage φC31-derived integration vector; Aprr | 5 |
| pQX17 | Fragment containing apr-oriT of pSET152 cloned in a fragment containing rep-sopABC of the F plasmid (PCR) | This work |
| pZQ104 | 3.5-kb BamHI fragment of pZL12 cloned in pQX17 | This work |
| pZQ107 | pZQ104 integrated in pZL12 via a 3.5-kb BamHI fragment of pZL12 | This work |
| 107c16 | 32.1-kb Sau3A1 fragment of pZL12 cloned in pHAQ31 (BamHI) | This work |
| pZQ140 | 3-kb Sau3A1 fragment of pZL12 cloned in pQC156 (BamHI) | This work |
| pZQ149 | 2.1-kb EcoRI fragment (PCR) of pZL12 cloned in pQC156 | This work |
| pZQ150 | 1.6-kb EcoRI fragment (PCR) of pZL12 cloned in pQC156 | This work |
| pZQ141 | 1.4-kb EcoRI fragment (PCR) of pZL12 cloned in pQC156 | This work |
| pZQ155 | 6.5-kb HindIII fragment (PCR) of pZL12 cloned in pQC578 | This work |
| pZQ157 | 6-kb HindIII fragment (PCR) of pZL12 cloned in pQC578 | This work |
| pZQ159 | 4.9-kb HindIII fragment (PCR) of pZL12 cloned in pQC578 | This work |
| pZQ158 | 2.4-kb HindIII fragment (PCR) of pZL12 cloned in pQC578 | This work |
| pZQ162 | 5.3-kb HindIII fragment (PCR) of pZL12 cloned in pQC578 | This work |
Isolation and recombinational cloning of Streptomyces large circular plasmids.
Isolation of Streptomyces large circular plasmids were done in accordance with the method of Kieser (17), with slight modification. About 0.5 g mycelium was resuspended in 10 ml lysozyme solution (2 mg/ml lysozyme, 10.3% sucrose, 25 mM Tris-HCl, 25 mM EDTA at pH 8.0) at 37°C for 30 min. Five milliliters of 0.3 M NaOH-2% SDS solution was added, mixed thoroughly, and incubated at 55°C for 30 min. After cooling, the DNA solution was extracted with 5 ml acid phenol-chloroform and then with 15 ml neutral phenol-chloroform and centrifuged at 12,000 rpm for 10 min. DNA was precipitated from the supernatant with isopropanol, washed twice with 70% ethanol, and dissolved in 100 μl Tris-EDTA (TE) buffer.
Recombinational cloning of pZL12 from Streptomyces into E. coli included the following steps. pZL12 was digested with BamHI, ligated to an E. coli BAC-derived pQX17 containing the apr selection marker and the oriT site, and introduced by transformation into E. coli DH5α. A plasmid (pZQ104) containing a 3.5-kb fragment of pZL12 was obtained. pZQ104 was introduced by transformation into E. coli ET12567 containing pUZ8002 and was mixed with 108 Streptomyces sp. 9R-2 spores in MS medium (18) containing 25 μg/ml nalidixic acid and 50 μg/ml apramycin at 30°C for 3 days. Apramycin-resistant colonies in 9R-2 were obtained by recombination between pQX17 containing the 3.5-kb pZL12 sequence and intact pZL12. The cointegrated plasmid was isolated and introduced by electroporation into E. coli DH5α.
Identification of a replication locus in pZL12.
A cosmid library for pZL12 was constructed in pHAQ31 (36) by using a Giga-pack III XL Gold packaging extract kit (Stratagene). DNA from 40 cosmids was mixed and introduced by transformation into S. lividans ZX7. Thiostrepton-resistant colonies were obtained, and a cosmid (107c16) containing a 32.1-kb pZL12 sequence was able to propagate in S. lividans ZX7. Various fragments of 107c16 were cloned in an E. coli plasmid (pQC156), and the resulting plasmids (pZQ140, pZQ141, pZQ149, and pZQ150) (Table 1) were introduced by transformation into ZX7. To compare the transformation frequencies of plasmids in different experiments, we used 0.1 ng DNA of Streptomyces plasmid pIJ702 (16) and took 1 × 106 transformants per μg DNA as a control frequency.
Identification of a locus for conjugal transfer of pZL12.
Various PCR fragments of pZL12 were cloned in pQC578, and the resulting plasmids (pZQ155, pZQ57, pZQ158, pZQ159, and pZQ162) (Table 1) were introduced by transformation into ZX7. About equal numbers (107) of spores of ZX7 containing these plasmids and ZX7 containing a chromosome-integrated pSET152 plasmid were mixed in MS medium at 30°C for 4 days. Spores were harvested, diluted in water, and plated on MS medium containing thiostrepton, MS medium containing apramycin, and MS medium containing thiostrepton plus apramycin. The frequency of plasmid transfer was defined as 100 times the ratio of the number of colonies in MS medium containing thiostrepton plus apramycin to the number of colonies in MS medium containing apramycin.
Curing of indigenous plasmid of strain 9R-2.
Strain 9R-2 was inoculated into 3 ml tryptone soy broth (TSB) liquid medium containing 0.002% SDS at 30°C for 24 h, and 0.1 ml culture was transferred to 3 ml fresh TSB at 37°C for 48 h. Mycelium was harvested, diluted in TSB (10 to 105 times), and plated in MS medium at 30°C for 4 days. Individual colonies were picked for detection of possible plasmids on a gel.
9R-2 culture containing partially purified phage particles and formation of plaques on 9R-2X.
To isolate suspected phage particles, ∼108 spores of 9R-2 were inoculated into 100 ml YMB liquid medium (yeast extract, 4 g; malt extract, 10 g; glucose, 4 g; trace element solution, 0.2 ml; H2O, 1,000 ml; pH 7.2) and incubated with shaking at 30°C for 48 h. Supernatant was obtained by centrifugation at 12,000 rpm for 15 min and was filtered through a 0.45-μm membrane. DNase I (1 μg/ml) and RNase (1 μg/ml) were added at room temperature for 30 min, and then 5.84 g NaCl was added and stirred on ice for 1 h. Cell debris was removed by centrifugation at 12,000 rpm for 10 min. Ten percent polyethylene glycol (PEG) was added to the supernatant and the mixture kept on ice for 3 h. The precipitated material was obtained by centrifugation at 12,000 rpm for 10 min. The pellet was dissolved by addition of 0.8 ml SM buffer for 1 h, extracted with chloroform for 30 s, and centrifuged at 3,000 rpm for 15 min to obtain an approximately 700-μl aqueous solution.
To detect possible plaques, an approximately 250-μl solution was plated on DNB medium and overlaid with 2 ml soft nutrient agar containing 108 spores of 9R-2X before incubation at 30°C for 60 h. The phage particles were soaked out of plagues by using liquid nutrient broth and plated onto DNB medium and overlaid with soft nutrient agar containing spores of 9R-2X to obtain phage particles. About 109 PFU/ml of phage was employed for observation by transmission electron microscopy.
Isolation and determination of proteins of the φZL12 virion.
About 104 PFU/ml of φZL12 solution was plated on DNB medium, overlaid with soft nutrient agar containing 108 spores of 9R-2X, and incubated at 30°C for 60 h. The complete lysate was soaked into 4 ml DNB solution at room temperature for 4 h and centrifuged at 55,000 × g for 75 min. The pellet was resuspended in 170-μl SM buffer, mixed with 5× loading buffer in boiling water for 3 min, and electrophoresed in a 15% SDS-polyacrylamide gel at 100 V for 1.5 h. Coomassie blue-stained bands were cut from the gel. Each band was digested with trypsin and characterized by using a model 4800 matrix-assisted laser desorption ionization-tandem time of flight (MALDI-TOF/TOF) analyzer (Applied Biosystems). The data were analyzed with the Turbo SEQUEST program in the BioWorks 3.0 software suite.
Isolation of φZL12 DNA and sequencing of a free end.
The complete plate lysate was prepared as described above. The pellet was digested with RNase (50 μg/ml) at 37°C for 20 min, then mixed with SDS solution (0.1 M Tris-HCl, pH 9.6, 0.05 M EDTA, pH 7.5, 0.5% SDS), and kept at 70°C for 30 min. Potassium acetate (1.5 M final concentration) was added, and after 15 min at 4°C, the mixture was centrifuged at 12,000 rpm for 10 min to collect the supernatant. After extraction with phenol-chloroform, precipitation with isopropanol, and washing with 70% ethanol, DNA was dissolved in TE buffer. φZL12 DNA was digested with ClaI, and a ca. 8-kb band was recovered from the gel. After treatment with T4 DNA polymerase, the 8-kb DNA was cloned in pBluescript II SK (EcoRV, blunt end) and sequenced with T7 and T3 primers.
“Lysogenization” by φZL12.
About 108 9R-2X spores were plated on R2YE medium, dried, and overlaid (marked) with 20 μl phage suspension (109 PFU/ml). After 5 days at 30°C, spores were collected from the marked area, washed twice with 25 mM sodium pyrophosphate, diluted in water, and plated on R2YE medium at 30°C before incubation for 4 days. About 100 colonies were observed after 104× dilution.
The supplemental material for this article consists of Table S1, Fig. S1, and Fig. S2.
Nucleotide sequence accession number.
The complete nucleotide sequence of pZL12 was deposited in GenBank under accession number GQ919031.
RESULTS AND DISCUSSION
Abundance of large circular plasmids among endophytic Streptomyces strains.
We isolated 560 endophytic actinomycetes strains from 87 Chinese medicinal herbs during 2005 and 2006 (unpublished data). By using a modified procedure for isolation of actinomycete large circular plasmids (see Materials and Methods), we detected 16 plasmids from 14 of 200 strains. Sequencing of their 16S rRNA genes showed that 13 plasmid-harboring strains resembled known Streptomyces species strains (>98% identity) and that 1 strain was a Mycobacterium sp. strain. Among 14 Streptomyces plasmids (Table 2), 9 were large (85 kb to 120 kb) (see Fig. S1 in the supplemental material), while 5 others were small (6.5 kb to 20 kb). Thus, large plasmids are frequent among the endophytic Streptomyces strains. By using this modified procedure, we isolated and detected pSV1 (∼163 kb) in an agarose gel (unpublished data).
TABLE 2.
Newly detected circular plasmids among endophytic Streptomyces strains
| Strain | Plant source | Size (kb) of plasmid detected (name) |
|---|---|---|
| 14R-10 | Saxifraga stolonifera | 120 (pZL1) |
| 14R-3-2 | Saxifraga stolonifera | 85 (pZL2); 11 (pZL3) |
| 13R-3A | Duchesnea indica | 110 (pZL4) |
| 21R-9-1A | Sambuci chinensis | 110 (pZL5) |
| 37L-3 | Ginkgo biloba | 100 (pZL6) |
| 9L-2A | Ajuga decumbens | 95 (pZL7) |
| 9R-4C | Ajuga decumbens | 92 (pZL11) |
| 9R-2 | Ajuga decumbens | 90 (pZL12) |
| 7R-9-2 | Ranunculus kaponicus | 92 (pZL13) |
| 41S-1 | Allium sativum | 20 (pZL14) |
| H2GS-3B | Taxus chinensis | 13 (pZL8) |
| 56LL-1 | Lonicera japonica | 13 (pZL9) |
| 75R-4B | Salvia miltiorrhiza | 6.5 (pZL10) |
Recombinational cloning and sequencing of pZL12 from endophytic Streptomyces sp. 9R-2.
Although large circular plasmids could be isolated and detected on gels, preparation on a large scale for shotgun cloning and sequencing was difficult. To clone and sequence pZL12 (∼90 kb), we developed a strategy of recombination in Streptomyces and transfer by electroporation into E. coli (see Materials and Methods; see also Fig. S2 in the supplemental material). An ∼100-kb pZL12/pQX17 cointegrated plasmid (pZQ107) was obtained. After BamHI digestion and electrophoresis, most bands resembled those of BamHI-digested pZL12 (data not shown), confirming that pZL12 was cloned in E. coli. By using a similar strategy, we cloned six Streptomyces large circular plasmids in E. coli (unpublished data), suggesting that this is a useful method.
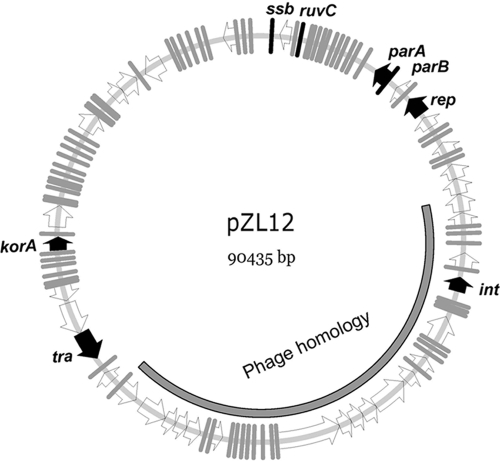
Sequencing of pZQ107 showed that pZL12 comprised 90,435 bp, with 69.5% G+C content. Of 112 open reading frames (ORFs) predicted by FramePlot 3.0 beta (see Table S1 in the supplemental material; also Fig. 1), 22 resembled genes of known function and 90 were hypothetical genes. Notably, 30 clustered genes (pZL12.39c-pZL12.68c) with the same transcription direction resembled known or hypothetical phage genes, including phage tail, capsid, and portal genes (e.g., pZL12.47c, pZL12.51c, pZL12.63c, and pZL12.66c). Another large putative operon in pZL12 contained 41 genes (pZL12.1c-pZL12.30 and pZL12.102c-pZL12.112c), some of which encoded Ssb (single-stranded DNA-binding proteins), ParAB, and RuvC, presumably having roles in inheritance and recombination. No genes in pZL12 resembled any known to be involved in replication. pZL12.71c, located within a gene cluster (pZL12.70c-pZL12.79c), encodes a cell division FtsK/SpoIIIE-like protein and is presumptively involved in DNA translocation or transfer.
FIG. 1.
Schematic map of pZL12. Predicted ORFs and their transcriptional directions are indicated by open arrowheads and some relevant genes mentioned in the text by filled arrowheads. A phage homology region in pZL12 is indicated.
Identification of a replication locus in pZL12.
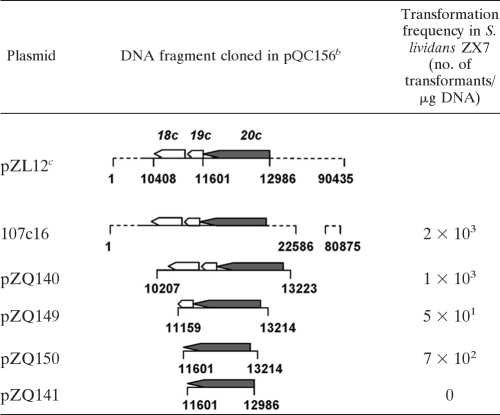
We constructed a cosmid library for pZL12 and identified a cosmid, 107c16, capable of propagation in S. lividans ZX7 (see Materials and Methods). Cosmid 107c16 was subcloned in pQC156 containing tsr and melC selection markers (30). As shown in Table 3, plasmids pZQ140, pZQ149, and pZQ150 containing only pZL12.20c (designated repA) and a 230-bp upstream sequence were able to propagate in ZX7, but deletion of the 230-bp sequence (pZQ141) abolished propagation.
TABLE 3.
Identification of a pZL12 locus for replication in Streptomycesa
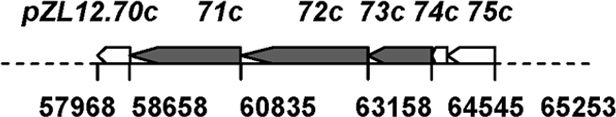
Plasmids were constructed in E. coli (see Materials and Methods) and introduced by transformation into ZX7. The positions of these cloned fragments on pZL12 and the transformation frequencies are shown. Relevant genes are indicated by open arrowheads, and a replication gene is indicated by filled arrowheads.
18c, 19c, and 20c represent ORFs pZL12.18c, pZL12.19c, and pZL12.20c, respectively.
pZL12 comprises 90,435 bp.
Identification of three genes for conjugal transfer of pZL12.
To investigate if pZL12.71c (encoding a cell division FtsK/SpoIIIE-like protein) and its adjacent genes might mediate plasmid transfer, various fragments were cloned in pQC578, which contains a replication origin of pSLA2, rlrA, and rorA genes for stable inheritance and the tsr selection marker (30). The resulting plasmids were introduced by transformation into ZX7. About equal numbers of spores of ZX7 containing these plasmids and ZX7 containing a chromosome-integrated pSET152, conferring apramycin resistance (5), were mixed in MS medium at 30°C for 4 days. As shown in Table 4, plasmids (pZQ155 and pZQ157) containing pZL12.71c-pZL12.73c (designated traA, traB, and traC, respectively) transferred at high frequencies between the two ZX7 strains. Deletion of traA, traC, or traB and traC (in pZQ162, pZQ159, or pZQ158, respectively) dramatically decreased transfer frequency. This indicated that traA, traB, and traC of pZL12 were major transfer genes for plasmid transfer.
TABLE 4.
Identification of a pZL12 locus for conjugal transfer in Streptomycesa
 |
Plasmids were constructed in E. coli and introduced by transformation into strain ZX7, and about equal amounts of transformed spores were mated with ZX7 containing pSET152 (see Materials and Methods). Spores were harvested, diluted, and plated on MS medium containing apramycin, thiostrepton, and apramycin/thiostrepton. The percent frequency of conjugal transfer was determined as the ratio of Thior Aprr colonies to Aprr colonies, multiplied by 100. Relevant genes are indicated by open arrowheads and conjugal transfer genes by filled arrowheads.
Formation of plaques on 9R-2 cured of pZL12 but not on 18 other Streptomyces strains.
As shown in Table S1 in the supplemental material, 19 genes of pZL12 resembled known or hypothetical genes of other bacteriophages. To investigate if pZL12 also functioned as a phage, pZL12 was first cured from 9R-2 to obtain 9R-2X (see Materials and Methods). An approximately 250-μl solution of 9R-2 culture containing partially purified phage particles was employed to infect 9R-2X (see Materials and Methods). About 80 plaques were observed in DNB medium containing the supernatant after being overlaid with soft nutrient agar containing 9R-2X spores, but no plaques arose with 9R-2. Soak-outs from the plaques were used to reinfect 9R-2X. As shown in Fig. 2A, turbid plaques were observed. Phage particles were further confirmed by transmission electron microscopy. As shown in Fig. 2B, phage particles had a more or less spherical (presumably icosahedral) head and a long, flexible, noncontractile tail. Thus, a phage (designated φZL12) was released into liquid culture of 9R-2 at a low frequency and was able to form turbid plaques on 9R-2 cured of pZL12.
FIG. 2.
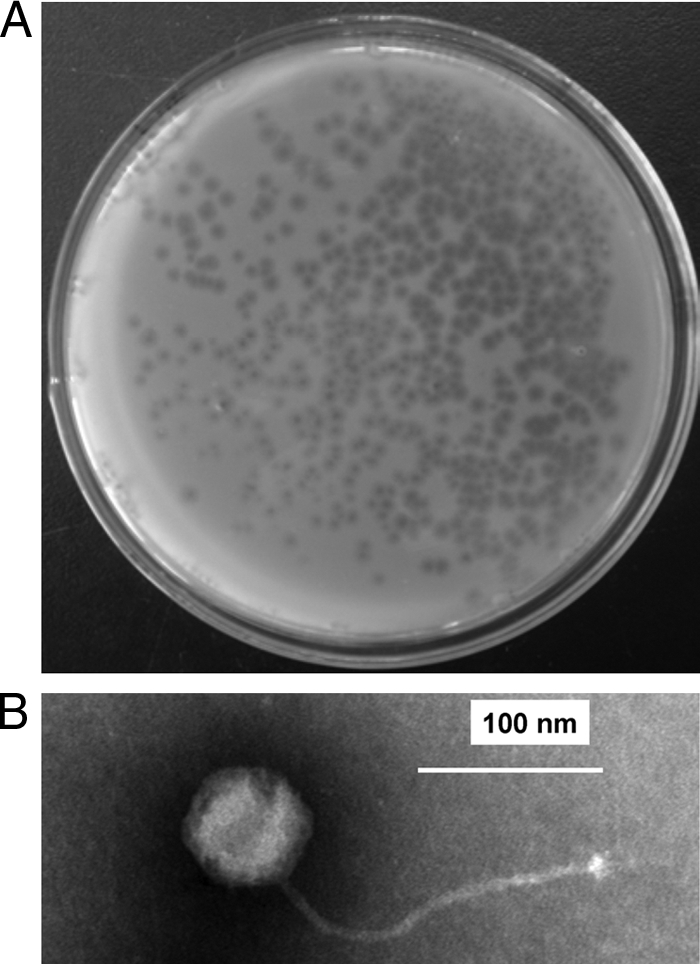
Plaques on a plate and a phage particle viewed by transmission electron microscopy. (A) Formation of plaques. Lytic φZL12 virions on a DNB plate were overlaid with soft nutrient agar containing 9R-2X spores. (B) Phage particle viewed by transmission electron microscopy. Bar, 100 nm.
To determine the host range of phage φZL12, 18 Streptomyces strains (Streptomyces coelicolor M145, S. lividans ZX7, S. violaceoruber SANK95570, S. venezuelae ISP5230, S. glaucescens GLA 4-26, S. avermitilis MMR630, S. hygroscopicus 5008, and 11 randomly selected endophytic Streptomyces strains) were examined. All these strains were resistant to infection by φZL12, although both high (109 PFU per ml) and low (103 PFU per ml) titers of φZL12 were used. Thus, φZL12 had a very narrow host range; only 9R-2X was infected and formed turbid plaques. Nevertheless, the fact that no plaque was observed does not mean that the phage does not infect the host. Perhaps in some particular host, it can only exist mainly in the lysogenic state, with very little phage produced from the lytic cycle (and thus no plaque).
Determination of φZL12 structural proteins.
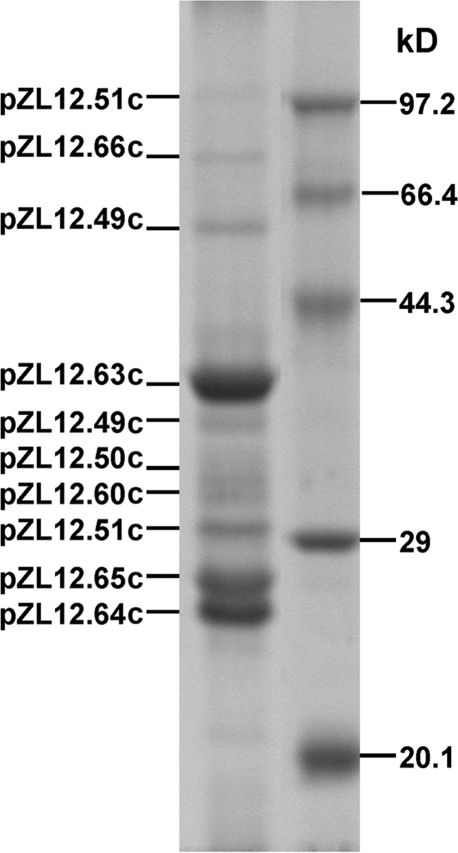
To understand the composition of phage φZL12 structural proteins, we isolated total proteins from phage particles, recovered bands from a denatured SDS-polyacrylamide gel, and employed MALDI-TOF/TOF mass spectrometry to determine each protein (see Materials and Methods). As shown in Fig. 3, 10 φZL12 proteins were confirmed from pZL12 genes (within a large operon of 30 genes, pZL12.39c-pZL12.68c), and two proteins (pZL12.49C and pZL12.51c) were present twice on gel. The most abundant φZL12 structural protein was the major capsid protein (pZL12.63c), and the second and third most abundant were pZL12.64c and pZL12.65c, respectively, both of unknown function. Phage tail (pZL12.51c) and portal (pZL12.66c) proteins were the two largest φZL12 structural proteins. Thus, major structural proteins of φZL12 virions (capsid, portal, and tail) are encoded by pZL12 genes.
FIG. 3.
Determination of φZL12 proteins on a gel. Phage φZL12 proteins were prepared and electrophoresed in a 15% SDS-polyacrylamide gel at 100 V for 1.5 h. Each band was determined by using a model 4800 MALDI-TOF/TOF analyzer (see Materials and Methods) and corresponded to the pZL12 genes shown.
Characteristics of φZL12 DNA.
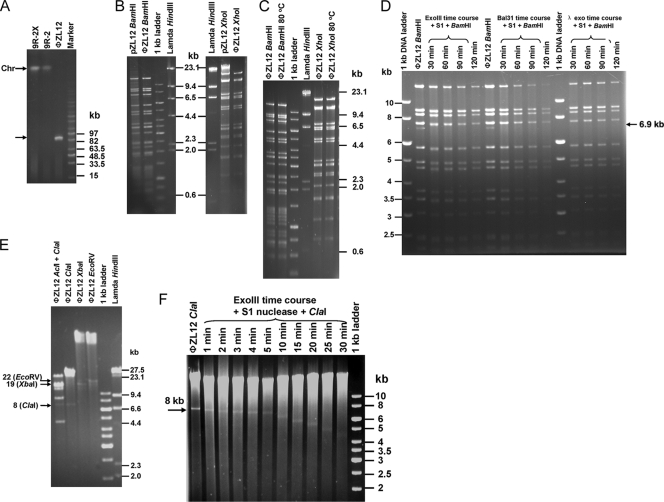
ΦZL12 DNA was isolated from soft nutrient agar containing 9R-2X spores infected with φZL12 (see Materials and Methods) and electrophoresed in a pulsed-field gel. As shown in Fig. 4A, an ∼90-kb band of φZL12 was observed, while the 90-kb circular plasmid pZL12 of strain 9R-2 could not enter the gel during electrophoresis. φZL12 DNA was digested completely with BamHI and XhoI and showed (Fig. 4B) that the expected sizes of bands of circular plasmid pZL12 were present on gel (except of some faint bands), suggesting that the φZL12 sequence was indeed from pZL12. Unlike for λ-digested DNA, the patterns of the digestion bands of φZL12 DNA were not changed on gel after heat treatment (80°C for 10 min) (Fig. 4C), suggesting that blunt or very short cohesive ends were in the linear φZL12 DNA. To further characterize the ends of the linear φZL12 DNA, we used double-stranded DNA end-specific enzymes E. coli exonuclease III (which digests duplex DNA in a 3′ to 5′ direction from a nick, a blunt end, or 3′-recessed end), λ exonuclease (which digests one strand of a DNA duplex from a 5′ phosphorylated end), and Bal31 nuclease (degrades both 3′ and 5′ termini of duplex DNA) and single-stranded DNA end-specific enzymes Bal31 nuclease and S1 nuclease. Aliquots of φZL12 DNAs were treated with E. coli exonuclease III, λ exonuclease, or Bal31 nuclease in a time course experiment and then with S1 nuclease and completely digested with BamHI or XhoI. As shown in Fig. 4D, all BamHI-digested bands were sensitive to the treatments with E. coli exonuclease III, λ exonuclease, and Bal31 nuclease, suggesting that they were equally acting as ends of the linear DNA. Similar results were obtained for the XhoI-digested bands (data not shown). Thus, like bacteriophage P1 (39), linear φZL12 DNA contains ends from largely random plasmid pZL12 sequences.
FIG. 4.
Characterization of φZL12 DNA. (A) Detection of genomic DNA by pulsed-field gel electrophoresis. Plug-embedded φZL12 DNA and mycelia of 9R-2 and 9R-2X were electrophoresed in a 1.0% agarose gel at 120 V, with a 10-s to 60-s switch time, at 14°C for 20 h. Linear chromosomes and linear φZL12 are indicated by arrowheads. (B) Plasmid pZL12 and phage φZL12 DNA were digested with BamHI and XhoI and electrophoresed in a 0.6% agarose gel at 60 V for 10 h. The λ HindIII and 1-kb ladder are markers. (C) φZL12 DNA was digested with BamHI and XhoI and was heated at 80°C for 10 min before being electrophoresed in a 0.6% agarose gel at 20 V for 20 h. (D) Aliquots of φZL12 DNAs (ca. 1 μg) were treated with 100 units of E. coli exonuclease III, 2 units of Bal31 nuclease, and 5 units of bacteriophage λ exonuclease in a time course experience, followed by treatment with 120 units of S1 nuclease at room temperature for 30 min. After ethanol precipitation, DNA was dissolved in TE buffer, followed by addition of BamHI for digestion and electrophoresis in a 0.6% agarose gel at 50 V for 15 h. (E) Detection of digested DNA by pulsed-field gel electrophoresis. φZL12 DNA was digested with AclI plus ClaI, ClaI, EcoRV, and XbaI and electrophoresed in a 1.0% agarose gel at 120 V, with a 10-s to 60-s switch time, at 14°C for 10 h. Relevant DNA bands are indicated by arrowheads. (F) Aliquots of φZL12 DNAs were treated with E. coli exonuclease III in a time course experiment, followed by addition of S1 nuclease. After ethanol precipitation, DNA was dissolved in TE buffer, followed by addition of ClaI for digestion and electrophoresis in a 0.6% agarose gel at 50 V for 10 h.
As shown in Fig. 4D, the appearance of a faint 6.9-kb BamHI band of φZL12 DNA was not expected from the plasmid pZL12 sequence. Similarly, besides the expected sizes of bands digested with XbaI (one site; 90.4 kb), EcoRV (one site), and ClaI (four sites; 31.6, 30, 28, and 0.6 kb), additional faint (e.g., a 5-kb band of ClaI-AclI digestion as a reference) bands (ca. 19, 22, and 8 kb for XbaI, EcoRV, and ClaI, respectively) were also detected on gel (Fig. 4E). φZL12 DNA was digested with E. coli exonuclease III in a time course experiment, and single-stranded DNA was removed with S1 nuclease and then digested with ClaI. As shown in Fig. 4F, this 8-kb band was sensitive to treatment with E. coli exonuclease III, suggesting that this band was not a contaminant but acted as a free end of φZL12 DNA. Cloning and sequencing of the 8-kb DNA in pBluescript II SK showed that one end was at bp 2679 of the pZL12 sequence. This “breakage end” in linear φZL12 DNA was also consistent with the appearance of additional, ca. 19- and 22-kb faint bands for XbaI (90.4, 71.9, and 18.5 kb) and EcoRV (90.4, 68.8, and 21.6 kb) on gel. Thus, besides ends of linear φZL12 from a random pZL12 sequence, there is also a hot end sequence in linear φZL12.
Cycle of autonomous plasmid pZL12 and lytic phage φZL12.
pZL12 was able to propagate as both an autonomous plasmid, pZL12, in 9R-2 and a plaque-forming phage, φZL12, in 9R-2X. To investigate how a lytic phage became an autonomous plasmid, we tried to obtain “lysogenic φZL12” in 9R-2X (see Materials and Methods). After 9R-2X was rescued from lysis with φZL12 virions and allowed to sporulate in R2YE medium, 10 colonies were randomly selected. Circular pZL12 was obtained from all 10 colonies, and no integrated copy of pZL12 was detected in the host chromosome by Southern hybridization with a φZL12 probe (data not shown). Thus, phage φZL12 is able to directly generate an autonomously circular plasmid, pZL12, again, completing a cycle of plasmid and phage.
In summary, φZL12 particles are released at a low frequency from 9R-2 liquid culture, and plaques are formed on 9R-2 cured of pZL12. 9R-2X spores are susceptible to efficient lysis by φZL12 virions, while some 9R-2X spores rescued from lysis contain a circular pZL12 plasmid obtained from largely random linearization of φZL12. Different frequencies of these states indicate that there is a regulatory mechanism for controlling a cycle of pZL12 and φZL12.
Comparisons with other bacterial phage-plasmids.
pZL12-φZL12 is the first example of a plasmid-phage in Streptomyces. Plasmids functioning as phages have also been reported to occur in other bacteria. Bacteriophage P1 infects and lysogenizes E. coli and several other enteric bacterial species. It lysogenizes as a circular, low-copy-number plasmid. Infective P1 virions contain a linear DNA with a terminal redundancy of 10 to 15 kb (21). The P1 package starts at the pac site and then undergoes a processive headful mechanism (39). The lysogenic stage and the package character of Streptomyces phage φZL12 may resemble those of P1. The plasmid prophage N15 contains a linear double-stranded DNA with covalently closed ends (telomeres). A unique mechanism for telomere resolution by protelomerase is required for conversion of circular phage DNA to linear plasmid (31). φZL12 DNA was sensitive to E. coli exonuclease III and λ exonuclease, indicating a linear DNA with free 3′ and 5′ ends, probably in a blunt or 5′-protruded structure.
Evolutionary implications.
In this work, we have identified the loci for replication, transfer, and phage formation on plasmid pZL12. Where do these functional components originate from? The repA gene of pZL12 resembles SCP1.85c and SCP1.158 of Streptomyces large linear plasmid SCP1 (3), which were experimentally confirmed as replication loci (our unpublished data). However, a few genes of pZL12 (e.g., pZL12.80) resemble a gene of SCP1 (e.g., SCP1.91c). The repA gene of pZL12 also resembles pCQ3.83 of the Streptomyces large circular plasmid pCQ3 (85,518 bp; GenBank accession number GQ983381) of the endophytic Streptomyces sp. W9 and some other Streptomyces chromosomal genes. The major transfer gene traA of pZL12 resembles a gene of Frankia sp. strain CcI3 containing a cell division FtsK/SpoIIIE domain. The traB and traC genes of pZL12 display no significant homology with other Streptomyces or actinomycete genes. These results suggest that the replication and transfer genes of pZL12 evolved independently from different sources (e.g., the chromosome or the linear or circular plasmids).
As for a phage component on pZL12, three major phage structural proteins (capsid, portal, and tail) encoded by pZL12 genes (pZL12.63c, pZL12.66c, and pZL12.51c) highly resemble the genes (pCQ3.40, pCQ3.37, and pCQ3.51) of plasmid pCQ3 and the genes (ShygA5_49102, ShygA5_49087, and ShygA5_48642) of S. hygroscopicus ATCC 53653. The liquid culture of a pCQ3-harboring strain infected the strain cured of pCQ3 and formed turbid plaques on plates (our unpublished data). These results suggest that the phage formation loci of pZL12 are closely related to those of pCQ3. The pZL12-harboring strain Streptomyces sp. 9R-2 was isolated from the herb Ajuga decumbens, while the pCQ3-harboring strain Streptomyces sp. W9 was from Artemisia annua.
Taken together, these results imply that the formation and evolution of pZL12 are very complicated and that recombinations between the chromosome and the circular and linear plasmids and phage, and even a host plant, may be involved in the process.
Supplementary Material
Acknowledgments
We are very grateful to David Hopwood and Keith Chater for critical reading of and useful comments on the manuscript. We thank Keith Chater, Yuezhong Li, and Xiaoming Ding for suggestions on phage experiments.
These investigations were supported by grants from the National Nature Science Foundation of China (30770045 and 30870067), the National “863” project (2007AA021503), and the Chinese Academy of Sciences project (KSCX2-YW-G-069) to Z. Qin.
Footnotes
Published ahead of print on 14 May 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aguilar, A., and D. A. Hopwood. 1982. Determination of methylenomycin A synthesis by the pSV1 plasmid from Streptomyces violaceus-ruber SANK 95570. J. Gen. Microbiol. 128:1893-1901. [DOI] [PubMed] [Google Scholar]
- 2.Bai, L., L. Li, H. Xu, K. Minagawa, Y. Yu, Y. Zhang, X. Zhou, H. G. Floss, T. Mahmud, and Z. Deng. 2006. Functional analysis of the validamycin biosynthetic gene cluster and engineered production of validoxylamine A. Chem. Biol. 13:387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley, S. D., S. Brown, L. D. Murphy, D. E. Harris, M. A. Quail, J. Parkhill, B. G. Barrell, J. R. McCormick, R. I. Santamaria, R. Losick, M. Yamasaki, H. Kinashi, C. W. Chen, G. Chandra, D. Jakimowicz, H. M. Kieser, T. Kieser, and K. F. Chater. 2004. SCP1, a 356,023 bp linear plasmid adapted to the ecology and developmental biology of its host, Streptomyces coelicolor A3(2). Mol. Microbiol. 51:1615-1628. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, M. J., J. M. Ward, T. Kieser, S. N. Cohen, and D. A. Hopwood. 1981. Excision of chromosomal DNA sequences from Streptomyces coelicolor forms a novel family of plasmids detectable in Streptomyces lividans. Mol. Gen. Genet. 184:230-240. [DOI] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Burke, J., D. Schneider, and J. Westpheling. 2001. Generalized transduction in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U. S. A. 98:6289-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, L., Z. Qiu, X. Dai, H. Tan, Y. Lin, and S. Zhou. 2004. Isolation of endophytic actinomycetes from roots and leaves of banana (Musa acuminata) plants and their activities against Fusarium oxysporum f. sp. cubense. World J. Microbiol. Biotechnol. 20:501-504. [Google Scholar]
- 8.Chater, K. F. 1986. Streptomyces phages and their application to Streptomyces genetics, p. 119-158. In S. W. Queener and L. E. Day (ed.), The Bacteria, vol. 9. Academic Press, Orlando, FL. [Google Scholar]
- 9.Farkasovská, J., L. Klucar, C. Vlcek, J. Kokavec, and A. Godány. 2007. Complete genome sequence and analysis of the Streptomyces aureofaciens phage mu1/6. Folia Microbiol. (Praha) 52:347-358. [DOI] [PubMed] [Google Scholar]
- 10.Fong, R., J. A. Vroom, Z. Hu, C. R. Hutchinson, J. Huang, S. N. Cohen, and C. Kao. 2007. Characterization of a large, stable, high-copy-number Streptomyces plasmid that requires stability and transfer functions for heterologous polyketide overproduction. Appl. Environ. Microbiol. 73:1296-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foor, F., G. P. Roberts, N. Morin, L. Snyder, M. Hwang, P. H. Gibbons, M. J. Paradiso, R. L. Stotish, C. L. Ruby, B. Wolanski, et al. 1985. Isolation and characterization of the Streptomyces cattleya temperate phage TG1. Gene 39:11-16. [DOI] [PubMed] [Google Scholar]
- 12.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haug, I., A. Weissenborn, D. Brolle, S. Bentley, T. Kieser, and J. Altenbuchner. 2003. Streptomyces coelicolor A3(2) plasmid SCP2*: deductions from the complete sequence. Microbiology 149:505-513. [DOI] [PubMed] [Google Scholar]
- 14.Hopwood, D. A., and T. Kieser. 1993. Conjugative plasmids of Streptomyces, p. 293-311. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, NY.
- 15.Hopwood, D. A. 2006. Soil to genomics: the Streptomyces chromosome. Annu. Rev. Genet. 40:1-23. [DOI] [PubMed] [Google Scholar]
- 16.Katz, E., C. J. Thompson, and D. A. Hopwood. 1983. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J. Gen. Microbiol. 129:2703-2714. [DOI] [PubMed] [Google Scholar]
- 17.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12(1):19-36. [DOI] [PubMed] [Google Scholar]
- 18.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 19.Kuhn, S. P., J. S. Lampel, and W. R. Strohl. 1987. Isolation and characterization of a temperate bacteriophage from Streptomyces galilaeus. Appl. Environ. Microbiol. 53:2708-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 21.Łobocka, M. B., D. J. Rose, G. Plunkett III, M. Rusin, A. Samojedny, H. Lehnherr, M. B. Yarmolinsky, and F. R. Blattner. 2004. Genome of bacteriophage P1. J. Bacteriol. 186:7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomovskaya, N. D., N. M. Mkrtumian, N. L. Gostimskaya, and V. N. Danilenko. 1972. Characterization of temperate actinophage phi C31 isolated from Streptomyces coelicolor A3(2). J. Virol. 9:258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomovskaya, N. D., K. F. Chater, and N. M. Mkrtumian. 1980. Genetics and molecular biology of Streptomyces bacteriophages. Microbiol. Rev. 44:206-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McHenney, M. A., and R. H. Baltz. 1988. Transduction of plasmid DNA in Streptomyces spp. and related genera by bacteriophage FP43. J. Bacteriol. 170:2276-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motamedi, H., E. Wendt-Pienkowski, and C. R. Hutchinson. 1986. Isolation of tetracenomycin C-nonproducing Streptomyces glaucescens mutants. J. Bacteriol. 167:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogata, S., H. Suenaga, and S. Hayashida. 1985. A temperate phage of Streptomyces azureus. Appl. Environ. Microbiol. 49:201-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omer, C. A., and S. N. Cohen. 1984. Plasmid formation in Streptomyces: excision and integration of the SLP1 replicon at a specific chromosomal site. Mol. Gen. Genet. 196:429-438. [DOI] [PubMed] [Google Scholar]
- 28.Pernodet, J. L., J. M. Simonet, and M. Guérineau. 1984. Plasmids in different strains of Streptomyces ambofaciens: free and integrated form of plasmid pSAM2. Mol. Gen. Genet. 198:35-41. [DOI] [PubMed] [Google Scholar]
- 29.Pettis, G. S., and S. N. Cohen. 1994. Transfer of the plJ101 plasmid in Streptomyces lividans requires a cis-acting function dispensable for chromosomal gene transfer. Mol. Microbiol. 13:955-964. [DOI] [PubMed] [Google Scholar]
- 30.Qin, Z., M. Shen, and S. N. Cohen. 2003. Identification and characterization of a pSLA2 plasmid locus required for linear DNA replication and circular plasmid stable inheritance in Streptomyces lividans. J. Bacteriol. 185:6575-6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravin, N. V. 2003. Mechanisms of replication and telomere resolution of the linear plasmid prophage N15. FEMS Microbiol. Lett. 221:1-6. [DOI] [PubMed] [Google Scholar]
- 32.Reuther, J., C. Gekeler, Y. Tiffert, W. Wohlleben, and G. Muth. 2006. Unique conjugation mechanism in mycelial streptomycetes: a DNA-binding ATPase translocates unprocessed plasmid DNA at the hyphal tip. Mol. Microbiol. 61:436-446. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schneider, J., F. Korn-Wendisch, and H. J. Kutzner. 1990. Phi SC623, a temperate actinophage of Streptomyces coelicolor Müller, and its relatives phi SC347 and phi SC681. J. Gen. Microbiol. 136:767-772. [DOI] [PubMed] [Google Scholar]
- 35.Smith, M. C., R. N. Burns, S. E. Wilson, and M. A. Gregory. 1999. The complete genome sequence of the Streptomyces temperate phage straight φC31: evolutionary relationships to other viruses. Nucleic Acids Res. 27:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia, H., J. Huang, M. Hu, M. Shen, P. Xie, L., Zhang, H. Wang, and Z. Qin. 2009. Construction of an ordered cosmid library of S. avermitilis for genetic modification of the industrial strains. Chin. J. Antibiot. 34:340-343. [Google Scholar]
- 37.Yamasaki, M., Y. Ikuto, A. Ohira, K. Chater, and H. Kinashi. 2003. Limited regions of homology between linear and circular plasmids encoding methylenomycin biosynthesis in two independently isolated streptomycetes. Microbiology 149(5):1351-1356. [DOI] [PubMed] [Google Scholar]
- 38.Yang, K., L. Han, J. He, L. Wang, and L. C. Vining. 2001. A repressor-response regulator gene pair controlling jadomycin B production in Streptomyces venezuelae ISP5230. Gene 279:165-173. [DOI] [PubMed] [Google Scholar]
- 39.Yarmolinsky, M. B., and N. Strernberg. 1988. Bacteriophage P1, p. 291-438. In R. Calender (ed.), The bacteriophages, vol. 1. Plenum Publishing Corporation, New York, NY. [Google Scholar]
- 40.Zhang, R., A. Zeng, P. Fang, and Z. Qin. 2008. Characterization of the replication and conjugation loci of Streptomyces circular plasmids pFP11 and pFP1 and their ability to propagate in linear mode with artificially attached telomeres. Appl. Environ. Microbiol. 74:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, X., Z. Deng, J. L. Firmin, D. A. Hopwood, and T. Kieser. 1988. Site-specific degradation of Streptomyces lividans DNA during electrophoresis in buffers contaminated with ferrous iron. Nucleic Acids Res. 16:4341-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.