Abstract
RpoS, Escherichia coli's general stress response sigma factor, regulates error-prone DNA polymerase IV (Pol IV) (encoded by the dinB gene). Pol IV is induced in stationary-phase cells, and thereafter, levels of the protein remain elevated for several days of continuous incubation. This induction and persistence in stationary-phase cells are dependent on RpoS. Data presented here show that this regulation is direct via the RpoS-directed transcription of the dinB gene. However, a loss of RpoS also results in a decrease in Pol IV-dependent mutation when Pol IV is overexpressed from an RpoS-independent promoter in exponentially growing cells. The loss of RpoS also increases cell sensitivity to 4-nitroquinoline-1-oxide, indicating that RpoS affects the ability of Pol IV to bypass DNA lesions. Thus, in addition to directly driving the transcription of the dinB gene in stationary-phase cells, RpoS regulates the activity of Pol IV in exponentially growing cells via a second, indirect pathway.
In their natural environments bacteria are constantly assaulted by a variety of stresses that include nutritional deprivation, temperature fluctuation, desiccation, and exposure to DNA-damaging agents such as UV light and reactive oxygen species. To survive such challenges, bacteria have evolved coordinated cellular defense mechanisms, two of which are the RpoS-regulated response to nutritional deprivation and the SOS response to DNA damage.
Sigma factors are the subunits of RNA polymerase that recognize the promoter regions of genes and direct the holoenzyme to them. Escherichia coli has seven sigma factors, each of which is responsible for gene regulation under specific conditions. The vegetative sigma factor RpoD (also called σD and σ70) directs the RNA polymerase holoenzyme (EσD) to transcribe housekeeping genes and the genes specifically expressed during the exponential phase. A second major sigma factor, RpoS (also called σS and σ38), regulates the cell's response to nutritional deprivation. RpoS is activated as cells enter stationary phase and is responsible, either directly or indirectly, for the regulation of as many as 500 genes (33, 40, 41). Because of their similar recognition sequences, RpoS and RpoD often direct the RNA polymerase holoenzyme to bind to the same promoters (17). However, promoter binding by the RpoS-containing RNA polymerase holoenzyme (EσS) is enhanced in stationary phase by the accumulation of accessory factors, such as ppGpp, and/or by changes in DNA topology (7, 17, 23, 24).
DNA damage in E. coli and its relatives results in the upregulation of more than 40 genes (11) that include the genes encoding DNA polymerase II (Pol II), Pol IV, and Pol V (8, 19). Pol IV, encoded by dinB, and Pol V, encoded by umuDC, are members of the Y family of specialized DNA polymerases (for a review, see reference 30). These polymerases are found in organisms from all three domains of life and can replicate damaged DNA, a process called translesion synthesis. Because of its ability to bypass many types of DNA lesions, Pol V has a clear role in promoting survival after DNA damage (30). In contrast, Pol IV's role is less clear. Pol IV can bypass some spontaneous DNA lesions, such as oxidized and alkylated bases (2, 44), and it is also efficient at bypassing deoxyguanosines with adducts at the N2 position (18, 27, 45). However, compared to Pol V, Pol IV's lesion bypass ability is limited (13).
Many members of the Y family of polymerases replicate undamaged DNA with fidelities that are orders of magnitude lower than that achieved by replicative DNA polymerases (21, 39). Therefore, it is likely that these polymerases are tightly regulated to avoid unwanted mutations. Regulation could be particularly crucial for Pol IV, whose cellular levels in E. coli are high even when there is no DNA damage (20); indeed, in the absence of DNA damage, only Pol I is more abundant than Pol IV. When the DNA is damaged and the SOS response is induced, Pol IV becomes the dominant DNA polymerase (30). While the overexpression of Pol IV dramatically increases spontaneous mutation rates, a loss of Pol IV has very little effect on mutation rates in normally growing wild-type cells (20, 22, 37, 43). In contrast, mutations in stationary-phase cells (adaptive mutations) are reduced up to 80% by a loss of Pol IV (10, 28). These results suggest that normally, Pol IV activity is strictly controlled.
Although the levels of the dinB transcript are low, the levels of Pol IV increase late in the stationary phase and remain elevated for several days thereafter. This induction and this persistence are dependent on RpoS (25). RpoS-dependent regulation occurs in addition to and independently of SOS regulation, making dinB a rare example of a gene that is regulated by both LexA, the SOS repressor, and RpoS (25, 32). The simplest hypothesis to explain how RpoS regulates Pol IV is that RpoS directs RNA polymerase to the dinB promoter. An alternative hypothesis is that RpoS activates one or more intermediates that regulate Pol IV. Here we present evidence that both mechanisms are active: RpoS directly regulates Pol IV levels in stationary-phase cells and indirectly regulates Pol IV's mutagenic activity in exponentially growing cells.
MATERIALS AND METHODS
Bacterial strains and media.
All strains used in this study are E. coli K-12 derivatives and are listed in Table 1. Genetic manipulations were performed as previously described (29). Cultures were grown in either Luria broth (LB; Difco) or M9-0.1% glycerol minimal medium (29). When required, minimal medium was supplemented with 100 μg/ml proline. LB agar medium contained 15 g/liter agar; LB top agar contained 8 g/liter agar. Antibiotics were added to rich medium at the following concentrations: carbenicillin (Cb) at 100 μg/ml, chloramphenicol (Cm) at 10 μg/ml, and tetracycline (Tc) at 20 μg/ml. Except for Cm, antibiotics were added to minimal medium at half of the above-mentioned concentrations. Strains GW1030 and PFB893 were grown at 30°C in minimal medium supplemented with 25 μg/ml Cb and 100 μg/ml each alanine, histidine, leucine, proline, and threonine.
TABLE 1.
E. coli strains and plasmid used in this study
| Strain or plasmid | Relevant phenotype or genotype | Reference |
|---|---|---|
| E. coli strains | ||
| FC36 | F−ara Δ(lac-proB)XIIIthi Rifr | 4 |
| FC40 | FC36/F′ φ(lacI33-lacZ) Pro+ | 4 |
| FC722 | FC40 with a Tcs allele on the episome | 9 |
| FC1413 | FC942 sulA11 lexA71::Tn5 | 25 |
| PFG266 | FC722 with pPFG96 | 38 |
| PFG419 | FC722 rpoS::Cm | This study |
| PFG420 | PFG419 with pPFG96 | This study |
| PFB505 | FC1413 dinB-lacZ translational fusion in λ att | This study |
| PFB593 | PFB505 rpoS::Cm | This study |
| PFB691 | FC36 with pPFV323 | This study |
| GW1030 | AB1157 recA441 sulA1 ΔlacUI69 dinB1::Mud(Ap lac) | 19 |
| PFB893 | GW1030 rpoS::Cm | This study |
| Plasmids | ||
| pPFG96 | dinB in pBAD24 | 38 |
| pBAD24 | Cloning vector with a pBR322 origin and araBAD promoter | 14 |
| pPFV323 | dinB-lacZ operon fusion in pRS415 | This study |
| pRS415 | lacZ operon fusion vector | 36 |
| pMC1403 | lacZ gene fusion vector | 5 |
Two different dinB-lacZ fusions were used in β-galactosidase assays. A translational fusion was constructed by amplifying the region starting 141 bp before the dinB start codon and including 115 bp of the dinB coding sequence using the following primers: forward primer 5′-CATCAGAATTCGGGATAAAGTGGTGCAGC-3′ and reverse primer 5′-ATTATGGATCCCGACGTTCGCGGCTGCCG-3′.
The amplified DNA was digested with the BamHI and EcoRI restriction enzymes (New England Biolabs, Inc.) and ligated into BamHI- and EcoRI-digested plasmid pMC1403 (5), resulting in a gene fusion consisting of the first 38 amino acids (of 351 amino acids) of Pol IV in frame with LacZ, under the control of the dinB promoter. To create strain PFB505, this fusion was recombined into the chromosomal lambda att site by using the lambda InCh method (3) and transduced by using P1 bacteriophage in strain PFG1413 (25), selecting for low-level Cb resistance (50 μg/ml) and screening for lacZ expression. The expression of the dinB-lacZ gene fusion both on the plasmid and on the chromosome was shown to be under LexA control (P. L. Foster, unpublished data). The second fusion strain was GW1030, carrying a chromosomal dinB::Mud(Apr lac) operon fusion, the expression of which is also under LexA control (19). An rpoS::Cm allele (25) was transduced into both fusion strains by using P1 bacteriophage.
Plasmid pPFV323 was constructed by amplifying the region starting 141 bp upstream of the dinB start codon and including 201 bp of the dinB coding sequence using the forward primer described above and a reverse primer with the sequence 5′-GTCGAGGATCCGTGGGCATAATTTGAGCG-3′. The amplified insert was digested with the BamHI and EcoRI restriction enzymes (New England Biolabs, Inc.) and ligated into BamHI- and EcoRI-digested plasmid pRS415 (35), creating a dinB-lacZ operon fusion. The expression of this fusion was shown to be under LexA control (our unpublished data).
β-Galactosidase assays.
β-Galactosidase assays were performed as previously described (29). To measure β-galactosidase activity in stationary-phase cells, cultures were grown overnight in LB broth plus the appropriate antibiotics and then diluted 105-fold into four tubes containing M9-glycerol minimal medium. These cultures were then grown for 48 h to saturation, and β-galactosidase assays were performed. To measure β-galactosidase activity in exponentially growing cells, cultures were grown overnight in LB broth plus the appropriate antibiotics and then diluted 103-fold into four tubes containing M9-glycerol medium plus antibiotics. These cultures were grown to an optical density at 600 nm (OD600) of ∼0.2, diluted, and grown to an OD600 of ∼0.2 twice more. This procedure yields four independent cultures containing nearly all exponentially growing cells.
Determination of the dinB transcription start site.
Strain PFB691, which carries plasmid pPFV323 with a dinB-lacZ operon fusion, was used to determine the transcription start site of the dinB gene. The culture was grown to saturation at 37°C in LB broth plus Cb. RNA was isolated from 1 ml of the culture by using TRIzol reagent (Invitrogen Corp.) according to the manufacturer's instructions. The quality of the RNA was confirmed by electrophoresis in a 1% agarose gel and by absorbance measurements at 260 and 280 nm. 5′ rapid amplification of cDNA ends (RACE) was performed by using the 5′ RACE system, version 2.0, kit (Invitrogen Corp.). The template-specific primers, corresponding to the lacZ gene, were as follows: Gsp (5′-GTAAAACGACGGCCAGTGAA-3′), Gsp 1 (5′-GATGTGCTGCAAGGCGATTA-3′), and Gsp 2 (5′-GGGTAACGCCAGGGTTTTCC-3′).
5′ RACE was performed by using both C tailing and A tailing of the cDNA. When the cDNA was C tailed, the 5′-RACE AUAP primer was used; when the cDNA was A tailed, the 3′ RACE adapter primer with the sequence 5′-GGCCACGCGTCGACTAGTAC(T)17-3′ was used. 5′ RACE products were sequenced by using an ABI3730 DNA analyzer.
Electromobility shift assay.
A 214-bp region including the dinB promoter region was amplified by using PCR with the following primers (the forward primer was labeled with an infrared dye, IRdye 700 [Li-Cor Biosciences]): 5′-IRdye 700-ATACTTTGGTCAGCATGGGG-3′ and 5′-GGGATTGTCGCGCATCTC-3′.
To reconstitute the RNA polymerase holoenzyme, 25-μl binding mixtures included RpoS or RpoD (obtained from K. Wassarman) at a concentration of 250 nM and core RNA polymerase (Epicentre Biotechnologies) at a concentration of 60 nM. The binding buffer included 200 mM Tris (pH 8.0), 30 mM KCl, 10 mM MgCl2, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), and 20 μg/ml bovine serum albumin (BSA). This mixture was incubated at 30°C for 45 min.
Gel shift reaction mixtures containing labeled probe at a concentration of 0.2 nM and RNA polymerase holoenzyme (1 μl of the above-described mixture) in 25 μl of the binding buffer were incubated for 20 min at room temperature. A 50% binding buffer-50% glycerol solution (5 μl) was then added to each reaction mixture, and 15 μl of each reaction mixture was loaded onto a 6.5% native acrylamide gel. The gel was run for 2 h at room temperature at approximately 100 V. The gel was visualized by using an Odyssey IR scanner (Li-Cor Biosciences).
Determination of growth-dependent mutation rates.
Cultures of each strain were grown overnight to saturation in LB broth plus the appropriate antibiotics. The cultures were then diluted 105-fold into LB broth plus the appropriate antibiotics, and 0.1-ml aliquots were distributed into 45 wells of a 96-well microtiter plate, which was then incubated for 48 h at 37°C. Next, 40 cultures per strain were diluted 10−1 into 90 μl of saline and plated in LB top agar plus Tc onto plates containing LB agar plus Tc. The rest of the cultures were used for determining the total number of cells plated. Only colonies appearing on the plates of LB plus Tc after 48 h were counted. Mutation rates and confidence limits were calculated by using the Ma-Sandri-Sarkar maximum likelihood estimator (MSS-MLE) maximum likelihood method (35), implemented by the FALCOR Web tool (found at http://www.mitochondria.org/protocols/FALCOR.html) (15). The number of cultures plated, 40, was sufficient to detect a 2-fold-or-greater change in the mutation rate with 95% significance.
Western blotting.
Western blotting was performed as previously described (1). E. coli strains were grown to an OD of ∼0.2 in LB broth plus Cb. One milliliter of each culture was centrifuged, and the pelleted cells were washed in 1× M9 salts, resuspended in 1× SDS-PAGE sample loading buffer, and boiled for 15 min. The total amount of protein in the samples was determined by Bradford assays (Bio-Rad Laboratories). Samples containing the amounts of total protein noted in the legend to Fig. 5 were diluted into 1× sample loading buffer containing bromophenol blue and loaded onto a 12% SDS polyacrylamide gel. Proteins were separated by electrophoresis and then electrotransferred onto an Immobilon-P membrane (pore size, 0.45 μm; Millipore Corp.). The membranes were probed with rabbit anti-Pol IV polyclonal antibody [obtained from H. Ohmori and clarified using acetone powder made from a Δ(dinB) strain according to a procedure described previously (16)]; bands were visualized by using alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Promega, Inc.) and Western-Light chemiluminescent reagent (Applied Biosystems). Bands were quantified by using ImageJ, version 1.36b, software (34).
FIG. 5.
RpoS does not affect the amount of Pol IV in strains overexpressing Pol IV from an exogenous promoter. (A) Western blot of total cell protein probed with anti-Pol IV antibody. For each pair of strains, the indicated amounts of protein were loaded onto the polyacrylamide gel. The strains overexpressing Pol IV were harvested in the exponential phase (see Materials and Methods), whereas the three control strains on the right were harvested in the stationary phase. (B) Quantification of the bands in A for the strains overexpressing Pol IV. The lighter bars correspond to the rpoS::Cm mutant strain, and the darker bars represent the wild-type strain. Wild type, FC722; PFG419, rpoS::Cm; wild type/pdinB++, PFG266; rpoS::Cm/pdinB++, PFG420.
Assays of sensitivity to 4-NQO.
Cultures of each strain were grown overnight in LB broth plus the appropriate drugs. Each culture was diluted 10-fold successively from 10−1 to 10−6, and 0.01 ml of each dilution was spotted onto both an LB agar plate and an LB plate supplemented with 10 μM 4-nitroquiniline-1-oxide (4-NQO). Plates were incubated overnight at 37°C in the dark.
RESULTS
The simplest hypothesis to explain how RpoS controls Pol IV levels in stationary-phase cells is that RpoS directs RNA polymerase to transcribe the dinB gene. An alternative hypothesis is that RpoS acts indirectly by regulating one or more intermediates that then affect Pol IV. It is also possible that both hypotheses are true. To determine which of these three possibilities prevails, we examined several of their predicted consequences.
RpoS regulates Pol IV transcription directly in stationary-phase cells.
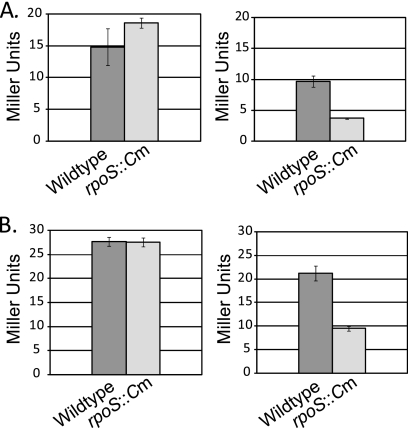
To determine if RpoS regulates the transcription of dinB in vivo, we made a translational fusion encoding the first 38 amino acids of the Pol IV protein followed by the lacZ coding sequence and recombined this fusion into the lambda att site on the chromosome. We also used the chromosomal dinB::Mud(Apr lac) transcriptional fusion described previously (19). We measured β-galactosidase activity in stationary-phase cells and exponentially growing cells of both wild-type and rpoS::Cm mutant strains containing these fusions. Because it was previously shown that levels of Pol IV increase in stationary-phase cells incubated in minimal-glycerol medium (25), the cultures to be assayed in stationary phase were grown in this medium. As shown in Fig. 1, RpoS is required for the transcription of dinB in stationary-phase cells but not in exponentially growing cells. This is true regardless of whether the cells were grown in minimal or rich medium (data not shown).
FIG. 1.
Loss of RpoS reduces expression of the dinB gene in stationary-phase cells only. β-Galactosidase assays were performed as previously described (29). The graphs on the left show the results of β-galactosidase assays of four independent isolates of each strain grown to an OD of ∼0.2 in M9 plus glycerol minimal medium. The graphs on the right show the results of β-galactosidase assays of four independent isolates of each strain grown for 48 h to saturation in M9 plus glycerol minimal medium (see Materials and Methods). Error bars are standard errors of the means (SEM). (A) Strains carried a translational fusion of the first 6 amino acids of dinB fused in frame to the lacZ gene and were grown at 37°C. Wild type, PFB505; rpoS::Cm, PFB593. (B) Strains carried the dinB::Mud(Apr lac) transcriptional fusion described previously (19) and were grown at 30°C. Wild type, GW1030; rpoS::Cm, PFB893.
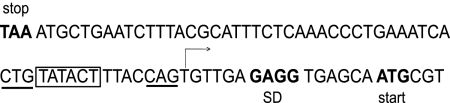
The transcription start site of dinB has been determined.
In order to test directly if EσS can transcribe dinB, we first determined the transcription start site of the gene. Because the dinB transcript is scarce (8; our unpublished results), we used a plasmid-borne operon fusion of lacZ to a DNA region that includes 141 bp upstream and 191 bp downstream of the dinB start codon. This plasmid has a high expression level. 5′ RACE using RNA isolated from stationary-phase cells identified the T shown in Fig. 2 as the transcription start site of the dinB gene. From this information, we deduced that the hexamer TATACT shown in Fig. 2 is the −10 region of the dinB promoter. This designation was confirmed by deleting the T in the center of the hexamer; this change resulted in a 5-fold decrease in β-galactosidase activity (data not shown). When 5′ RACE was repeated with RNA isolated from a strain lacking RpoS, the transcription start site was the same; thus, at least in stationary-phase cells, transcription initiates from the same position whether or not RpoS is present.
FIG. 2.
The transcription start site of dinB was determined. The transcription start site of dinB determined by 5′ RACE is indicated by an arrow, and the −10 promoter element is boxed. The previously identified LexA recognition sequences are underlined, and the dinB translation start site, a possible Shine-Dalgarno (SD) sequence, and the stop codon of the upstream gene mbhA are shown in boldface type and labeled (32).
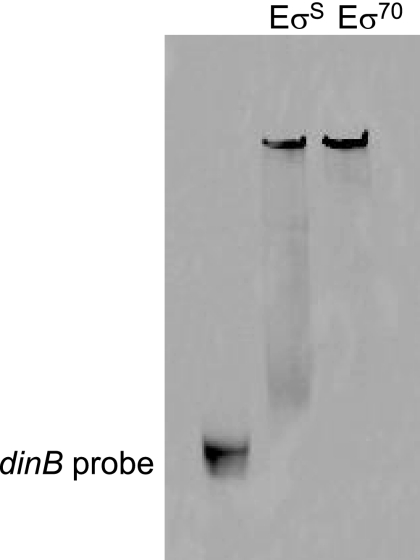
EσS binds the promoter region of the dinB gene.
An electromobility shift assay demonstrated that both EσD and EσS bind to the dinB promoter (Fig. 3). In addition, an in vitro transcription assay showed that both holoenzymes can transcribe from the dinB promoter in vitro (data not shown).
FIG. 3.
Both EσS and Eσ70 bind to the dinB promoter. A 214-bp region containing the promoter of the dinB gene and labeled with an infrared dye, IRdye 700 (Li-Cor Biosciences), was incubated with RNA polymerase holoenzyme containing either σS or σ70. The reaction mixture was electrophoresed on a nondenaturing polyacrylamide gel and visualized with an Odyssey IR scanner.
RpoS affects Pol IV mutagenic activity in exponentially growing cells when Pol IV is overexpressed from an exogenous promoter.
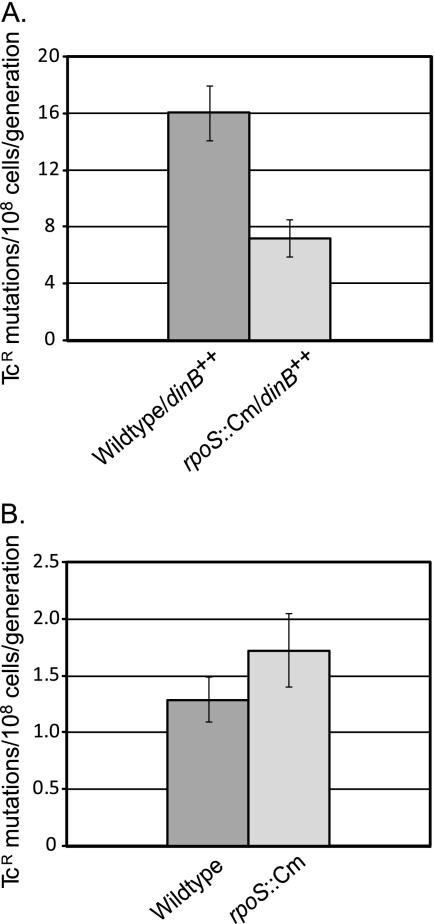
When Pol IV is overexpressed, the mutation rate in normally growing cells can be increased as much as 100-fold (21, 38). We took advantage of this result to determine whether RpoS affects Pol IV's mutagenic activity even when dinB is under the control of an RpoS-independent promoter. To measure growth-dependent mutation rates, we assayed the reversion to tetracycline resistance (Tcr) of strain FC722, which carries an episome with an allele of the tetA gene that has a +1 frameshift mutation (9). To overproduce Pol IV, we transformed this strain with plasmid pPFG96, carrying dinB under the control of the araBAD promoter; this plasmid results in the increased expression of Pol IV even in the absence of induction by arabinose (38). As shown in Fig. 4 A, when strains were carrying this plasmid, the loss of RpoS resulted in a statistically significant 2.5-fold decrease in the mutation rate to Tcr. This phenotype is clearly dependent on Pol IV, because when Pol IV was not overexpressed, the loss of RpoS did not affect the mutation rate (Fig. 4B). This result could mean either that cells lacking RpoS have less Pol IV or that Pol IV is present in a less active state. A Western blot of these strains harvested in the exponential phase indicated that the amount of Pol IV being expressed from the plasmid was not affected by the presence or absence of RpoS (Fig. 5). We used several concentrations of total protein to ensure that the film was not saturated and that the relative amount of Pol IV was clear. The Western blot shows that the loss of RpoS had no effect on the amount of Pol IV expressed from an exogenous promoter in exponentially growing cells; thus, RpoS must affect Pol IV's activity.
FIG. 4.
Loss of RpoS decreases the growth-dependent mutation rate of cells overexpressing Pol IV from an exogenous promoter but not of cells not overexpressing Pol IV. The mutation rates were determined by 40-culture fluctuation tests and were calculated by the MSS maximum likelihood method; error bars indicate 95% confidence levels (15, 35). (A) Wild type, PFG266; rpoS::Cm, PFG420. (B) Wild type, FC722; rpoS::Cm, PFG419.
Loss of RpoS increases the sensitivity of cells to 4-NQO.
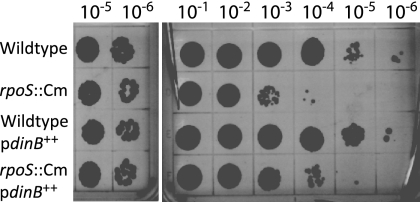
Because Pol IV can efficiently bypass the N2-dG adduct that is produced by 4-NQO, sensitivity to 4-NQO is a good assay for Pol IV's translesion activity (18). As shown in Fig. 6, the loss of RpoS increased the 4-NQO sensitivity of cells whether Pol IV was expressed from its own promoter on the chromosome or expressed at higher levels from an RpoS-independent exogenous promoter. Thus, RpoS is required for the full expression of both Pol IV's translesion activity and its mutagenic activity.
FIG. 6.
Loss of RpoS increases cell sensitivity to 4-NQO whether or not Pol IV is overexpressed from an exogenous promoter. Spot tests measuring sensitivity to 10 μM 4-NQO were performed as described in Materials and Methods; only the 10−5 and 10−6 dilutions from the untreated LB plate are shown. Wild type, FC722; rpoS::Cm, PFG419; wild-type/pdinB++, PFG266; rpoS::Cm/pdinB++, PFG420.
DISCUSSION
In wild-type cells the levels of both the Pol IV protein and its transcript increase in stationary phase (25) and thereafter remain elevated for several days (25, 46). However, in the absence of RpoS the levels of Pol IV rapidly decline in stationary-phase cells upon continued incubation (25). These results indicate that RpoS regulates Pol IV either directly, by targeting RNA polymerase to the dinB promoter, or indirectly, by regulating other factors that positively regulate Pol IV. These two hypotheses are not mutually exclusive, and the data presented here support both a direct role (in stationary-phase cells) and an indirect role (in exponentially growing cells) for RpoS.
It is not common for genes to be regulated by both RpoS and the SOS repressor LexA; indeed, only one gene (sbmC) other than dinB has been shown to have this dual regulation (31). Because of this rarity and the fact that the induction of dinB in stationary-phase cells occurs late (25), it seemed possible that the RpoS-dependent regulation of Pol IV was indirect. We looked for an RpoS-regulated intermediate by screening both an overexpression library and a knockout library for the suppression of the low-adaptive-mutation phenotype of an rpoS mutant strain. We failed to find an intermediate, suggesting that there is no single RpoS-regulated gene that itself regulates Pol IV under the conditions that pertain to adaptive mutation. Additionally, cells overexpressing dinB from the arabinose-inducible promoter araBAD have the same amount of Pol IV whether or not RpoS is present (Fig. 5). This result indicates that the regulation of Pol IV by RpoS is dependent upon the dinB promoter.
RpoS and RpoD, the vegetative sigma factor, often recognize the same promoters (17); the region 5′ to the dinB coding sequence could contain one of these dually recognized promoters. To determine if RpoS directs transcription from the dinB promoter, we needed to define the promoter elements. The location of the dinB transcription start site determined by 5′ RACE (Fig. 2) indicated that the boxed TATACT is the −10 element of the promoter. We confirmed this by showing that the deletion of the second T of the hexamer greatly diminished the transcription of dinB in vivo (data not shown). We also used 5′ RACE to show that the same transcription start site is used whether or not RpoS is present, indicating that, at least for stationary-phase cells, there is only one dinB transcript regardless of which sigma factor directs its transcription.
In our initial attempts to determine if RpoS drives Pol IV transcription in stationary-phase cells, we first fused the lacZ gene directly to the dinB promoter. However, we found that this fusion was regulated by neither RpoS nor LexA. The reason for this lack of regulation is currently under investigation, but as a result, the fusion could not be used to study dinB transcription. Therefore, we used two other fusions: (i) a translational fusion that placed LacZ in frame with the first 6 amino acids of Pol IV and (ii) the original dinB::Mud(Apr lac) transcriptional fusion that demonstrated that dinB was induced by DNA damage as part of the SOS response (19). Both of these fusions indicated that RpoS directs dinB expression in stationary-phase cells but not in exponentially growing cells (Fig. 1). To strengthen the argument that the RpoS regulation is direct, we used an electromobility shift assay to show that both EσS and EσD can bind the dinB promoter (Fig. 3). These results support the hypothesis that the transcription of dinB is driven by RpoD in exponential-phase cells and by RpoS in stationary-phase cells.
Interestingly, although we previously found that the amount of the Pol IV protein in rpoS+ cells is larger in stationary-phase cells than in exponential-phase cells (25), the level of transcription of the dinB-lacZ fusions appeared to be lower in the stationary phase than during the exponential phase. This difference indicates that the dinB mRNA may be more stable or translated more efficiently during the stationary phase, resulting in more protein. Such an additional regulation of Pol IV at the posttranscriptional level is a subject for future investigations.
In addition to driving the transcription of dinB in stationary-phase cells, our results indicate that RpoS regulates Pol IV activity in exponentially growing cells via an indirect mechanism. The overexpression of Pol IV increases the mutation rate of exponentially growing cells (21, 38). When Pol IV was overexpressed from an exogenous, RpoS-independent promoter, it seemed unlikely that RpoS would affect the mutation rate. However, this expectation was incorrect; in the absence of RpoS, the Pol IV-dependent mutation rate was decreased almost 3-fold (Fig. 4A). RpoS did not have this effect when Pol IV was not overexpressed, indicating that this RpoS phenotype is exerted via Pol IV (Fig. 4B). Western blot analysis revealed that the level of Pol IV being expressed from the exogenous promoter was not decreased by the loss of RpoS (Fig. 5), suggesting that RpoS affects Pol IV's mutagenic activity. Cells lacking RpoS are also more sensitive to 4-NQO whether or not Pol IV is overexpressed (Fig. 6); thus, RpoS also affects Pol IV's translesion synthesis activity. Since RpoS is a transcriptional regulator, this regulation is most likely via the expression of another gene or genes whose products are required for full Pol IV activity. We are currently attempting to identify this pathway.
Previous results have demonstrated that complex cellular pathways regulate both the amount and activity of Pol IV (6, 8, 12, 19, 25, 26, 38, 42). The results presented here clarify that RpoS regulates Pol IV directly by altering its amount and indirectly by altering its activity and that these regulatory modes are in effect at different times during the cell cycle. This regulation may not only promote cell survival but also provide a way for cells to accumulate advantageous mutations during nutrient limitation and under other stressful conditions.
Acknowledgments
We thank K. M. Wassarman for providing us with the purified RpoS and RpoD proteins and D. Boyd, M. J. Casadaban, J. H. Miller, R. W. Simons, and G. C. Walker for bacterial strains, bacteriophage, and plasmids. We are grateful to members of our laboratory for advice and patience and especially K. M. Hetrick for constructing pPFV323.
This work was supported by USPHS NIH grant GM065175 to P.L.F. and U.S. NSF IGERT training grant 0504627/206251A to K.A.M.S.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1988. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 2.Bjedov, I., C. N. Dasgupta, D. Slade, S. Le Blastier, M. Selva, and I. Matic. 2007. Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics 176:1431-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadaban, M. J., J. Chou, and S. N. Cohen. 1980. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 143:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, S. E., and G. C. Walker. 2010. The transcription elongation factor NusA is required for stress-induced mutagenesis in Escherichia coli. Curr. Biol. 20:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colland, F., M. Barth, R. Hengge-Aronis, and A. Kolb. 2000. Sigma factor selectivity of Escherichia coli RNA polymerase: role for CRP, IHF and Lrp transcription factors. EMBO J. 19:3028-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courcelle, J., A. Khodursky, B. Peter, P. O. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wildtype and SOS-deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster, P. L. 1997. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J. Bacteriol. 179:1550-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, P. L. 2000. Adaptive mutation in Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 65:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg, E., G. Walker, W. Siede, R. Wood, R. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis. ASM Press, Washington, DC.
- 12.Godoy, V. G., D. F. Jarosz, S. M. Simon, A. Abyzov, V. Ilyin, and G. C. Walker. 2007. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell 28:1058-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman, M. F. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71:17-50. [DOI] [PubMed] [Google Scholar]
- 14.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall, B. M., C. X. Ma, P. Liang, and K. K. Singh. 2009. Fluctuation analysis CalculatOR: a Web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25:1564-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Hengge-Aronis, R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter sigmaS-selective? Curr. Opin. Microbiol. 5:591-595. [DOI] [PubMed] [Google Scholar]
- 18.Jarosz, D. F., V. G. Godoy, J. C. Delaney, J. M. Essigmann, and G. C. Walker. 2006. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439:225-228. [DOI] [PubMed] [Google Scholar]
- 19.Kenyon, C. J., and G. C. Walker. 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 77:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, S. R., K. Matsui, M. Yamada, P. Gruz, and T. Nohmi. 2001. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266:207-215. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S. R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. U. S. A. 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuban, W., P. Jonczyk, D. Gawel, K. Malanowska, R. M. Schaaper, and I. J. Fijalkowska. 2004. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J. Bacteriol. 186:4802-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusano, S., Q. Ding, N. Fujita, and A. Ishihama. 1996. Promoter selectivity of Escherichia coli RNA polymerase holoenzymes. J. Biol. Chem. 271:1998-2004. [DOI] [PubMed] [Google Scholar]
- 24.Lacour, S., and P. Landini. 2004. SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186:7186-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Layton, J. C., and P. L. Foster. 2005. Error-prone DNA polymerase IV is regulated by the heat shock chaperone GroE in Escherichia coli. J. Bacteriol. 187:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenne-Samuel, N., R. Janel-Bintz, A. Kolbanovskiy, N. E. Geacintov, and R. P. Fuchs. 2000. The processing of a Benzo(a) pyrene adduct into a frameshift or a base substitution mutation requires a different set of genes in Escherichia coli. Mol. Microbiol. 38:299-307. [DOI] [PubMed] [Google Scholar]
- 28.McKenzie, G. J., P. L. Lee, M. J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7:571-579. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Nohmi, T. 2006. Environmental stress and lesion-bypass DNA polymerases. Annu. Rev. Microbiol. 60:231-253. [DOI] [PubMed] [Google Scholar]
- 31.Oh, T. J., I. L. Jung, and I. G. Kim. 2001. The Escherichia coli SOS gene sbmC is regulated by H-NS and RpoS during the SOS induction and stationary growth phase. Biochem. Biophys. Res. Commun. 288:1052-1058. [DOI] [PubMed] [Google Scholar]
- 32.Ohmori, H., E. Hatada, Y. Qiao, M. Tsuji, and R. Fukuda. 1995. dinP, a new gene in Escherichia coli, whose product shows similarities to Umuc and its homologs. Mutat. Res. 347:1-7. [DOI] [PubMed] [Google Scholar]
- 33.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580-591. [DOI] [PubMed] [Google Scholar]
- 34.Rasband, W. S. 1997. ImageJ. National Institutes of Health, Bethesda, MD.
- 35.Sarkar, S., W. T. Ma, and G. H. Sandri. 1992. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica 85:173-179. [DOI] [PubMed] [Google Scholar]
- 36.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 37.Strauss, B. S., R. Roberts, L. Francis, and P. Pouryazdanparast. 2000. Role of the dinB gene product in spontaneous mutation in Escherichia coli with an impaired replicative polymerase. J. Bacteriol. 182:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stumpf, J. D., and P. L. Foster. 2005. Polyphosphate kinase regulates error-prone replication by DNA polymerase IV in Escherichia coli. Mol. Microbiol. 57:751-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, M. J., P. Pham, X. Shen, J. S. Taylor, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 40.Vijayakumar, S. R., M. G. Kirchhof, C. L. Patten, and H. E. Schellhorn. 2004. RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J. Bacteriol. 186:8499-8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, A. B., and P. L. Foster. 2007. The Escherichia coli histone-like protein HU has a role in stationary phase adaptive mutation. Genetics 177:723-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolff, E., M. Kim, K. Hu, H. Yang, and J. H. Miller. 2004. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J. Bacteriol. 186:2900-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada, M., T. Nunoshiba, M. Shimizu, P. Gruz, H. Kamiya, H. Harashima, and T. Nohmi. 2006. Involvement of Y-family DNA polymerases in mutagenesis caused by oxidized nucleotides in Escherichia coli. J. Bacteriol. 188:4992-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, W., and R. Woodgate. 2007. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 104:15591-15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. U. S. A. 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]