Abstract
Poly(A) polymerase I (PAP I) is the enzyme responsible for the addition of poly(A) tails onto RNA molecules in Escherichia coli. Polyadenylation is believed to facilitate the destruction of such RNAs by the mRNA degradosome. Recently, it was discovered that the stationary-phase regulatory protein SprE (RssB) has a second function in the control of polyadenylation that is distinct from its known function in the regulated proteolysis of RpoS. In the work presented herein, we used a targeted proteomic approach to further investigate SprE's involvement in the polyadenylation pathway. Specifically, we used cryogenic cell lysis, immunopurifications on magnetic beads, and mass spectrometry to identify interacting partners of PAP I-green fluorescent protein. We provide the first in vivo evidence that PAP I interacts with the mRNA degradosome during both exponential and stationary phases and find that the degradosome can contain up to 10 different proteins under certain conditions. Moreover, we demonstrate that the majority of these PAP I interactions are formed via protein-protein interactions and that SprE plays an important role in the maintenance of the PAP I-degradosome association during stationary phase.
Polyadenylation in Escherichia coli is important for proper RNA metabolism and, therefore, proper gene expression. Poly(A) polymerase I (PAP I) catalyzes the addition of poly(A) tails onto the 3′ ends of RNA molecules, which ultimately facilitates their destruction (25, 32, 39). PAP I requires the RNA chaperone Hfq for efficient recognition of RNA substrates that contain 3′ hairpin structures but not for unstructured RNA products (35). It was recently shown that ∼90% of transcripts are polyadenylated in exponentially growing cells, demonstrating that polyadenylation is a frequent and fundamental process (33). A second enzyme involved in the polyadenylation of RNA species is polynucleotide phosphorylase (PNPase) (34). PNPase is a reversible enzyme, as it has a 3′ → 5′ exoribonuclease activity and a 5′ → 3′ template-independent poly(A) polymerase activity (19, 34, 47).
Polyadenylation of mRNAs has been shown in vitro to stimulate mRNA degradation (2). RNA degradation in vivo is not completely understood; however, one of the key players in this process is the RNA degradosome. The degradosome, a multiprotein complex involved in RNA degradation, is composed of four core components—RNase E, PNPase, RhlB, and enolase (11, 31, 42, 43). The C-terminal domain of RNase E serves as a scaffold for proteins of the degradosome (8, 9, 48). The ATP-dependent helicase RhlB unwinds complex secondary structures, perhaps to facilitate exonucleolytic degradation by PNPase (10, 16, 43). Other DEAD-box helicases, such as RhlE, SrmB, and CsdA, have also been reported to interact with the degradosome, although at a site distinct from the RhlB docking site (23, 40). The role of the glycolytic enzyme enolase in the degradosome is not fully understood; however, it is required for a proper response to phosphosugar stress, perhaps indicating that it has a regulatory role (37). The loss of RhlB or enolase contacts with the degradosome results in reduced degradation activity, and mutations that alter the scaffolding properties of RNase E result in a reduced growth rate (26). Other proteins that copurify with the degradosome are the chaperones DnaK and GroEL and polyphosphate kinase (PPK) (3, 31). The physiological significance of these copurifications is unclear.
It has been speculated that the polyadenylation and RNA degradation processes are functionally related. Indeed, there is in vitro evidence that PAP I and RNase E interact (44). The degradosome localizes to the inner membrane, and it is anchored there by the N-terminal portion of RNase E (22, 27). Thus, it has been proposed that RNA degradation may be a compartmentalized phenomenon. PAP I also localizes to the inner membrane during exponential growth (7, 21). A common subcellular localization of both PAP I and the degradosome supports the idea that these two processes are coordinated.
Recently, it was discovered that the adaptor protein SprE (RssB) has a second function in the regulation of polyadenylation (7) that is distinct from its well-characterized function in the regulation of RpoS stability (20). During exponential growth, SprE plays a role in the normal maintenance of the poly(A) levels and, therefore, the stability of mRNAs. In exponential phase, PAP I is membrane localized; however, upon entry into stationary phase, PAP I is localized in a few punctate spots in the cytoplasm (7). Polyadenylation in stationary phase is poorly understood, but SprE appears to be required for the change in the intracellular localization of PAP I. This observation suggests that SprE has a function in the polyadenylation pathway during stationary phase that involves PAP I. However, the exact mechanism by which SprE performs this second function in either growth phase is unknown. Here, we use a targeted proteomics approach to probe SprE's function in this pathway. Specifically, we provide in vivo evidence that PAP I and the degradosome interact and that SprE is required for the maintenance of this association during stationary phase.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
All strains are listed in Table 1. Standard microbial techniques were used for strain construction (46). Luria broth (LB) was prepared as described previously (46) and was supplemented with appropriate antibiotics as needed. The antibiotic concentrations used were 125 μg/ml ampicillin and 50 μg/ml kanamycin. Bacteria were grown at 37°C with aeration, and growth was monitored by measuring the optical density at 600 nm (OD600). The ppcnB-GFP plasmid has been described previously (7). This plasmid expresses the PAP I-green fluorescent protein (GFP) fusion protein under the control of the native pcnB promoters. The plasmid pZS*11GFP contains the GFP protein under the control of the anhydrotetracycline (ATC)-inducible promoter PLtetO-1 (29). ATC was added at a concentration of 200 ng/ml for induction of this construct.
TABLE 1.
Strains used in this study
Preparation of cells for immunopurifications.
A single colony was inoculated into a 5-ml overnight culture in LB containing the appropriate antibiotic. The culture was diluted 1:1,000 (vol/vol) in 2 liters fresh medium and grown to an OD600 of 0.4 to 0.5 for exponential-phase samples or overnight to an OD600 of 4 to 5 for stationary-phase samples. Cells were subjected to centrifugation using two 10-min steps at 4,000 × g in a Beckman centrifuge (Model J-6B) and two 15-min spins at 3,300 × g in a Sorvall GLC-2B (Dupont Instruments). The pellets were weighed, and 100 μl of 20 mM HEPES, pH 7.5, 1.2% (wt/vol) polyvinyl-pyrrolidone (Sigma), 1/100 (vol/vol) protease inhibitor cocktail (Sigma) were added per gram of cells (wet weight). Cell pellets were frozen in liquid nitrogen as described previously (18).
Conjugation of magnetic beads.
Polyclonal anti-GFP antibody was prepared, purified, and conjugated to M270 Epoxy Dynabeads (Dynal Invitrogen) using 7 μg of anti-GFP per mg of beads, as described previously (18). Eight to 12 mg of beads was used for each immunoaffinity purification, depending on the amount of starting material (3 to 7 g cells, as optimized).
Cell grinding and lysis.
Cryogenic cell lysis was carried out as described previously (18). Briefly, lysis was performed by cryogenic grinding using a Retsch MM 301 mixer mill (Retsch, Newtown, PA) for 10 cycles of 3 min at 30 Hz inside 25-ml jars (Retch), with one 20-mm stainless steel grinding ball per jar (McMaster, NJ). Following optimization tests, the appropriate lysis buffer in terms of protein recovery was found to be 20 mM HEPES, pH 7.4, 0.11 M potassium acetate (KOAc), 2 mM MgCl2, 0.1% Tween 20 (vol/vol), 1 μM ZnCl2, 1 μM CaCl2, 1% Triton X-100, 0.5% deoxycholate, 150 mM NaCl, 1:100 protease inhibitor cocktail (Sigma), and 1:200 phenylmethylsulphonyl fluoride (PMSF). Where applicable, RNase A was added to a final concentration of 0.1 mg/ml and the mixture incubated for 15 min at room temperature. The efficiency of the RNase treatment was assessed under the same lysis buffer conditions as those utilized for the isolation of PAP I (see Fig. S2 in the supplemental material). Lysis buffer was added to the frozen cell powder (5 ml of buffer/g of cells), and the mixture was homogenized for 15 s using a PT 10-35 GT Polytron (Kinematica). The resulting lysate was centrifuged for 10 min at 4,000 × g at 4°C, and the soluble fraction was used for the affinity purification experiments.
Immunoaffinity purification.
Immunoaffinity purifications were carried out by incubation of the conjugated magnetic beads with the cell lysate at 4°C for 1 h, as described previously (18). Since the volume of buffer used was relatively large (30 to 40 ml), the magnetic beads were collected by passing the lysate over a magnet (Magcraft) in two steps, to ensure that all of the beads were recovered. Isolated proteins were eluted from the beads using a freshly prepared solution of 0.5 N NH4OH, 0.5 mM EDTA, as described previously (18), and the supernatant was frozen on liquid nitrogen and dried by vacuum centrifugation (Speed Vac Plus SC110A with a GP110 gel pump; Thermo Scientific). The dried samples were suspended in lithium dodecyl sulfate (LDS)-PAGE sample buffer (Invitrogen), alkylated with 100 mM iodoacetamide, and resolved on a 4 to 12% NuPAGE Novex Bis-Tris gel (Invitrogen) according to the manufacturer's instructions. The gels were stained with GelCode Blue stain reagent (Thermo Scientific).
Protein digestion.
After being stained, entire gel lanes were cut into approximately 25 slices, diced into small pieces, and destained in 200 μl 50% (vol/vol) acetonitrile (ACN) and 50 mM ammonium bicarbonate (ABC) for 20 min with agitation at 4°C. The gel pieces were then dehydrated in 100% ACN and digested with 12.5 ng/μl sequencing-grade modified trypsin (Promega) or metalloendopeptidase Lys-N (Seikagaku Corp., Tokyo, Japan) in 50 mM ABC overnight at 37°C. Peptides were extracted from the gel slices on reverse-phase resin (Poros 20 R2; Applied Biosystems). The samples were loaded onto ZipTips with C18 resin (Millipore), washed twice with 20 μl 0.1% (vol/vol) trifluoroacetic acid (TFA), and eluted with 20 mg/ml DHB (2,5-dihydroxybenzoic acid) in 50% (vol/vol) methanol, 20% (vol/vol) ACN, and 0.1% (vol/vol) TFA directly onto a matrix-assisted laser desorption ionization (MALDI) target.
Relative quantification of isolated proteins using guanidination.
Guanidination was carried out as described previously (5, 49), with minor modifications. After in-gel digestion, the reaction was stopped by adding TFA to 0.1% final concentration. The liquid component was transferred to a clean tube, and the remaining gel pieces were incubated with 70% ACN, 0.1% TFA in two steps (10 and 5 min) with shaking at room temperature. The resulting soluble fraction was combined with the previous liquid and dried for 1 h by vacuum centrifugation. The dried pellets were suspended in 20 μl of 7N NH4OH and 1.5 μl of either [14N]O-methylisourea or [15N]O-methylisourea (prepared as described below) and allowed to incubate overnight at room temperature with shaking. The reaction was stopped by adding an equal volume (20 μl) of 1% (vol/vol) TFA, and then the suspension was dried by vacuum centrifugation. The pellets were suspended in 25 μl of reverse-phase solution (as described above) as a 1:1 mixture of Poros beads in 5% formic acid, 0.2% TFA with 50 mM ABC and were agitated overnight at 4°C. The corresponding gel slices from the wild-type and ΔsprE lanes were loaded onto the same ZipTip and processed as described above. This enabled the relative levels of the peptides from the two lanes to be directly compared from the intensities of the peaks in the mass spectra, as the heavy-isotope labeled ΔsprE peptides were shifted by +2 Thomson (Th) compared to the wild-type samples. The overall shift from guanidination was 42 and 44 Th. The +2-Th-shifted peaks following the guanidination with [15N]O-methylisourea represent a mixture of the monoisotopic peak of the ΔsprE peptides and the third isotopic peak from the wild-type sample. Therefore, this was corrected by subtracting the theoretical isotopic peak abundance generated by the program Isotopident (http://education.expasy.org/student_projects/isotopident/htdocs/). The reverse experiment, in which the ΔsprE peptides were derivatized with [14N]O-methylisourea and the wild-type peptides with [15N]O-methylisourea, was performed to confirm the findings. The samples from each strain were normalized by OD600 prior to the immunopurifications, and the peptides from PAP I-GFP served as an internal calibrator to adjust for differences in input amounts. At least 4 tandem mass spectrometry (MS/MS)-confirmed peptides were used for the relative quantification of each protein.
Preparation of [14N]O-methylisourea and [15N]O-methylisourea.
The [14N]O-methylisourea and [15N]O-methylisourea were prepared as described previously (49). Briefly, 60 mg of [14N]urea or [15N]urea (Sigma Aldrich) was dissolved in 95 μl of dimethyl sulfate and incubated at 50°C for 5 h with agitation. An amount of 250 μl of acetone was added, and the solution was mixed for 1 h at room temperature. One milliliter of diethyl ether was added, the mixture was inverted a few times, and the top layer was discarded. This step was repeated twice, and the remaining ∼100 μl was dissolved into 900 μl of high-pressure liquid chromatography-grade water to make an approximately 0.5 M solution of N-O-methylisourea hemisulfate.
Mass spectrometry.
For all experiments performed, the proteins isolated were analyzed by MALDI MS and MS/MS analyses using a MALDI LTQ Orbitrap XL mass spectrometer (Thermo Electronics, Bremen, Germany), as previously described (28). MS spectra were acquired using both manual and automated modes of acquisition and detection at the Orbitrap level. MALDI linear trap quadrupole (LTQ) MS/MS analyses were carried out by collision-induced dissociation (CID). For automatic MS/MS acquisition, the mass spectrometer was operated in a data-dependent mode, and the 50 most prominent ions were selected for CID fragmentation. The mass spectra were visualized and processed in Qual Browser (version 2.0.7; Thermo Fisher Scientific). Proteins were identified by database searching for Escherichia coli (52,568 sequences) in the most recent NCBI nonredundant protein database, using the computer algorithm XProteo (http://www.xproteo.com). For hypothesis-driven MS and MS/MS analyses, proteins of interest were theoretically digested into monoisotopic masses using the computer program PROWL (http://prowl.rockefeller.edu/prowl-cgi/sequence.exe) and the mass spectra were manually examined for matching masses. Only candidates indicated by XProteo with significant scores and confirmed by MS/MS analyses for at least 2 peptides were considered to be present in the isolations. The reproducibility of the protein isolation in at least triplicate experiments and the absence of the protein in control immunopurifications were conditions imposed for a candidate to be considered associated with the isolated protein.
RESULTS
Strategy for isolating PAP I-GFP complexes.
We wish to understand the mechanism by which SprE participates in the mRNA polyadenylation pathway. Since SprE and PAP I appear to be functionally linked (7), we thought it possible that SprE may affect this pathway through protein-protein interactions with either PAP I or an interacting partner. To this end, we sought to characterize PAP I's interactions in the presence and absence of SprE.
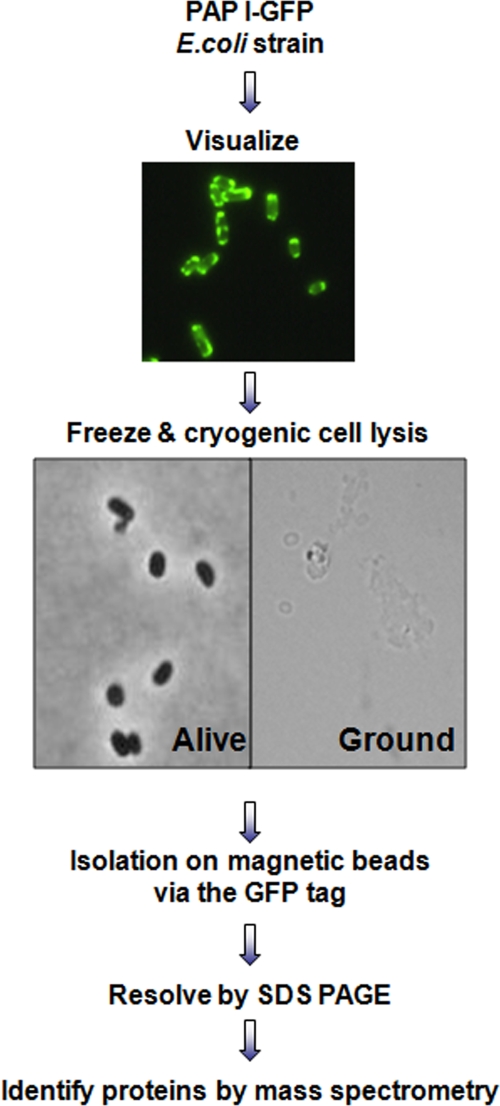
To identify the interacting partners of PAP I, we followed the strategy outlined in Fig. 1. A plasmid producing a functional PAP I-GFP hybrid protein (7) was transformed into wild-type E. coli. This plasmid, ppcnB-GFP, contains a functional pcnB-GFP fusion under the control of the pcnB native promoters (7). The endogenous levels of PAP I are relatively low (2), and it has been shown that this enzyme is the limiting factor for polyadenylation (32). Thus, increasing the levels of PAP I increases polyadenylation in a dose-dependent manner. Continually increasing the PAP I levels eventually causes a growth defect and even cell death (32). Therefore, the assessment of the functionality of the overproduced PAP I-GFP is critical. We previously demonstrated that the overproduced PAP I-GFP leads to a 6-fold increase in poly(A) levels in the cell and does not confer a growth defect (7). Therefore, the PAP I-GFP is functional and provides an opportunity to study PAP I interacting partners by enrichment via the GFP tag. Cells expressing PAP I-GFP were harvested, rapidly frozen in liquid nitrogen, and cryogenically lysed as described in Materials and Methods. Following the cryogenic lysis, a limited number of cells remained intact, but the majority of cells (>95%) were lysed and had a ghost-like appearance (Fig. 1). This procedure was previously shown to help maintain and isolate protein complexes close to their native form (18). The lysis conditions were optimized for efficient recovery of PAP I-GFP. Following immunopurification on magnetic beads coated with high-affinity polyclonal anti-GFP antibodies and separation by SDS-PAGE, the isolated proteins were identified by mass spectrometry using a MALDI LTQ Orbitrap (28).
FIG. 1.
Strategy to isolate PAP I-GFP complexes. An E. coli strain that expressed a GFP-tagged PAP I was constructed as described in Materials and Methods. PAP I-GFP was visualized by fluorescence microscopy to determine intracellular localization. Cells were frozen and subjected to cryogenic cell lysis, which allowed for efficient lysis of >95% of cells, as determined by light microscopy. Proteins were isolated on magnetic beads coated with high-purity anti-GFP antibodies, resolved by SDS-PAGE, and identified by MS and MS/MS analyses using a MALDI LTQ Orbitrap mass spectrometer.
PAP I interacts with the degradosome.
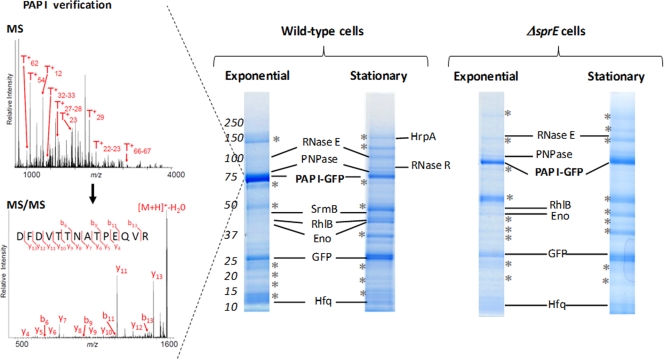
Following the strategy outlined above, we immunopurified PAP I from wild-type cells (sprE+) in both growth phases. Only the proteins reproducibly isolated in triplicate experiments are indicated (Fig. 2). A list of the proteins isolated is provided in Table S1 in the supplemental material. As shown in Fig. 2, PAP I-GFP is present as the main Coomassie blue-stained band in these isolations. Moreover, the RNA chaperone Hfq, which is a known interacting partner of PAP I, was also clearly present, thus serving as an internal positive control (35). In a control experiment, cells in which only GFP was expressed from a plasmid were subjected to the same immunopurification procedure (see Fig. S1 in the supplemental material). Comparison of the results of the experiments with PAP I-GFP and only GFP allowed us to distinguish the proteins obtained through nonspecific associations with the anti-GFP antibodies or the magnetic beads (see Table S2 and Fig. S1 in the supplemental material). The prominent bands visible in Fig. 2 and marked with asterisks represent such nonspecific associations (see Table S2 in the supplemental material).
FIG. 2.
PAP I-GFP interactions in the presence and absence of SprE. Representative immunopurifications of VC239 (MC4100/ppcnB-GFP), or VC244 (ΔsprE/ppcnB-GFP), where cells were grown to either exponential or stationary phase, are shown. Interacting partners that were not detected in control immunopurifications (see Fig. S1 and Table S2 in the supplemental material) and were reproducibly isolated in triplicate experiments are labeled. Prominent bands marked with asterisks represent nonspecific associations as determined from control experiments; these proteins are listed in Table S2 in the supplemental material. The left inset illustrates examples of MS (top left) and MS/MS (bottom left) analyses for the identification of PAP I from wild-type cells. Selected singly charged PAP I peptides derived from trypsin digestion are indicated (T+n, where n is the peptide number).
Interestingly, our isolations demonstrated that PAP I associates with the RNA degradosome, as all of the major degradosome components were isolated from cells in both growth phases (Fig. 2; also see Table S1 in the supplemental material) and were absent in our control experiments. The core components of the degradosome included the scaffold protein RNase E, the exonuclease PNPase, the RNA helicase RhlB, and enolase (Eno) (9, 10). In addition, SrmB, a DEAD-box helicase reported to associate with the degradosome (23) and PAP I (44), was present as well. In a separate experiment, the cells were inadvertently left to incubate at 4°C for an extended period of time, in which case the DEAD-box helicase CsdA was additionally isolated (data not shown; see Table S1 in the supplemental material). CsdA is a helicase that has been previously reported to associate with the degradosome during cold shock (40). RNase E was previously reported to interact with several of the degradosome-associated proteins that we observed in the PAP I isolation, and the in vivo interaction of PAP I and PNPase had been reported as well. However, these interactions were typically only detected as pairwise connections. This is the first report demonstrating that all of these proteins may be part of the same macromolecular assembly.
The majority of the proteins isolated with PAP I were present in both exponential and stationary phases. The DEAH-box helicase HrpA and the riboexonuclease RNase R, two putative members of the PAP I-degradosome complex, seemed enriched during stationary phase (as judged by their observed protein sequence coverage and scores from database searching; see Table S1 in the supplemental material). In agreement with these observations, RNase R expression is known to be induced during stationary phase (1).
Since PAP I and many of the other degradosome proteins have RNA binding activity, we next asked if the interactions detected were formed through RNA bridges and not protein-protein interactions. To address this possibility, the PAP I-GFP immunopurifications from wild-type cells were performed in the presence of RNase A in the lysis buffer. Under these conditions, the same interacting partners of PAP I-GFP were identified (see Fig. S2 and Table S3 in the supplemental material). The interaction with HrpA may have been reduced by this treatment, as judged by protein sequence coverage and database scores. However, its presence was still confirmed by MS/MS analysis. Our data suggest that the majority of proteins that are immunopurified with PAP I-GFP associate via protein-protein interactions and not via RNA bridges.
In summary, these results provide strong evidence that PAP I-GFP associates in vivo with the RNA degradosome in cells in both growth phases and that, under certain conditions, the degradosome may contain at least 10 different proteins.
PAP I-degradosome association is SprE dependent during stationary phase.
Since SprE is involved in the regulation of polyadenylation, we wondered if a possible mechanism by which it participates in this pathway is by affecting or altering, directly or indirectly, the interactions of PAP I. The PAP I-GFP interacting partners in wild-type and ΔsprE cells in exponential phase were nearly identical (Fig. 2; also see Table S4 in the supplemental material). Only the DEAD-box helicase SrmB was not isolated from the ΔsprE cells. However, significant differences were observed in the PAP I-GFP isolations from ΔsprE and wild-type cells during stationary phase (Fig. 2). PAP I-GFP interactions with PNPase, RhlB, and enolase, which were all identified in exponential phase in both strains, were absent in stationary phase in the ΔsprE cells. In addition, the interaction with RNase R, which was enriched during stationary phase in wild-type cells, was not identified in ΔsprE cells during stationary phase (Fig. 2). While still isolated with PAP I-GFP in ΔsprE cells, RNase E was present at lower levels than in wild-type cells, as judged by comparing the mass spectrometry data from wild-type and ΔsprE cells. RNase E was detected with a lower protein sequence coverage of 17% in ΔsprE cells in stationary phase (versus approximately 40% sequence coverage in ΔsprE cells in exponential phase and wild-type cells in both stationary and exponential cells) and, therefore, had a dramatically reduced score from database searching (see Tables S1 and S4 in the supplemental material). The only association that seemed to remain constant was that with Hfq. All of the degradosome proteins were easily confirmed by mass spectrometry in the flowthrough from both the wild-type and ΔsprE immunoprecipitations, being detected on the one-dimensional SDS-PAGE at the correct molecular masses (see Table S5 in the supplemental material). This is in agreement with our previous findings that PAP I and PNPase levels are not affected by the absence of SprE (7). These results indicated that the degradosome proteins were absent not because of degradation but because of loss of association with PAP I-GFP.
Relative quantification of SprE-dependent PAP I associations.
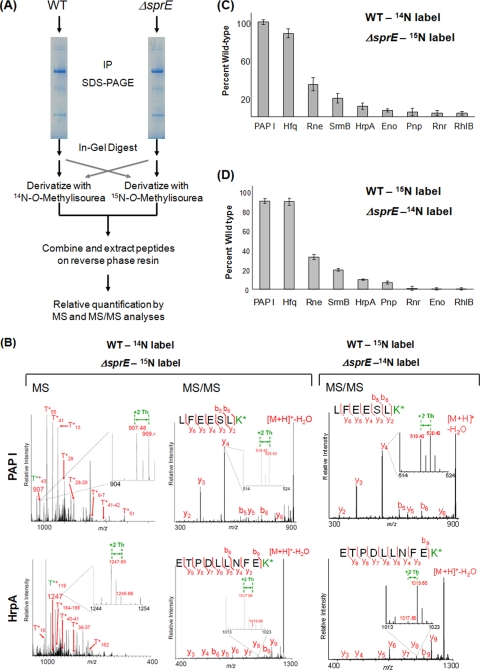
We next sought to quantify the SprE dependence of the PAP I associations and confirm the decreased levels of degradosome proteins in ΔsprE cells. We used guanidination (5, 49) in conjunction with differential isotopic labeling to relatively quantify the amounts of proteins isolated with PAP I from wild-type or ΔsprE cells. Guanidination, which converts lysine residues into homoarginines, increasing gas-phase basicity and the sensitivity of detection in MALDI MS analyses, was previously shown to be an efficient and economical mean for protein relative quantification when using [14N]O-methylisourea and [15N]O-methylisourea (5, 49). The samples from wild-type and ΔsprE cells were derivatized with the differentially labeled reagents at the peptide level, after enzymatic digestion (Fig. 3A).
FIG. 3.
Relative quantification of PAP I's interacting partners using guanidination. (A) Workflow of the guanidination procedure, described in detail in Materials and Methods. After in-gel digestion with trypsin, the guanidination reaction was carried out using differentially labeled N-O-methylisourea. The wild-type peptides were labeled with 14N, which shifts the mass of lysine-containing peptides by +42 Th, and the ΔsprE peptides were labeled with 15N, for a shift of +44 Th. After the samples were mixed, relative quantification was performed by comparing the intensities of the light and heavy monoisotopic peaks. A parallel experiment was performed using reverse labeling. IP, immunopurification. (B) Sample spectra of guanidinated PAP I and HrpA at the MS and MS/MS level. On the MS panels, representative arginine-containing peptides are labeled. The T*+ designation represents the tryptic fragments containing modified lysine residues. The enlarged regions show the peptide of interest, with the +2-Th shift between heavy and light peaks. The insets in the MS/MS spectra illustrate the +2-Th shift between y ions. K* refers to homoarginine. (C) The relative quantification of PAP I interacting partners in the ΔsprE background. Since the +2-Th-shifted peak represents a mixture of the monoisotopic ΔsprE peptides and the third isotope of the wild-type peaks, all intensities of the heavy peptides were adjusted using the formula (ΔsprEobs − ISOexp) + [PAP IWTobs − (PAP IΔSprEobs − ISOexp)], where ΔsprEobs is the intensity of any peptide from the ΔsprE samples (obs, observed) and ISOexp is the predicted isotopic peak intensity (exp, expected). All intensities were adjusted to that of PAP I to correct for differences in starting material (see Fig. S3 in the supplemental material). (D) Results of the reverse labeling experiment, where wild-type peptides were derivatized with [15N]O-methylisourea and ΔsprE peptides with [14N]O-methylisourea. The isotopic correction described in Fig. S3 in the supplemental material was therefore not necessary. The intensities of the peaks from the wild-type peptides were set at 100%, and the intensities of the ΔsprE peptides are reported relative to that. All intensities were adjusted to that of PAP I to correct for differences in starting material. Error bars show standard deviations. WT, wild type.
The results of these relative quantification studies confirmed our previous findings, showing that the absence of SprE during stationary phase led to a dramatic decrease in the PAP I-GFP interacting partners, with the exception of Hfq (Fig. 3C and D). RNase E (Rne) was the most abundant degradosome protein in the PAP I immunopurifications from ΔsprE cells in stationary phase, even though it was reduced by 65% compared to the level in the wild type. All other interactions were reduced even more significantly, with SrmB reduced by 80% and the other proteins reduced by 85% or more. Examples of the MS and MS/MS relative quantifications of PAP I and HrpA are shown in Fig. 3B. For each peptide present in the isolation from ΔsprE cells, the levels were adjusted to take into account that the +2-Th shift is a mixture of the monoisotopic peak from the ΔsprE peptides and the third isotopic peak from the wild-type peptides (see Fig. S3 in the supplemental material). It should be noted that even before the corrections were performed, drastic reductions (RNase E by 50% and all others by >70%) in the relative amounts of the degradosome proteins were observed. To correct for starting sample amounts between the two strains, all values were then normalized to the levels of PAP I in the immunopurifications. To further confirm these observations, the reverse experiments were performed, where the ΔsprE peptides were derivatized with [14N]O-methylisourea and the wild-type peptides with [15N]O-methylisourea, thus eliminating the need for the isotopic correction (Fig. 3D). Furthermore, this reverse derivatization experiment was repeated in conjunction with incorporating enzymatic digestion with Lys-N (4, 6), which was shown to improve peptide fragmentation by MALDI ion trap CID. The data from these three separate experiments are consistent (Fig. 3C and D; also see Fig. S5 in the supplemental material), thus confirming the reduced interactions of PAP I and the degradosome proteins. Altogether, our results indicate that SprE is required to maintain the PAP I-degradosome association during stationary phase.
SprE's effect on PAP I associations is distinct from its RpoS degradation function.
The striking difference in PAP I associations in wild-type and ΔsprE cells during stationary phase (Fig. 3) led us to question why SprE was not observed in our initial PAP I isolations (Fig. 2). To ask if SprE was present at low levels and below the initial threshold of detection in our PAP I purification, we utilized a hypothesis-driven MS/MS approach (13) to probe for the presence of SprE peptides in the PAP I immunopurifications. Using this manual approach for sequence verification by MS/MS, one SprE peptide was reproducibly (in all six experiments) detected only in stationary phase (see Fig. S5 in the supplemental material). The presence of this peptide indicates that SprE is present at low levels in these immunopurifications.
To test whether we could increase this association, we next performed a similar experiment in the presence of an rssA2 allele. This allele leads to the overproduction of chromosomal SprE to levels that are 4-fold higher than endogenous levels and does not cause a growth defect (7). This elevated level of SprE did not enhance the association with PAP I, as SprE was still only present at trace levels. The PAP I associations with the degradosome were still present (see Fig. S6 and Table S6 in the supplemental material). Altogether, these data indicate that SprE is present at low levels in the PAP I isolations from stationary phase cells, which may reflect a weak or transient association with PAP I or one of its interacting partners.
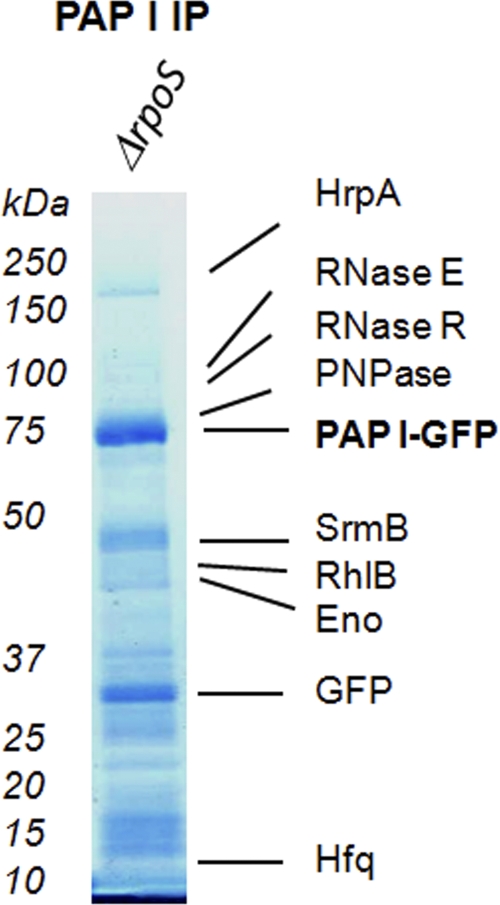
SprE directly controls the stability of RpoS, the stationary-phase master regulator, which controls a large regulon of stress-response proteins (20). One possibility is that any phenotype observed to be associated with the deletion of SprE is a consequence of the stabilization of RpoS. Previously, we demonstrated that SprE's function in the regulation of PAP I-dependent polyadenylation and mRNA stability during exponential phase was independent of the RpoS degradation pathway. We further showed that the observed change in intracellular localization during stationary phase was also independent of RpoS (7). Therefore, to rule out an indirect effect from increased levels of RpoS, we performed PAP I-GFP immunopurifications in the absence of RpoS. All of the PAP I-GFP interacting partners were identified in this experiment, including the degradosome members and the previously observed SprE peptide (Fig. 4; also see Table S7 in the supplemental material). These results indicate that SprE's involvement in the polyadenylation pathway is separate and distinct from its RpoS degradation function.
FIG. 4.
Results of PAP I-GFP immunopurification (IP) from ΔrpoS cells. A control PAP I-GFP immunopurification was performed from VC252 (ΔrpoS/ppcnB-GFP) during stationary phase. Interacting partners are labeled, and a complete list of proteins is given in Table S7 in the supplemental material.
DISCUSSION
Polyadenylation is known to enhance RNA degradation, and this has led to the prediction that PAP I and Hfq may be associated with the RNA degradosome. We have confirmed and extended this prediction. We have shown that PAP I and Hfq are associated with the RNA degradosome in vivo in both exponential and stationary phase (Fig. 5). Our results further show that, besides PAP I and Hfq (35), the degradosome contains not only the four core components RNase E, PNPase, RhlB, and enolase but also three additional proteins, the RNA helicases SrmB (23), HrpA (24, 38), and RNase R (1, 14, 15, 41). Indeed, under certain conditions, such as cold shock, the RNA degradosome may contain as many as 10 different proteins. While previous reports have provided insights into various components and features of the degradosome (9, 30, 31), our results provide, to our knowledge, the most comprehensive characterization of the in vivo degradosome to date.
FIG. 5.
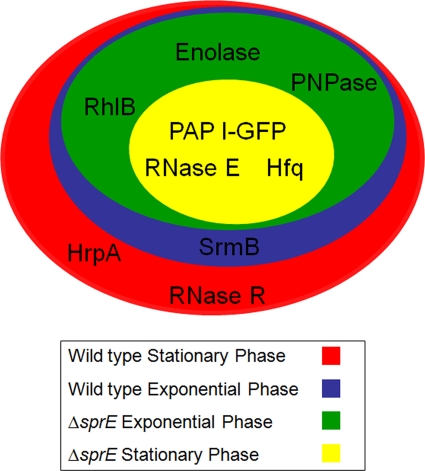
Summary of PAP I-GFP interacting partners. A summary of the results of the four immunopurifications illustrated in Fig. 2 represented as a Venn diagram. During stationary phase, in the absence of SprE, the only interacting partners of PAP I-GFP isolated were RNase E and Hfq. During exponential phase, the rest of the degradosome components were isolated, with the exception of SrmB. In a wild-type background, SrmB could be isolated in exponential phase, along with the other degradosome components. HrpA and RNase R were additionally isolated during stationary phase in the wild-type case.
Strikingly, we have discovered that the adaptor protein SprE is required to maintain the association of PAP I and Hfq with the degradosome during stationary but not exponential growth phase (Fig. 5). Previously, we had shown that SprE was required for the change in the intracellular localization of PAP I that occurs during stationary phase (7). We suspect that these two observations are functionally related and likely reflect an important role for SprE in stationary-phase physiology through its effects on polyadenylation and mRNA stability.
In stationary phase, in the absence of SprE, PAP I remains membrane associated (7) and, as we have shown here, in a tight complex with Hfq, but the association of PAP I and Hfq with the degradosome is lost. Although it is clear that the core degradosome members are not degraded, we do not know if they remain associated with each other in a ΔsprE background, nor do we know their cellular localization during stationary phase. Performing immunopurifications and localization studies with a GFP-tagged variant of another complex member would address these issues. In addition, we do not yet understand the phenotypic consequences of these changes in degradosome membership. Microarray analysis might reveal SprE-dependent changes in the stability of important mRNAs during stationary phase.
We showed that the PAP I-degradosome association is independent of the alternate sigma factor RpoS and, therefore, is distinct from SprE's known function in the regulated proteolysis of RpoS. So, how does SprE regulate the PAP I-degradosome association in stationary phase? The simplest explanation is that SprE is the glue that holds the complex together under these conditions. If this were true, then we should have detected SprE in the complex during stationary phase. While the evidence supporting its presence in this complex is weak (only one SprE peptide confirmed by MS/MS analyses), we think this is real because it was reproducibly detected (in all six experiments) only during stationary phase. The fact that the PAP I-degradosome association was still maintained in exponential phase in the absence of SprE indicates that SprE is not the main scaffold for this interaction. Thus, we think that SprE may interact with the degradosome, but probably it is not the glue holding it together. It seems more likely that its interaction is weak or transient.
It is important to note that the PAP I isolations were performed under particularly stringent lysis buffer conditions (i.e., incorporating high concentrations of both Triton X-100 and deoxycholate detergents and salt). Our previous experience has indicated that the lysis buffer conditions are critical for obtaining a balance between the efficient isolation of a tagged protein and the maintenance of interactions (18). During our optimizations of the PAP I isolation, we noticed that the efficient purification of PAP I required stringent conditions, likely due to the necessity to penetrate the E. coli cell wall and obtain access to the cytoplasmic PAP I-GFP. As a comparison, this lysis condition was more stringent than those we utilized for isolating members of the nuclear pore complex in yeast (18), postsynaptic densities from mice cerebella (45), or cytomegalovirus virion assemblies in human cells (36), all known to present special challenges. We have previously utilized these harsh lysis conditions only when accessing virally induced vesicles in mammalian cells (17). While these stringent conditions demonstrate that the observed interactions between PAP I and the degradosome members are strong, it is likely that they have also led to the disruption of weak-affinity or transient interactions. Other approaches incorporating reversible cross-linking and reciprocal isolations could be implemented to stabilize such interactions, potentially revealing the presence of SprE.
In summary, our targeted proteomic approach incorporating a GFP-tagged PAP I, cryogenic cell lysis, affinity purification, and mass spectrometry has allowed us to capture a more complete picture of the RNA degradation machinery. Our results provide the first evidence for the in vivo association of PAP I and Hfq with the degradosome and uncover a role for SprE in maintaining this important complex during stationary phase.
Supplementary Material
Acknowledgments
This work was supported by a grant from NIGMS (GM065216) to T.J.S. and by award number DP1DA026192 from the National Institute On Drug Abuse and Princeton University Start-up funding to I.M.C.
Footnotes
Published ahead of print on 14 May 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andrade, J. M., F. Cairrao, and C. M. Arraiano. 2006. RNase R affects gene expression in stationary phase: regulation of ompA. Mol. Microbiol. 60:219-228. [DOI] [PubMed] [Google Scholar]
- 2.Blum, E., A. J. Carpousis, and C. F. Higgins. 1999. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 274:4009-4016. [DOI] [PubMed] [Google Scholar]
- 3.Blum, E., B. Py, A. J. Carpousis, and C. F. Higgins. 1997. Polyphosphate kinase is a component of the Escherichia coli RNA degradosome. Mol. Microbiol. 26:387-398. [DOI] [PubMed] [Google Scholar]
- 4.Boersema, P. J., N. Taouatas, A. F. Altelaar, J. W. Gouw, P. L. Ross, D. J. Pappin, A. J. Heck, and S. Mohammed. 2009. Straightforward and de novo peptide sequencing by MALDI-MS/MS using a Lys-N metalloendopeptidase. Mol. Cell. Proteomics 8:650-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brancia, F. L., S. G. Oliver, and S. J. Gaskell. 2000. Improved matrix-assisted laser desorption/ionization mass spectrometric analysis of tryptic hydrolysates of proteins following guanidination of lysine-containing peptides. Rapid Commun. Mass Spectrom. 14:2070-2073. [DOI] [PubMed] [Google Scholar]
- 6.Carabetta, V. J., T. Li, A. Shakya, T. M. Greco, and I. M. Cristea. 2010. Integrating Lys-N proteolysis and N-terminal guanidination for improved fragmentation and relative quantification of singly-charged ions. J. Am. Soc. Mass Spectrom. 21:1050-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carabetta, V. J., B. K. Mohanty, S. R. Kushner, and T. J. Silhavy. 2009. The response regulator SprE (RssB) modulates polyadenylation and mRNA stability in Escherichia coli. J. Bacteriol. 191:6812-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpousis, A. J. 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61:71-87. [DOI] [PubMed] [Google Scholar]
- 9.Carpousis, A. J., V. Khemici, S. Ait-Bara, and L. Poljak. 2008. Co-immunopurification of multiprotein complexes containing RNA-degrading enzymes. Methods Enzymol. 447:65-82. [DOI] [PubMed] [Google Scholar]
- 10.Carpousis, A. J., V. Khemici, and L. Poljak. 2008. Assaying DEAD-box RNA helicases and their role in mRNA degradation in Escherichia coli. Methods Enzymol. 447:183-197. [DOI] [PubMed] [Google Scholar]
- 11.Carpousis, A. J., G. Van Houwe, C. Ehretsmann, and H. M. Krisch. 1994. Copurification of E. coli RNase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889-900. [DOI] [PubMed] [Google Scholar]
- 12.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 13.Chang, E. J., V. Archambault, D. T. McLachlin, A. N. Krutchinsky, and B. T. Chait. 2004. Analysis of protein phosphorylation by hypothesis-driven multiple-stage mass spectrometry. Anal. Chem. 76:4472-4483. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, Z. F., and M. P. Deutscher. 2002. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 277:21624-21629. [DOI] [PubMed] [Google Scholar]
- 15.Cheng, Z. F., and M. P. Deutscher. 2003. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl. Acad. Sci. U. S. A. 100:6388-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coburn, G. A., X. Miao, D. J. Briant, and G. A. Mackie. 1999. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 13:2594-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cristea, I. M., J. W. Carroll, M. P. Rout, C. M. Rice, B. T. Chait, and M. R. MacDonald. 2006. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 281:30269-30278. [DOI] [PubMed] [Google Scholar]
- 18.Cristea, I. M., R. Williams, B. T. Chait, and M. P. Rout. 2005. Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4:1933-1941. [DOI] [PubMed] [Google Scholar]
- 19.Del Favero, M., E. Mazzantini, F. Briani, S. Zangrossi, P. Tortora, and G. Deho. 2008. Regulation of Escherichia coli polynucleotide phosphorylase by ATP. J. Biol. Chem. 283:27355-27359. [DOI] [PubMed] [Google Scholar]
- 20.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 21.Jasiecki, J., and G. Wegrzyn. 2005. Localization of Escherichia coli poly(A) polymerase I in cellular membrane. Biochem. Biophys. Res. Commun. 329:598-602. [DOI] [PubMed] [Google Scholar]
- 22.Khemici, V., L. Poljak, B. F. Luisi, and A. J. Carpousis. 2008. The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 70:799-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khemici, V., I. Toesca, L. Poljak, N. F. Vanzo, and A. J. Carpousis. 2004. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 54:1422-1430. [DOI] [PubMed] [Google Scholar]
- 24.Koo, J. T., J. Choe, and S. L. Moseley. 2004. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 52:1813-1826. [DOI] [PubMed] [Google Scholar]
- 25.Kushner, S. R. 2002. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184:4658-4665. (Discussion, 184:4657.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leroy, A., N. F. Vanzo, S. Sousa, M. Dreyfus, and A. J. Carpousis. 2002. Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol. Microbiol. 45:1231-1243. [DOI] [PubMed] [Google Scholar]
- 27.Liou, G. G., W. N. Jane, S. N. Cohen, N. S. Lin, and S. Lin-Chao. 2001. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc. Natl. Acad. Sci. U. S. A. 98:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, Y., T. Li, F. Yu, T. Kramer, and I. M. Cristea. 2010. Resolving the composition of protein complexes using a MALDI LTQ Orbitrap. J. Am. Soc. Mass Spectrom. 21:34-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mauri, P., and G. Deho. 2008. A proteomic approach to the analysis of RNA degradosome composition in Escherichia coli. Methods Enzymol. 447:99-117. [DOI] [PubMed] [Google Scholar]
- 31.Miczak, A., V. R. Kaberdin, C. L. Wei, and S. Lin-Chao. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. U. S. A. 93:3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohanty, B. K., and S. R. Kushner. 1999. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 34:1094-1108. [DOI] [PubMed] [Google Scholar]
- 33.Mohanty, B. K., and S. R. Kushner. 2006. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 34:5695-5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase functions both as a 3′→5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:11966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohanty, B. K., V. F. Maples, and S. R. Kushner. 2004. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 54:905-920. [DOI] [PubMed] [Google Scholar]
- 36.Moorman, N. J., R. Sharon-Friling, T. Shenk, and I. M. Cristea. 2010. A targeted spatial-temporal proteomic approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol. Cell. Proteomics 9:851-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morita, T., H. Kawamoto, T. Mizota, T. Inada, and H. Aiba. 2004. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 54:1063-1075. [DOI] [PubMed] [Google Scholar]
- 38.Moriya, H., H. Kasai, and K. Isono. 1995. Cloning and characterization of the hrpA gene in the terC region of Escherichia coli that is highly similar to the DEAH family RNA helicase genes of Saccharomyces cerevisiae. Nucleic Acids Res. 23:595-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Hara, E. B., J. A. Chekanova, C. A. Ingle, Z. R. Kushner, E. Peters, and S. R. Kushner. 1995. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92:1807-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prud'homme-Genereux, A., R. K. Beran, I. Iost, C. S. Ramey, G. A. Mackie, and R. W. Simons. 2004. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a “cold shock degradosome.” Mol. Microbiol. 54:1409-1421. [DOI] [PubMed] [Google Scholar]
- 41.Purusharth, R. I., F. Klein, S. Sulthana, S. Jager, M. V. Jagannadham, E. Evguenieva-Hackenberg, M. K. Ray, and G. Klug. 2005. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J. Biol. Chem. 280:14572-14578. [DOI] [PubMed] [Google Scholar]
- 42.Py, B., H. Causton, E. A. Mudd, and C. F. Higgins. 1994. A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 14:717-729. [DOI] [PubMed] [Google Scholar]
- 43.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169-172. [DOI] [PubMed] [Google Scholar]
- 44.Raynal, L. C., and A. J. Carpousis. 1999. Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol. 32:765-775. [DOI] [PubMed] [Google Scholar]
- 45.Selimi, F., I. M. Cristea, E. Heller, B. T. Chait, and N. Heintz. 2009. Proteomic studies of a single CNS synapse type: the parallel fiber/purkinje cell synapse. PLoS Biol. 7:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 47.Soreq, H., and U. Z. Littauer. 1977. Purification and characterization of polynucleotide phosphorylase from Escherichia coli. Probe for the analysis of 3′ sequences of RNA. J. Biol. Chem. 252:6885-6888. [PubMed] [Google Scholar]
- 48.Vanzo, N. F., Y. S. Li, B. Py, E. Blum, C. F. Higgins, L. C. Raynal, H. M. Krisch, and A. J. Carpousis. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12:2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warwood, S., S. Mohammed, I. M. Cristea, C. Evans, A. D. Whetton, and S. J. Gaskell. 2006. Guanidination chemistry for qualitative and quantitative proteomics. Rapid Commun. Mass Spectrom. 20:3245-3256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.