Abstract
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 responds to the host-produced epinephrine and norepinephrine, and bacterially produced autoinducer 3 (AI-3), through two-component systems. Further integration of multiple regulatory signaling networks, involving regulators such as the LysR-type transcriptional regulator (LTTR) QseA, promotes effective regulation of virulence factors. These include the production of flagella, a phage-encoded Shiga toxin, and genes within the locus of enterocyte effacement (LEE) responsible for attaching and effacing (AE) lesion formation. Here, we describe a new member of this signaling cascade, an LTTR heretofore renamed QseD (quorum-sensing E. coli regulator D). QseD is present in all enterobacteria but exists almost exclusively in O157:H7 isolates as a helix-turn-helix (HTH) truncated isoform. This “short” isoform (sQseD) is still able to regulate gene expression through a different mechanism than the full-length K-12 E. coli “long” QseD isoform (lQseD). The EHEC ΔqseD mutant exhibits increased expression of all LEE operons and deregulation of AE lesion formation. The loss of qseD in EHEC does not affect motility, but the K-12 ΔqseD mutant is hypermotile. While the lQseD directly binds to the ler promoter, encoding the LEE master regulator, to repress LEE transcription, the sQseD isoform does not. LTTRs bind to DNA as tetramers, and these data suggest that sQseD regulates ler by forming heterotetramers with another LTTR. The LTTRs known to regulate LEE transcription, QseA and LrhA, do not interact with sQseD, suggesting that sQseD acts as a dominant-negative partner with a yet-unidentified LTTR.
Enterohemorrhagic Escherichia coli (EHEC) is the causative agent of outbreaks of hemorrhagic colitis and hemolytic uremic syndrome (HUS) throughout the world. Of the multiple pathogenic serotypes of clinical importance, O157:H7, a serotype that is believed to have evolved recently from an O55:H7 atypical enteropathogenic E. coli (aEPEC) strain, is by far the most prevalent and virulent (18, 34, 40). EHEC strains are part of a larger group of enteric pathogens that includes enteropathogenic E. coli (EPEC), a rabbit EPEC strain, and Citrobacter rodentium, all of which are able to cause attaching and effacing (AE) lesions on intestinal epithelial cells (80).
The genes necessary for the formation of these characteristic AE lesions are chromosomally located within the locus of enterocyte effacement (LEE) pathogenicity island (PAI) (49). The LEE is composed of 41 genes, including the LEE-encoded regulator gene (ler) that activates transcription of all LEE genes (50). The majority of the LEE genes are arranged into five major operons that encode both structural proteins that form a type three secretion system (TTSS) and several of the secreted effectors, such as the translocated intimin receptor (Tir), EspH, and Map, which are translocated through the TTSS into host cells (15, 20, 32, 36). Once translocated, Tir embeds itself in the host membrane, where its extracellular domain serves as a docking point for its cognate bacterial receptor, intimin (also encoded within the LEE), thus allowing for intimate attachment and formation of the characteristic AE lesions on eukaryotic cells (36). Tir, EspH, and Map have also been shown to regulate the length and number of pedestals, as well as eukaryotic filopodium formation, by altered actin assembly (2, 37, 77).
EHEC is able to sense and respond to biotic cues from its environment, such as the human host produced catecholamines epinephrine and norepinephrine, through two two-component systems, QseBC and QseEF (11, 57). EHEC also senses abiotic environmental cues, such as phosphate and sulfate levels, through QseEF (57). Additionally, quorum-sensing (QS) signaling cascades have evolved to sense microbial population density and diversity through the recognition of bacterially produced autoinducers (AI) AI-2 and AI-3 (72, 74). AI-2, the enzymatic product of LuxS, is proposed to promote interspecies signaling in a broad range of bacterial species (75), whereas the breadth of AI-3 signaling has not been as extensively characterized (81). Through the interpretation and integration of these multiple regulatory signaling networks that often involve intracellular regulatory proteins, EHEC is able to regulate the expression of its multiple virulence factors. These factors include the LEE TTSS, Shiga toxin (Stx), the causative agent of HUS, and the expression of flagella through its master regulator flhDC, which allows for bacterial motility (72).
Regulation of the LEE is highly complex and includes the involvement of multiple regulatory proteins and pathways (6, 9, 15, 21, 24, 25, 27, 29, 30, 50, 53, 57, 60, 63, 66, 68-70, 73, 78, 85). Transcription of the ler gene, encoding the master regulator of the LEE (50), is directly activated by the LysR-type transcriptional regulator (LTTR) QseA (67, 68) and negatively activated by the RNA regulatory chaperone Hfq (25, 65). Other posttranscriptional regulatory factors, such as ClpXP, have been shown to increase transcription of LEE3 by inhibiting its repression by GrlR and by increasing the degradation of the EHEC global regulator stationary-phase sigma factor RpoS (17, 29, 70). RpoS is itself under a high level of posttranscriptional control by Hfq (8), translational control by Hfq and the LTTR LrhA (54), and posttranslational control by RssB (45). RssB, whose activity is modulated by LrhA, has been shown to sequester RpoS and targets it for ClpXP degradation (22, 55). Finally, IraD, an RssB antiadaptor protein, is induced in response to DNA damage and prevents RssB-mediated ClpXP degradation of RpoS, thereby altering cellular responses to stress (51, 52, 76).
LTTRs are the largest family of prokaryotic DNA binding regulatory proteins (64). LTTRs control many diverse regulatory pathways, including biofilm formation (4), motility (46), and TTSS (68), in a variety of bacterial groups, including E. coli, Salmonella enterica serovar Typhimurium (44), and Yersinia enterocolitica (5). LTTRs are about 300 amino acids in length and contain two functional domains, an amino-terminal helix-turn-helix (HTH) DNA binding transcriptional regulatory domain and a carboxy-terminal coinducer binding oligomerization domain (48). LTTRs can act as either transcriptional activators or repressors, generally depending upon the location of the HTH domain. In addition, some dual-functioning LTTRs have been described (28). In the classical model for the LTTR's dependent transcription, the LysR protein binds the promoter of the lysR gene and represses its transcription while simultaneously binding the upstream promoter of the divergently transcribed target gene, where, in the presence of coinducer, tetramerization occurs, and transcription is initiated. Recently, the regulatory prowess of LTTRs has been demonstrated to extend beyond the local genetic level, and both the scope and the manner in which these global transcriptional regulators were thought to function have been drastically altered (14). Although LTTRs are known to require homodimer and thereby tetramerization formation in order to regulate transcription, heterodimerization with additional LTTRs has also been predicted (16, 43, 64). Additionally, novel classes of LTTRs, which require octamerization instead of dimerization to alter target gene transcription, are constantly being discovered (61).
Here, we report the identification of an LTTR, YjiE, herein renamed QseD (quorum-sensing E. coli regulator D), which, although prevalent in enterobacteria, seemingly exists almost exclusively in EHEC O157:H7 isolates in a helix-turn-helix (HTH) truncated isoform. This truncated “short” isoform (sQseD) confers EHEC with altered cellular regulatory consequences compared with the full-length K-12 E. coli “long” isoform (lQseD). In EHEC, QseD downregulates the LEE and iraD transcription and alters AE lesion formation, while in K-12 QseD represses motility.
MATERIALS AND METHODS
Strains and plasmids.
The bacterial and Saccharomyces cerevisiae strains, and plasmids utilized in this study are listed in Table 1. All E. coli strains were grown aerobically in Luria-Bertani (LB) broth or Dulbecco's modified Eagle's medium (DMEM) at 37°C unless otherwise stated. All yeast strains were grown aerobically in yeast minimal medium at 30°C as previously described (3) unless otherwise stated. Where appropriate, media were supplemented with ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), kanamycin (50 μg/ml), or tetracycline (10 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| 86-24 | Wild-type EHEC strain (serotype O157:H7) | 23a |
| DH5α | supE44 lacU169 (80 lacZ M15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Stratagene |
| VS94 | 86-24 luxS mutant | 71 |
| BH68 | 86-24 qseD mutant | This study |
| BH85 | BH68 complemented with pBH22 | This study |
| BH86 | BH68 complemented with pBH23 | This study |
| BW25113 (K-12) | rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 13 |
| D7 | BW25113 qseD mutant; Keio collection; yjiE7 | 5a |
| BH51 | D7 complemented with plasmid pBH22 | This study |
| BH52 | D7 complemented with plasmid pBH23 | This study |
| D166 | Michigan State E. coli strain TW05550 | NIAID STEC Center |
| TB226A | Michigan State E. coli strain TW04257 | NIAID STEC Center |
| B2F1 | Michigan State E. coli strain TW07506 | NIAID STEC Center |
| VP08 | Michigan State E. coli strain TW04584 | NIAID STEC Center |
| DEC 16A | Michigan State E. coli strain TW02918 | NIAID STEC Center |
| 78/92 | Michigan State E. coli strain TW05608 | NIAID STEC Center |
| BH294-10 | Michigan State E. coli strain TW02898 | NIAID STEC Center |
| BH262C-8 | Michigan State E. coli strain TW02897 | NIAID STEC Center |
| D55 | Michigan State E. coli strain TW04549 | NIAID STEC Center |
| VP12 | Michigan State E. coli strain TW04588 | NIAID STEC Center |
| 97-3250 | Michigan State E. coli strain TW07814 | NIAID STEC Center |
| 493/89 | Michigan State E. coli strain TW06555 | NIAID STEC Center |
| DEC 5B | Michigan State E. coli strain TW02719 | NIAID STEC Center |
| DEC 5A | Michigan State E. coli strain TW00587 | NIAID STEC Center |
| DEC 5C | Michigan State E. coli strain TW01959 | NIAID STEC Center |
| DEC 5D | Michigan State E. coli strain TW00947 | NIAID STEC Center |
| DEC 5E | Michigan State E. coli strain TW00962 | NIAID STEC Center |
| 5905 | Michigan State E. coli strain TW05353 | NIAID STEC Center |
| G5101 | Michigan State E. coli strain TW05356 | NIAID STEC Center |
| 413 | Michigan State E. coli strain TW05359 | NIAID STEC Center |
| EDL931 | Michigan State E. coli strain TW02303 | NIAID STEC Center |
| EDL932 | Michigan State E. coli strain TW02299 | NIAID STEC Center |
| EDL933 | Michigan State E. coli strain TW02302 | NIAID STEC Center |
| L40 | Yeast reporter strain; MATα trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ | 79 |
| B2HPC | L40 transformed with pLX-YopJ plus pVP-MEKK (yeast 2-hybrid positive control) | 53a |
| B2HNC | L40 transformed with pLX-Laminin plus pVP16 (yeast 2-hybrid negative control) | 53a |
| BH129 | L40 transformed with pVPSA plus pLXSA | This study |
| BH130 | L40 transformed with pVPSA plus pLXLA | This study |
| BH131 | L40 transformed with pVPSA plus pLXSD | This study |
| BH132 | L40 transformed with pVPSA plus pLXLD | This study |
| BH133 | L40 transformed with pVPLA plus pLXSA | This study |
| BH134 | L40 transformed with pVPLA plus pLXLA | This study |
| BH135 | L40 transformed with pVPLA plus pLXSD | This study |
| BH136 | L40 transformed with pVPLA plus pLXLD | This study |
| BH137 | L40 transformed with pVPSD plus pLXSA | This study |
| BH138 | L40 transformed with pVPSD plus pLXLA | This study |
| BH139 | L40 transformed with pVPSD plus pLXSD | This study |
| BH140 | L40 transformed with pVPSD plus pLXLD | This study |
| BH141 | L40 transformed with pVPLD plus pLXSA | This study |
| BH142 | L40 transformed with pVPLD plus pLXLA | This study |
| BH143 | L40 transformed with pVPLD plus pLXSD | This study |
| BH144 | L40 transformed with pVPLD plus pLXLD | This study |
| BH157 | L40 transformed with pVPSL plus pLXSL | This study |
| BH158 | L40 transformed with pVPSL plus pLXLL | This study |
| BH159 | L40 transformed with pVPSL plus pLXSD | This study |
| BH160 | L40 transformed with pVPSL plus pLXLD | This study |
| BH161 | L40 transformed with pVPLL plus pLXSL | This study |
| BH162 | L40 transformed with pVPLL plus pLXLL | This study |
| BH163 | L40 transformed with pVPLL plus pLXSD | This study |
| BH164 | L40 transformed with pVPLL plus pLXLD | This study |
| BH165 | L40 transformed with pVPSD plus pLXSL | This study |
| BH166 | L40 transformed with pVPSD plus pLXLL | This study |
| BH167 | L40 transformed with pVPLD plus pLXSL | This study |
| BH168 | L40 transformed with pVPLD plus pLXLL | This study |
| BH29 | BH68 complemented with pBH24 | This study |
| BH31 | BH68 complemented with pBH26 | This study |
| Plasmids | ||
| pACYC184 | Cloning vector | NEB |
| pBAD-MycHIS (A) | C-terminal Myc-His expression vector | Invitrogen |
| pBH22 | K-12 qseD region inserted into Tet of pACYC184 | This study |
| pBH23 | 8624 qseD region inserted into Tet of pACYC184 | This study |
| pKD46 | λ Red helper plasmid | 13 |
| pKD3 | λ Red template plasmid | 13 |
| pCP20 | λ Red resolvase plasmid | 13 |
| pVP16 | Yeast library plasmid | 79 |
| pLex-ADE | Yeast bait plasmid; pBTM116 with the insertion of ADE2 at the PvuII site | 79 |
| pLX-YopJ | pLEX-ADE expressing YopJ | 53a |
| pVP-MEKK | pVP-16 expressing MEKK | 53a |
| pLX-Laminin | pLEX-ADE expressing laminin | 53a |
| pVPSA | pVP16 expressing “short” QseA | This study |
| pVPLA | pVP16 expressing “long” QseA | This study |
| pVPSD | pVP16 expressing “short” QseD | This study |
| pVPLD | pVP16 expressing “long” QseD | This study |
| pVPSL | pVP16 expressing “short” LrhA | This study |
| pVPLL | pVP16 expressing “long” LrhA | This study |
| pLXSA | pLEX-ADE expressing “short” QseA | This study |
| pLXLA | pLEX-ADE expressing “long” QseA | This study |
| pLXSD | pLEX-ADE expressing “short” QseD | This study |
| pLXLD | pLEX-ADE expressing “long” QseD | This study |
| pLXSL | pLEX-ADE expressing “short” LrhA | This study |
| pLXLL | pLEX-ADE expressing “long” LrhA | This study |
| pET28 | N-terminal His-tagged T7 expression construct | Novagen |
| pBH24 | K-12 QseD in pBAD-MycHIS (A) | This study |
| pBH26 | 86-24 QseD in pBAD-MycHIS (A) | This study |
| pMK208 | QseA-His (N-terminal) in pET28 | This study |
Recombinant DNA techniques.
Standard methods were used to perform plasmid purification, PCR, restriction digestion, ligation, transformation, and gel electrophoresis (62).
Isogenic mutant construction and complementation.
Construction of the isogenic qseD mutant was carried out as previously described (13). Briefly, 86-24 cells containing pKD46 were electroporated with a qseD PCR product generated by primers QseDλRedF and QseDλRedR (Table 2), using pKD3 as a template. After electroporation, cells were incubated at 22°C overnight (OVN) in super optimal broth with catabolite repression (SOC) medium. Cells were then plated on medium containing chloramphenicol (30 μg/ml) and incubated at 22°C overnight once again. Resulting colonies were patched for chloramphenicol resistance and ampicillin sensitivity and PCR verified for the absence of qseD. The chloramphenicol cassette was then resolved using pCP20-mediated recombination, with additional patching for chloramphenicol and ampicillin sensitivity. The generated qseD mutant, BH68, was then complemented with either pBH22 (lQseD) or pBH23 (sQseD) for complementation studies or pBH24 (lQseD) or pBH26 (sQseD) for protein expression and purification. Plasmids pBH22 and pBH23 were constructed from the PCR product of primer set QseDCompF/R, using K-12 or EHEC as a template, and this product was digested with SalI and BamHI and inserted into pACYC184. Plasmids pBH24 and pBH26 were constructed from the PCR product of the primer sets LQseDMycHISF/R and SQseDMycHISF/R, using K-12 or EHEC as a template, and this product was digested with HindIII and NcoI and inserted into pACYC184.
TABLE 2.
Oligonucleotides used in this study
| Primer name | Sequence |
|---|---|
| QseDλRedF | 5′-TTGTCAGCCGCCCTAATGGACGTAGAATGCCCCCAAGGGCGGCTGACAGAGTAAAACGTAGTGTAGGCTGGAGCTGCTTC-3′ |
| QseDλRedR | 5′-CTAACGTTTGATCTGGTCTGGGATAATGGCGGTTGGCGCAGTGCGACGCTTGAGAATGTCCATATGAATATCCTCCTTAG-3′ |
| QseDCompF | 5′-GCGGTCGACTCAGCTAAGCACAATCTC-3′ |
| QseDCompR | 5′-CGGATCCCAAAGACGGCAAAGCCTG-3′ |
| QseDRTF | 5′-CGGAGTATGCCATCCAACAA-3′ |
| QseDRTR | 5′-TCGTCCCGATTCAGCACAA-3′ |
| KptARTF | 5′-GCGATAAAAGCGTTTTAGTTATTCCA-3′ |
| KptARTR | 5′-GAAGTCGAATGCCCCTGAAC-3′ |
| YjiHRTF | 5′-TTTCGCCGCATATCAAACC-3′ |
| YjiHRTR | 5′-GCGACGCCGGAAGAGAA-3′ |
| YjiGRTF | 5′-GCTGCCAAACGTGGTGATG-3′ |
| YjiGRTR | 5′-CGAGCAGGCCGGTAATTTT-3′ |
| IadaRTF | 5′-GCTAATATGGCGGCAGAATCC-3′ |
| IadaRTR | 5′-TGTCGCCCATGTGGAACAC-3′ |
| IraDRTF | 5′-AATGCTGTACCACGACGATGAA-3′ |
| IraDRTR | 5′-GCGCCAACCGCCATTA-3′ |
| YjiCRTF | 5′-GCCCTTTCGATCCTGTTGAG-3′ |
| YjiCRTR | 5′-GCGTAACCTGGAACATTGCA-3′ |
| FliCRTF | 5′-TCCATCGACAAATTCCGTTCT-3′ |
| FliCRTR | 5′-TGGTGACTGCGGAATCCA-3′ |
| MotARTF | 5′-GAAGAGATTGAGACGCACGAAA-3′ |
| MotARTR | 5′-CGACCAGCGCCAGACTGT-3′ |
| LQseDMycHISF | 5′-CCCCATGGATGACTGTGGTGCG-3′ |
| LQseDMycHISR | 5′-GCGAAGCTTGCTAAGCACAATCTCCAG-3′ |
| SQseDMycHISF | 5′-CCCCATGGTGACGCCGCTGCAACTC-3′ |
| SQseDMycHISR | 5′-GCGAAGCTTGCTAAGCACAATCTCCAG-3′ |
| QseDOperF | 5′-CGGCGGCATCTGGCTGATAATG-3′ |
| QseDOperR | 5′-GTTCAGGCGGGTATTCCGCTGG-3′ |
| QseDSequF | 5′-CGAAATTCTGCCAGGCAATGACGCAGAC-3′ |
| QseDSequR | 5′-GCTGGTATTGCCGTCAGCTGCG-3′ |
| VPLEXlQseDF | 5′-GGGGGATCCTTATGGATGACTGTGGTGCG-3′ |
| VPLEXsQseDF | 5′-GGGGGATCCTTGTGACGCCGCTGCAACTC-3′ |
| VPslQseDR | 5′-GGGGCGGCCGCTCAGCTAAGCACAATCTC-3′ |
| LEXslQseDR | 5′-GGGCTGCAGTCAGCTAAGCACAATCTCCAG-3′ |
| VPLEXlQseAF | 5′-GGGGGATCCTTATGGAACGACTAAAACGCATGTCGG-3′ |
| VPLEXsQseAF | 5′-GGGGGATCCTTGGCTGCCGTCGTATGCTTCATGAAG-3′ |
| VPslQseAR | 5′-GGTGCGGCCGCTTACTTCTCTTTCCCGCG-3′ |
| LEXslQseAR | 5′-GGGCTGCAGTTACTTCTCTTTCCCGCGCCC-3′ |
| VPLEXlLrhAF | 5′-GGGGGATCCTTATGATAAGTGCAAATCGTCCG-3′ |
| VPLEXsLrhAF | 5′-GGTGGATCCTTGCCAGGAAAATCCTGCG-3′ |
| VPslLrhAR | 5′-GGTGCGGCCGCTTACTCGATATCCCTTTCAATC-3′ |
| LEXslLrhAR | 5′-GGGCTGCAGTTACTCGATATCCCTTTCAATCAAC-3′ |
RNA extraction and quantitative real-time RT-PCR.
Cultures were grown aerobically in LB medium at 37°C overnight, diluted 1:100 in LB (for flhD, fliC, and motA) or DMEM (for ler, escV, escC, espA, stx2a, and nleA), and grown aerobically at 37°C. RNA from three biological replicate cultures of each strain was extracted at lag phase (optical density at 600 nm [OD600] of 0.2), mid-log phase (OD600 of 0.5), late log phase (OD600 of 1.0), and stationary/death phase (OD600 of 1.5) using a RiboPure bacterial RNA isolation kit (Ambion) according to the manufacturer's guidelines. Primers were designed using Primer Express version 1.5 (Applied Biosystems) (Table 2) with the primers for the LEE genes, and rpoA (82), stx2a, and nleA (27) have previously been described. Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed in a one-step reaction using an ABI 7500 sequence detection system (Applied Biosystems). The amplification efficiencies for all primer sets were validated by standard curves with various concentrations of the RNA template. To ensure template specificity, products were heated to 95°C for 15 s, cooled to 60°C, and heated to 95°C while fluorescence was monitored. Relative quantification analysis was used to compare gene expression in BH68, BH85, and BH86 with that in 86-24 Escherichia coli. The parameters for cDNA generation and amplification were as follows: 1 cycle at 48°C for 30 min, 1 cycle at 95°C for 10 min, and 40 cycles at 95°C for 15 s and 60°C for 1 min. The rpoA (RNA polymerase subunit A) gene was used as the endogenous control. Each 20-μl reaction mixture consisted of 10 μl of 2× SYBR master mix, 0.1 μl of Multiscribe reverse transcriptase (Applied Biosystems), 0.1 μl of RNase inhibitor (Applied Biosystems), 2 μl of target RNA (50 ng/μl), and 8 μl of double-distilled water (ddH2O). Expression is shown in graphs as n-fold change in expression level compared with wild-type (WT) levels. The error bars in the figures represent the standard deviations of the ΔΔCT value. The Student t test was performed to assess statistical significance. A P value of less than 0.05 was considered significant.
Motility assays.
Assays were performed as previously described (12). Briefly, static overnight (OVN) cultures were stabbed on tryptone soft agar (1% tryptone, 0.25% NaCl, and 0.3% agar) motility plates and incubated at 37°C, with motility halos being measured at 8 h.
Microarrays.
Microarrays and analysis were performed as previously described (35). The GeneChip E. coli Genome 2.0 array (Affymetrix) was used to compare the gene expression in strain BH68 to that in 86-24 and to compare that in D7 to that in strain BW25113. The array includes 10,208 probe sets for all 20,366 genes present in the following four strains of E. coli: K-12 MG1655 (laboratory strain), CFT073 (uropathogenic strain), EDL933 (O157:H7 enterohemorrhagic strain), and Sakai (O157:H7 enterohemorrhagic strain) (Affymetrix). The RNA-processing, labeling, hybridization, and slide-scanning procedures were preformed as described in the Affymetrix Gene Expression Technical Manual (www.affymetrix.com). The output scan from each single replicate array was obtained using GCOS version 1.4 according to the manufacturer's instructions. Data were normalized using robust multiarray analysis at the RMAExpress website (http://rmaexpress.bmbolstad.com/). The resulting data were compared to determine features whose expression levels were increased or decreased in response to inactivation of the qseD gene. Custom analysis scripts were written in Perl to complete multiple array analyses. We note that the isolate used in these studies has not been sequenced and thus is not fully contained on the array and that differences in genome content are evident. Expression data can be accessed using accession number GS20413 at the NCBI GEO database.
Whole-cell lysate immunoblotting.
Whole-cell lysates from WT EHEC 86-24, BH68, BH85, BH86, BW25113 (K-12), D7, BH51, and BH52 were prepared by sonication from strains grown in LB (FliC) or DMEM (EspA and EspB) to an OD600 of 1.0. SDS-PAGE and immunoblotting were completed as previously described (62). Protein concentration was determined using a NanoDrop spectrophotometer (Thermo Scientific). Samples were probed by Western blot analysis using polyclonal antisera against EspA, EspB, and FliC (a gift from James B. Kaper, University of Maryland) and monoclonal antisera against RpoA (Neoclone). Ponceau Red staining was used to visualize bovine serum albumin (BSA) loading controls.
AE lesion FAS.
Fluorescent-actin staining (FAS) was performed as previously described (39). Briefly, overnight cultures that were grown aerobically in LB at 37°C were diluted 1:100 and used to infect confluent monolayers of HeLa cells grown on glass coverslips at 37°C and 5% CO2. Cell infections were allowed to progress for 6 h at 37°C and 5% CO2. At 6 h, the coverslips were washed, fixed, permeabilized with 0.2% Triton X-100, and treated with fluorescein isothiocyanate (FITC)-labeled phalloidin to visualize actin accumulation, and propidium iodide (PI) was added to stain bacteria. Samples were visualized by immunofluorescence with a Zeiss Axiovert microscope. The entire field of at least six coverslips from each strain was examined, and images of AE lesions were taken and processed using ImageJ.
Protein purification.
One liter each of LB medium was inoculated from BH29, BH31, and MK208 LB overnight growths at 1:100 and grown at 37°C to an OD600 of 0.6. The cultures were then induced with either 400 μM IPTG (Sigma) or 0.2% arabinose and grown for 6 h at 25°C. Cells were harvested, suspended in lysis buffer (50 mM phosphate buffer [pH 8], 1 M NaCl, and 20 mM imidazole), and lysed by homogenization. The cell lysates were centrifuged and loaded onto to a Ni2+-nitrilotriacetic acid (NTA)-agarose gravity column (Qiagen). The column was washed with lysis buffer, and protein was eluted with elution buffer (50 mM phosphate buffer [pH 8], 1 M NaCl, 250 mM imidazole). Fractions containing purified protein were confirmed by SDS-PAGE and concentrated for further use.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were used as previously described (27) to explore the possible binding of either isoform of QseD to the ler promoter. Briefly, a ler DNA probe was generated with primers Ler-173 and Ler-42 (67), using 86-24 as a template. DNA probes were then end labeled with [γ-32P]ATP (NEB) using T4 polynucleotide kinase, using standard procedures (62). End-labeled fragments were run on a 5% polyacrylamide gel, excised, and purified using a PCR purification kit (Qiagen). EMSAs were performed by adding increasing amounts of purified QseA (MK208) or QseD (BH29 and BH31) protein to end-labeled probe (20 ng) in binding buffer [500 μg/ml BSA (NEB), 300 ng/μl poly(dI-dC), 30 mM Tris-HCl (pH 7.4), 1 mM EDTA, 50 mM KCl, 5 mM NaCl] for 30 min at 22°C. In competitions, both proteins (QseA and QseD) were mixed and incubated on ice for 20 min before addition to probe and binding buffer. Immediately before the loading, a 5% Ficoll solution was added to the mixtures. The reactions were electrophoresed for approximately 14 h at 65 V on a 5% polyacrylamide gel, dried, and exposed to Kodak X-OMAT film.
Y2H analysis.
A yeast two-hybrid (Y2H) analysis was used as previously described (79) to test for possible protein-protein interaction between QseD and the LysR-like family members QseA and LrhA. Briefly, the yeast report strain L40 was transformed (23) with all combinations of the short (“s”) and long (“l”) variants of QseD, QseA, and LrhA in both the bait (pLEX-ADE) and the library (pVP16) vectors. Plasmids pVPLD and pLXLD were constructed from the PCR products of the primer VPLEXlQseDF with VPslQseDR or LEXslQseDR, using K-12 as a template, and these products were digested by BamHI with NotI or PstI and inserted into pVP16 or pLEX-ADE. Plasmids pVPSD and pLXSD were constructed from the PCR products of the primer VPLEXsQseDF with VPslQseDR or LEXslQseDR, using EHEC as a template, and these products were digested by BamHI with NotI or PstI and inserted into pVP16 or pLEX-ADE. Plasmids pVPLA and pLXLA were constructed from the PCR products of the primer VPLEXlQseAF with VPslQseAR or LEXslQseAR, using K-12 as a template, and these products were digested by BamHI with NotI or PstI and inserted into pVP16 or pLEX-ADE. Plasmids pVPSA and pLXSA were constructed from the PCR products of the primer VPLEXsQseAF with VPslQseAR or LEXslQseAR, using EHEC as a template, and these products were digested by BamHI with NotI or PstI and inserted into pVP16 or pLEX-ADE. Plasmids pVPLL and pLXLL were constructed from the PCR products of the primer VPLEXlLrhAF with VPslLrhAR or LEXslLrhAR, using K-12 as a template, and these products were digested by BamHI with NotI or PstI and inserted into pVP16 or pLEX-ADE. Plasmids pVPSL and pLXSL were constructed from the PCR products of the primer VPLEXsLrhAF with VPslLrhAR or LEXslLrhAR, using EHEC as a template, and these products were digested by BamHI with NotI or PstI and inserted into pVP16 or pLEX-ADE. Dual transformants were selected on yeast minimal medium lacking leucine, and tryptophan and then assayed for protein-protein interactions using the integrated lacZ and HIS3 reporters. Association of the two protein fusions was determined by growth on yeast minimal medium lacking histidine. In this manner, all combinations of QseD, QseA, and LrhA (see Table 6) were screened for protein-protein interactions.
TABLE 6.
Summary of yeast two-hybrid results
| Strain | Construct | Growth on His− medium | Result for blue coloniesa |
|---|---|---|---|
| BH125 | VP-LXSA | ||
| BH126 | VP-LXLA | ||
| BH127 | VP-LXSD | + | |
| BH128 | VP-LXLD | ||
| BH129 | VPSA-LXSA | Yes | +++ |
| BH130 | VPSA-LXLA | Yes | +++ |
| BH131 | VPSA-LXSD | ++ | |
| BH132 | VPSA-LXLD | ||
| BH133 | VPLA-LXSA | Yes | +++ |
| BH134 | VPLA-LXLA | Yes | +++ |
| BH135 | VPLA-LXSD | ++ | |
| BH136 | VPLA-LXLD | ||
| BH137 | VPSD-LXSA | ||
| BH138 | VPSD-LXLA | ||
| BH139 | VPSD-LXSD | Yes | +++ |
| BH140 | VPSD-LXLD | Yes | +++ |
| BH141 | VPLD-LXSA | ||
| BH142 | VPLD-LXLA | ||
| BH143 | VPLD-LXSD | + | |
| BH144 | VPLD-LXLD | ||
| BH155 | VP-LXSL | Yes | ++ |
| BH156 | VP-LXLL | ++ | |
| BH157 | VPSL-LXSL | Yes | +++ |
| BH158 | VPSL-LXLL | Yes | +++ |
| BH159 | VPSL-LXSD | ||
| BH160 | VPSL-LXLD | ++ | |
| BH161 | VPLL-LXSL | Yes | +++ |
| BH162 | VPLL-LXLL | Yes | +++ |
| BH163 | VPLL-LXSD | + | |
| BH164 | VPLL-LXLD | ||
| BH165 | VPSD-LXSL | Yes | ++ |
| BH166 | VPSD-LXLL | ++ | |
| BH167 | VPLD-LXSL | Yes | ++ |
| BH168 | VPLD-LXLL | ++ |
The symbols indicate the intensity and incubation time required for chromogenic substrate (X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside]) conversion, as follows: +++, intense and requiring under 30 min; ++, somewhat intense and requiring 2 to 3 h; and +, faint and requiring overnight incubation.
RESULTS
Identification of the quorum-sensing regulated LysR QseD.
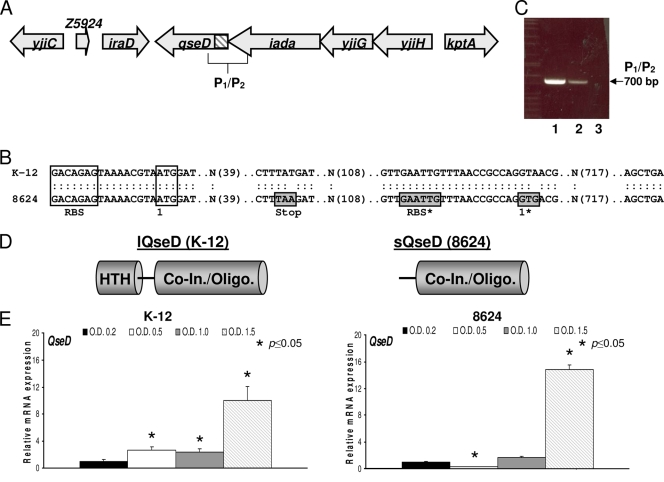
The gene yjiE encodes a 34.7-kDa putative LysR-type transcriptional regulator (LTTR) that is found at the end of a four-gene operon (Fig. 1 A). This gene was originally identified in a microarray study as being repressed by QseBC and in a spotted array as being transcriptionally regulated by LuxS in a quorum-sensing-dependent manner (27, 71). yjiE is found throughout the Enterobacteriaceae family, but it contains a point mutation generating a stop codon and was predicted to be a pseudogene almost exclusively in EHEC O157:H7 and its suggested parent EPEC O55:H7 (Fig. 1B and Table 3). Upon closer examination, we noticed that downstream of the stop codon in EHEC there was a compensatory mutationthat generated an alternative translational start site (site 1) following a ribosome binding site (RBS) (Fig. 1B). Therefore, to confirm that yjiE was still transcribed as part of a functional operon in EHEC, we performed RT-PCR using a primer set, P1/P2, that flanked the untranslated region (one primer within the upstream iadA gene and the second within yjiE) and that should generate an ∼700-bp product. PCR products of the predicted size were observed with the use of both extracted genomic DNA (gDNA; positive PCR control) and cDNA generated from RNA as a template, thus confirming that yjiE is still cotranscribed within its operon in EHEC (Fig. 1C). We therefore reasoned that yjiE may still generate a protein product in EHEC, although truncated and missing an HTH domain compared to K-12 E. coli, which could lead to altered gene regulation (Fig. 1D).
FIG. 1.
In EHEC 86-24, the intact qseD operon encodes a truncated QseD protein. (A) Cartoon representation of the qseD operon (untranslated region shaded) and the surrounding genes. (B) Comparison of the qseD sequences of EHEC 86-24 and K-12 E. coli, where boxed regions include the ribosome binding site (RBS) and the translational start site, and boxed and shaded regions include the stop codon, the alternative RBS, and the translational start codon in EHEC 86-24. (C) RT-PCR using the P1/P2 primer set and either cDNA (1), gDNA (2), or RNA (3) harvested and/or synthesized from EHEC 86-24 as a PCR template. (D) Cartoon representation of the full-length QseD (lQseD) and truncated QseD (sQseD) protein products from K-12 E. coli and EHEC 86-24, respectively. In EHEC, loss of the translated helix-turn-helix (HTH) domain does not appear to affect translation of the cofactor recognition/oligomerization domain (Co-In./Oligo.). (E) qRT-PCR of qseD expression levels in DMEM at lag phase (OD600 of 0.2), mid-log phase (OD600 of 0.5), late log phase (OD600 of 1.0), and stationary phase (OD600 of 1.5) in K-12 E. coli and EHEC 86-24.
TABLE 3.
Strain/serotype distribution of QseD isoformsa
| Organism group | Strain | Serotype | QseD isoform |
|---|---|---|---|
| EHEC | 86-24 | O157:H7 | S |
| EHEC | Sakai | O157:H7 | S |
| EHEC | EDL933 | O157:H7 | S |
| EHEC | “Spinach” | O157:H7 | S |
| EHEC | EC4115 | O157:H7 | S |
| EHEC | TW14588 | O157:H7 | S |
| EHEC | TW05356 G5101 | O157:H7 | S |
| EHEC | TW02303 EDL931 | O157:H7 | S |
| EHEC | TW02299 EDL932 | O157:H7 | S |
| EHEC | TW02302 EDL933 | O157:H7 | S |
| EHEC | TW05359 413 | O157:HNM | S |
| EHEC | TW06555 493/89 | O157:H- | S |
| EHEC | TW02719 DEC 5B | O55:H7 | S |
| EHEC | TW00587 DEC 5A | O55:H7 | S |
| EHEC | TW01959 DEC 5C | O55:H7 | S |
| EHEC | TW00947 DEC 5D | O55:H7 | S |
| EHEC | TW05353 5905 | O55:H7 | S |
| UPEC | UTI89 | O18:K1:H7 | S* |
| APEC | O1 | O1:K1:H7 | S* |
| K-12 E. coli | MG1655 | OR:H48:K- | L |
| K-12 E. coli | BW25113 | OR:H48:K- | L |
| “Commensal” E. coli | HS | O9:H4 | L |
| EHEC | TW02897 BH262C-8 | O142:H10 | L |
| EHEC | TW05550 D166 | O121:H- | L |
| EHEC | TW02918 DEC 16A | O113:H21 | L |
| EHEC | 11128 | O111:H- | L |
| EHEC2 | TW04257 TB226A | O111:H- | L |
| EHEC2 | TW07506 B2F1 | O111:H- | L |
| EHEC2 | TW05608 78/92 | O111:H- | L |
| EHEC | 12009 | O103:H2 | L |
| EHEC | TW02898 BH294-10 | O76:H7 | L |
| EHEC | TW00962 DEC 5E | O55:H7 | L |
| EHEC | TW07814 97-3250 | O26:H11 | L |
| EHEC | 11368 | O26:H11 | L** |
| EHEC | TW04588 VP12 | O26:H- | L |
| EHEC | TW04584 VP08 | O26:H- | L |
| EPEC | E2348/69 | O127:H6 | L |
| EPEC | TW04549 D55 | O127:H- | L |
| EPEC | 11128 | O111:H- | L |
| aEPEC (atypical) | E110019 | O111:H9 | L |
| ETEC | B7A | O148:H28 | L |
| ExPEC | S88 | O45:K1:H7 | L |
| ExPEC | UMN026 | O17:K52:H18 | L |
| ExPEC | IAI39 | O7:K1 | L |
| EAEC | 55989 | L | |
| UPEC | 536 | O6:K15:H31 | L |
| UPEC | CTF073 | O6:K2:H1 | L** |
| Shigella flexneri 5 | 8401 | L | |
| Shigella boydii | CDC 3083-94 | L | |
| Shigella flexneri 2a | 301 | L | |
| Shigella boydii | Sb227 | L |
S, short; L, long; *, alternative A → T stop codon generating a point mutation upstream of the one herein described for all other short QseDs that should also generate the same truncated protein product; **, homologous QseD protein products with an extended C terminus; EHEC2, EHEC cluster 2; ExPEC, extraintestinal pathogenic E. coli.
To confirm that transcription of the yjiE gene was regulated in a cell density-dependent manner, we performed quantitative real-time RT-PCR (qRT-PCR) using cDNA synthesized from RNA extracted from both WT K-12 E. coli and WT EHEC during lag phase growth (OD600 of 0.2), mid-log-phase growth (OD600 of 0.5), late-log-phase growth (OD600 of 1.0), and stationary-phase growth (OD600 of 1.5), all qRT-PCRs were normalized against rpoA as an internal control. Transcription of yjiE in K-12 E. coli increases throughout growth, peaking during stationary phase, while in EHEC transcript levels drop temporarily during mid-log phase and then similarly peak during stationary phase (Fig. 1E). These results suggest that this gene is regulated in a cell density-dependent manner, and thus we renamed yjiE qseD (quorum-sensing E. coli regulator D).
QseD alters gene transcription.
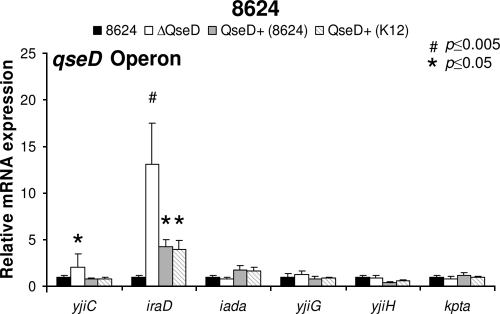
LTTRs are known to positively and/or negatively regulate transcription at both the local and the global genetic levels. To determine QseD's regulatory role in EHEC gene expression and/or pathogenesis, we constructed a nonpolar mutation in qseD and examined its effect on the expression of genes cotranscribed with or adjacent to qseD in the genome by qRT-PCR. QseD does not autoregulate the transcription of its own operon; however, it does repress the transcription of the adjacent genes yjiC and iraD, as their transcripts were upregulated 2-fold and 13-fold in the qseD mutant, respectively (Fig. 2). While YjiC has no predicted homology to any known protein, IraD has recently been demonstrated to prevent degradation of stress-alternative sigma factor RpoS by RssB sequestration, leading to an altered bacterial stress response and increased mutation rates (76, 84).
FIG. 2.
QseD regulates the qseD operon and surrounding genes in E. coli. qRT-PCR of the qseD operon (iadA, yjiG, and yjiH) and the surrounding genes (kptA, yjiC, and yjiD) in EHEC 86-24, the ΔqseD mutant, the ΔqseD mutant complemented in trans with qseD (86-24), and the ΔqseD mutant complemented in trans with qseD (K-12) grown in DMEM (OD600 of 1.0).
An E. coli K-12 qseD mutant was also constructed in order to further broaden the scope of our analysis and to compare and contrast the effects of the HTH truncation in EHEC on genetic regulation. To search for targets of QseD regulation, a transcriptome approach (data can be assessed using GEO database number GS20413) using the Affymetrix E. coli 2.0 microarrays was used to compare expression profiles of both WT EHEC and WT K-12 E. coli to their corresponding isogenic qseD mutants. These arrays contain ∼10,000 probe sets, covering the genomes of two sequenced EHEC strains (EDL933 and Sakai), K-12 strain MG1655, uropathogenic E. coli (UPEC) strain CFT073, and 700 probes to intergenic regions that can carry nonannotated ORFs or small regulatory RNAs. In EHEC during growth in DMEM, a condition known to induce virulence gene expression, 477 probe sets were upregulated (93 EHEC specific), and 505 were downregulated (128 EHEC specific) in the qseD mutant (Table 4). The largest number of genes with altered expression were also found in the E. coli K-12 strain MG1655 genome (43%), which represent a common E. coli backbone conserved among E. coli pathovars (Table 5) (56). In comparison, in K-12 E. coli grown in DMEM, there were considerably fewer genes with altered expression levels, with 109 genes upregulated and 135 genes downregulated in the qseD mutant. This trend was partially reversed when the same mutant was grown in LB, where there were 150 genes upregulated and 321 genes downregulated. Taken together, these data suggest that the truncated QseD EHEC isoform has a greater regulatory repertoire than full-length QseD and that a larger proportion of these genes are on the conserved E. coli backbone.
TABLE 4.
Comparison of WT EHEC 86-24 and K-12 BW25113 to their respective ΔqseD mutants under various growth conditionsa
| Growth condition | No. of genes showing: |
||||
|---|---|---|---|---|---|
| Increase | Marginal increase | Decrease | Marginal decrease | No change | |
| EHEC-DMEM | 477 | 368 | 505 | 272 | 8,586 |
| K-12-DMEM | 109 | 337 | 135 | 517 | 9,110 |
| K-12-LB | 150 | 515 | 321 | 908 | 8,314 |
“Increase” and “decrease” represent changes in expression level of at least 2-fold. “Marginal increase” and “marginal decrease” represent changes that are either less than 2-fold or designated as such by the Affymetrix analysis software program GCOS (version 1.4). Both EHEC and K-12 were grown to an OD600 of 1.0 in either low-glucose DMEM or LB medium.
TABLE 5.
Pathovar distribution of altered gene expression in ΔqseD mutants under various growth conditionsa
| Growth condition and result | No. of genes |
||||
|---|---|---|---|---|---|
| MG1655 | EDL933 | Sakai | CTF073 | Intergenic | |
| EHEC 86-24-DMEM | |||||
| Decrease | 151 | 128 | 29 | 114 | 54 |
| Marginal Decrease | 93 | 80 | 19 | 48 | 30 |
| Increase | 222 | 93 | 21 | 85 | 30 |
| Marginal increase | 213 | 85 | 11 | 32 | 26 |
| No change | 3,391 | 1,401 | 293 | 2,207 | 1,157 |
| K-12 BW25113-DMEM | |||||
| Decrease | 88 | 9 | 6 | 16 | 11 |
| Marginal decrease | 383 | 27 | 8 | 47 | 41 |
| Increase | 79 | 4 | 0 | 15 | 10 |
| Marginal increase | 240 | 10 | 4 | 42 | 40 |
| No change | 3,280 | 1,737 | 355 | 2,366 | 1,195 |
| K-12 BW25113-LB | |||||
| Decrease | 124 | 32 | 7 | 67 | 89 |
| Marginal decrease | 589 | 53 | 13 | 96 | 156 |
| Increase | 121 | 9 | 0 | 12 | 7 |
| Marginal increase | 431 | 9 | 9 | 26 | 17 |
| No change | 2,805 | 1,684 | 344 | 2,285 | 1,028 |
“Increase” and “decrease” represent changes in expression level of at least 2-fold. “Marginal increase” and “marginal decrease” represent changes that are either less than 2-fold or designated as such by the Affymetrix analysis software program GCOS (version 1.4). Both EHEC and K-12 were grown to an OD600 of 1.0 in either low-glucose DMEM or LB medium.
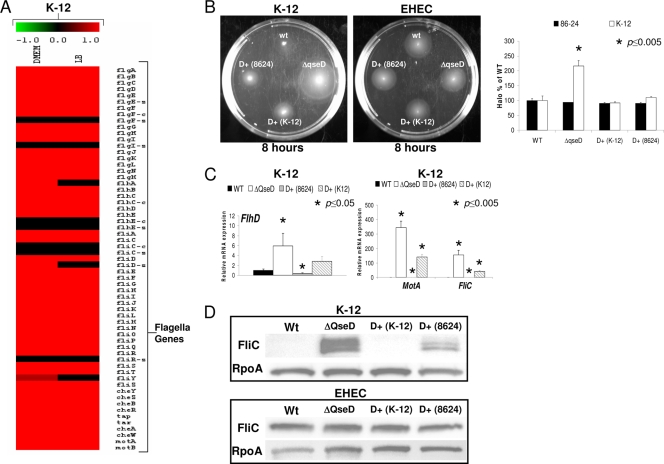
QseD also differentially regulates genetic pathways in EHEC compared to K-12 E. coli. One of the most striking examples of this differential regulation is the flagellar regulon, which while unaltered in the EHEC qseD mutant is upregulated in the K-12 qseD mutant (Fig. 3 A). Array findings were validated by observation of increased motility halos for the K-12, but not the EHEC, qseD mutants in tryptone soft agar motility plates (Fig. 3B). In congruence with the motility studies, we observed increased transcription of the flhD, motA, and fliC flagellar genes by qRT-PCR in the K-12 qseD mutant (Fig. 3C), as well as increased levels of FliC on whole-cell lysates of the K-12 qseD mutant, but not the EHEC qseD mutant, by Western blot analysis (Fig. 3D). The observation that both isoforms of QseD (short EHEC QseD and long K-12 QseD) were able to complement the K-12 qseD mutant in trans (Fig. 3) suggest that both isoforms are still able to regulate transcription of the flagellar regulon in K-12. The differential regulation between the flagellar regulons of EHEC and K-12 by QseD may not be a result of the different isoforms of this protein but could be a result of the presence of an insertion sequence (IS) in the flhDC regulatory region of K-12 E. coli, which is absent in EHEC. The presence of this IS has been shown to alter flhDC transcription and consequently motility in E. coli K-12 (7).
FIG. 3.
QseD affects motility in K-12 E. coli but not in EHEC 86-24. (A) Heat maps generated from microarray analysis depicting the differential regulations of the flagellar regulon in K-12 ΔqseD versus WT K-12 E. coli. (B) Motility plates of WT K-12 E. coli and WT EHEC 86-24 and their corresponding ΔqseD mutants, ΔqseD mutants complemented in trans with qseD (K-12), and ΔqseD mutants complemented in trans with qseD (86-24). Graphs plotting the measurement of the diameters of motility halos of three separate experiments are shown. (C) qRT-PCR of flhD, motA, and fliC in WT K-12 E. coli, the ΔqseD mutant, the ΔqseD mutant complemented in trans with qseD (K-12), and the ΔqseD mutant complemented in trans with qseD (86-24) grown in LB (OD600 of 1.0). (D) Western blots of FliC from WT K-12 E. coli and WT EHEC 86-24 and their corresponding ΔqseD mutants, ΔqseD mutants complemented in trans with qseD (K-12), and ΔqseD mutants complemented in trans with qseD (86-24).
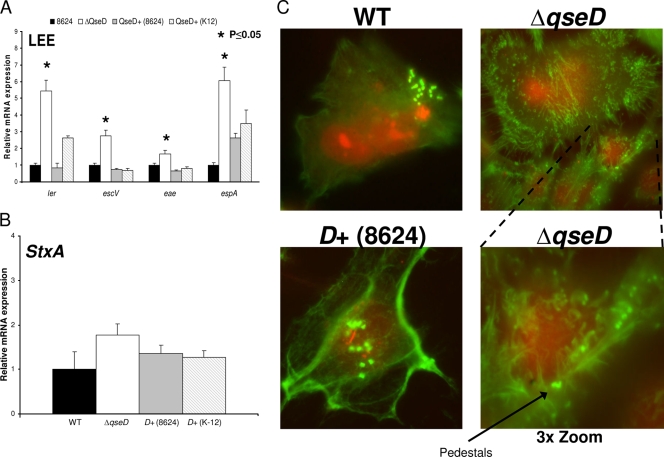
In addition to differentially regulating the flagellar regulon between EHEC and K-12, QseD also repressed the expression of pathogenesis-specific pathways in EHEC, absent in K-12, such as the expression of the LEE genes (Fig. 4). The EHEC qseD mutant exhibited upregulation of the expression of the ler (located within the LEE1 operon), escV (LEE3), espA (LEE4), and eae (LEE5) genes but no effect on stx2a (encoding Shiga toxin) gene expression (Fig. 4A and B). Here again, both isoforms of QseD were able to complement this mutation in trans.
FIG. 4.
QseD regulates the LEE pathogenicity island but not Stx in EHEC 86-24. (A) qRT-PCR of ler, escV, escC, and espA in WT EHEC 86-24, the ΔqseD mutant, the ΔqseD mutant complemented in trans with qseD (86-24), and the ΔqseD mutant complemented in trans with qseD (K-12) grown in DMEM (OD600 of 1.0). (B) qRT-PCR of stx2a in WT EHEC 86-24, the ΔqseD mutant, the ΔqseD mutant complemented in trans with qseD (86-24), and the ΔqseD mutant complemented in trans with qseD (K-12) grown in DMEM (OD600 of 1.0). (C) FAS assays depicting formation of AE lesions on HeLa cell monolayers by WT EHEC, the EHEC ΔqseD mutant, and the EHEC ΔqseD mutant complemented in trans with qseD (86-24). The bottom panel shows the EHEC ΔqseD mutant at 3× zoom.
As QseD represses transcription of ler, and therefore LEE expression in vitro, we then assessed AE lesion formation in the EHEC qseD mutant by fluorescence microscopy, as previously described (38). We observed that while the mutant was still able to form actin pedestals, deregulation of normal AE lesion formation was occurring. HeLa cells infected with the EHEC qseD mutant exhibited increased cellular filopodium formation, compared to cells infected with the WT and complemented strains (Fig. 4C). For HeLa cells infected by EHEC, it has been reported that the LEE-encoded TTSS effectors Tir, EspH, and Map can lead to alterations in filopodium formation and pedestal elongation (37, 77). Given that the expression of the genes encoding these effectors is controlled by Ler (19) and that transcription of ler is repressed by QseD, the alterations in the cellular architecture of HeLa cells infected by the qseD mutant could be due to overexpression of these secreted effectors in this mutant. These results suggest that QseD is a repressor of EHEC virulence expression.
Differential mechanisms of ler transcriptional regulation by lQseD and sQseD.
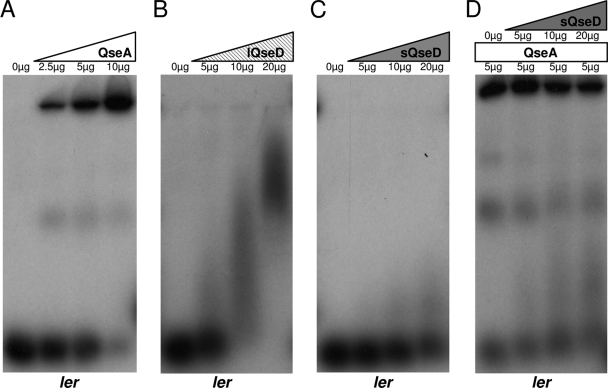
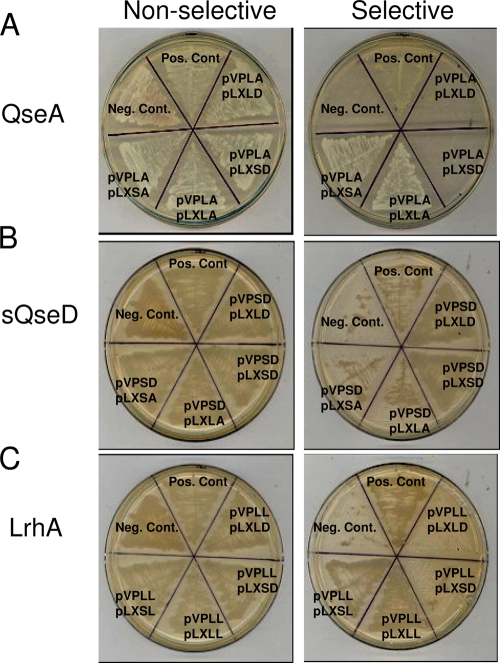
To investigate how both the long and short (HTH domain-truncated) forms of QseD are able to regulate LEE transcription in EHEC, we performed EMSAs to look for potential direct DNA binding. We hypothesized that QseD may bind to the same regulatory region of the ler promoter (positions −173 to −42) as the LTTR QseA (67). We observed that both purified QseA and lQseD directly bound to the ler promoter but that purified sQseD did not (Fig. 5 A to C). These results were expected, given that sQseD lacks the HTH domain necessary for DNA binding. While the downregulation of LEE transcription by lQseD can be accounted for by direct binding to the ler promoter, a mechanism for the downregulation of LEE transcription by sQseD was not apparent. LTTRs are known to require dimer formation in order to bind DNA and regulate transcription. To date, most studies assessed the need of homodimerization of LTTRs for function, but heterodimerization with additional LTTRs has been predicted in silico (16, 43, 64). We therefore hypothesized that HTH-truncated sQseD might be more promiscuous in its protein-protein interactions and that potential heterodimers with known LEE-regulating LTTRs, such as QseA (67) and LrhA (26), could account for LEE regulation in the absence of direct DNA binding by sQseD. However, we did not observe any significant inhibition of binding to ler by QseA when QseA was preincubated with purified sQseD (Fig. 5D). To test this hypothesis, we assessed whether sQseD could form heterodimers with LhrA and/or QseA using a yeast two-hybrid (Y2H) system. The yeast two-hybrid data showed that LhrA, QseA, and QseD form homodimers, which is an expected result, but that sQseD or LQseD could not form heterodimers with either LhrA nor QseA (Table 6 and Fig. 6 A to C). These results agree with our inability to identify interaction partners between QseA and LhrA in sQseD affinity-tagged pulldown experiments (data not shown). We therefore concluded that sQseD does not form protein-protein interactions with these known LEE LTTR regulators and must therefore regulate LEE transcription through interactions with another, yet-unidentified LTTR.
FIG. 5.
K-12 E. coli lQseD binds to the ler promoter. Shown are EMSAs of the ler promoter with purified QseA (A), K-12 E coli lQseD (B), EHEC sQseD (C), and an attempt to prevent QseA binding of ler by heterodimer formation with sQseD (D).
FIG. 6.
EHEC 86-24 sQseD and K-12 E. coli lQseD do not interact with QseA. Shown are representative yeast two-hybrid nonselective (His+) plates and selective (His−) plates, depicting the potential protein-protein interactions of the LysR-like proteins QseA (A), sQseD (B), and LrhA (C).
DISCUSSION
Genetic regulation in human pathogens is a complex process, and regulation in EHEC is no exception. EHEC contains multiple levels of hierarchical signal transduction pathways that converge with intracellular regulatory proteins in order to alter genetic expression. In EHEC, virulence factor expression is regulated through the sensing of epinephrine, norepinephrine, and AI-3 by the two two-component sensor kinases QseC and QseE (11, 57). These signals are then integrated using intracellular regulatory proteins, such as the LRRT QseA, in a complex process where numerous aspects remain unresolved.
Here, we describe QseD, a cell-to-cell-communication-regulated LTTR in EHEC. QseD expression is most highly induced in stationary phase. In EHEC, QseD represses expression of iraD, the LEE, and alters AE lesion formation (Fig. 2 and 4). However, in K-12 E. coli, but not in EHEC, QseD downregulates expression of the flagellar regulon, decreasing motility (Fig. 3). Flagellar expression is regulated in a hierarchical fashion, coupled with this machine's assembly in E. coli (10). The decision to make a flagellum starts at the level of regulation of the flhDC genes, encoding the flagellar master regulators. Most of the transcriptional regulation of the flagellar regulon occurs at the flhDC regulatory region (10). It should be noted that there are considerable differences in the flhDC regulatory regions between EHEC and K-12, with K-12 having an IS inserted within this region, which alters flhDC transcription and consequently motility (7). While we originally hypothesized that the genetic regulatory differences in the two E. coli backgrounds could be explained by the lack of an HTH domain on the EHEC sQseD isoform, complementation studies demonstrated that both isoforms of the protein were able to complement flagellar gene expression in K-12, where differences in the flhDC regulatory region may be the underlying reason of this differential regulation between EHEC and K-12.
This trend in which both isoforms of QseD are able to rescue QseD-regulated phenotypes extends to regulation of iraD (Fig. 2) and the LEE (Fig. 3) in EHEC, arguing that although O157:H7 E. coli possess a truncated form of QseD, this regulatory protein is still regulating targets similar to those regulated by full-length QseD. It is worth noting that genetic complementation with sQseD (Fig. 3C and 4A) for some of these genes is more efficient than that with lQseD. The reasons underlying these differences are unknown at this time. However, one has to take into consideration that unlike the lQseD isoform, the sQseD isoform lacks a DNA binding HTH domain and does not control gene expression by directly binding to its target genes (Fig. 5). We have therefore proposed a model whereby selection for a truncated QseD isoform was compensated for by the ability of this isoform to interact with other LTTRs. We first tested whether LEE gene regulation by sQseD occurred through its interaction with other known LEE-regulating LTTRs, such as QseA (67) and LhrA (26). However, sQseD failed to interact with both of these LTTRs (Fig. 6), suggesting that this regulation might occur through interactions with other, yet-unidentified LTTRs involved in LEE regulation. Considering that LTTRs is one of the most widespread classes of transcriptional regulators in bacteria, and that E. coli has over 60 LTTRs (31, 64) encoded within its genome, it is plausible that sQseD's interacting partner has not been identified yet. As depicted in the QseD regulatory model in Fig. 7, while lQseD is able to directly bind to the ler promoter, sQseD must interact with an additional positive ler regulatory protein (designated LysR-X) in a dominant-negative manner, thereby preventing transcription activation (Fig. 7). While direct protein-protein interactions between multiple LTTRs has never been proven, heterodimerization with additional LTTRs or members of an alternative regulatory protein family has been predicted (16, 41-43).
FIG. 7.

Model of the regulatory role of lQseD and sQseD in EHEC 86-24. In non-O157:H7 EHEC strains, the presence of full-length “long” QseD (lQseD) represses ler and LEE transcription. In the absence of lQseD, an as-yet-unidentified LysR protein (LysR-X) activates LEE transcription. However, in O157:H7 EHEC strains, the presence of truncated “short” QseD (sQseD) represses ler and LEE transcription, presumably through dominant-negative interactions with LysR-X, resulting in incomplete DNA remodeling and transcriptional activation.
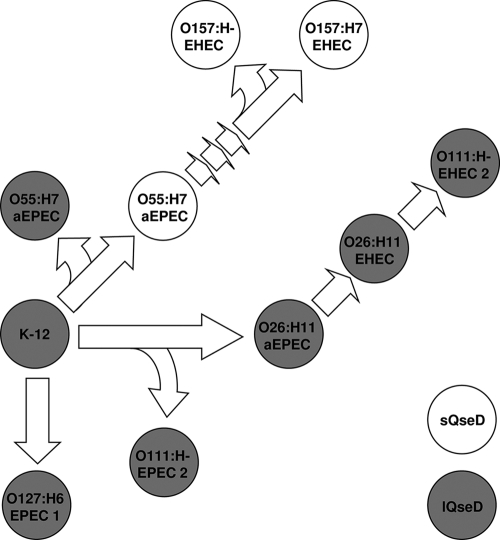
Evolutionary selection for a truncated LTTR is not unheard of, although to date, such proteins have been reported to lack regulatory function (83). As demonstrated by its prevalence almost exclusively in the O157:H7 serotype, sQseD must have been selected for late in an evolutionary branch close to the origin of this now highly prevalent and virulent EHEC serotype (Fig. 8 and Table 3). This evolutionary selection could afford QseD a higher level of plasticity concerning gene regulation by interacting with one or more LTTRs and expanding the breath of its regulatory cascade. This hypothesis is compelling, considering that sQseD was shown to regulate a higher number of genes than lQseD in our transcriptome studies (Tables 4 and 5). Although interaction of the lQseD with other LTTRs is also a possibility, the lack of the HTH domain in sQseD might facilitate the formation of heterotetramers. After compilation of a collection of sequenced E. coli isolates, in which QseD isoforms were characterized (Table 3), it was suggested that QseD truncation occurred in between the emergence of O157:H7 EHEC from its predicted ancestor O55:H7 atypical EPEC (aEPEC) (Fig. 8). EHEC is believed to have diverged from the common laboratory E. coli K-12 strain approximately 4.5 million years ago (58).
FIG. 8.
Evolution and prevalence of the various isoforms of QseD in E. coli. Shown is a cartoon representation of the evolution of EPEC and EHEC from the prototypical nonpathogenic K-12 ancestor. Solid (gray) strains represent the presence of lQseD, and open (white) strains represent the presence of sQseD (designed using original data from reference 18).
The evolutionary advantage gained by O157:H7 isolates due to the truncation of QseD still remains to be determined. However, one possibility is that through the regulation of iraD, a positive regulator of RpoS stability that was identified in a screen for genes that increased the error rate of replication in E. coli (84), QseD differentially regulates EHEC's ability to respond to cellular stress. A pathogen's ability to respond to stress is critical, and in EHEC, this ability has been linked to pathogenesis through multiple pathways, including improvement of survival in the acidic environments, such as those encountered in the host stomach (1), and an altered RpoS response (17), which may lead to additional levels of regulation of LEE and hemolysin expression (47). It should also be noted that qseD is located at the end of an operon containing four genes (including yjiG and yjiH, which are predicted to form an inner membrane spanning channel, and iadA, which is an isoaspartyl dipeptidase involved in the sensing and cleavage of damaged, degraded, or misfolded proteins) that is well suited for dealing with cellular stress (Fig. 1) (33, 59).
The regulation of virulence in enteric pathogens such as EHEC, which often involves the integration of multiple signals, is extremely complex and hierarchical. While we have identified and added a new member to this regulatory family, the true complexity of QseD's regulatory function and network remains to be foreseen. Only through the elucidation of these virulence-regulating networks will we gain the knowledge essential for understanding of EHEC pathogenesis and necessary for development of new tools and treatments against this deadly human pathogen.
Acknowledgments
We thank Beth Whittam and the NIAID STEC Center (Michigan State University, East Lansing, MI) for providing STEC strains for qseD analysis, Kim Orth (University of Texas Southwestern Medical Center, Dallas, TX) for providing the yeast two-hybrid stains and control plasmids, Christopher Parker (University of Texas Southwestern Medical Center, Dallas, TX) for his assistance with microarray analysis and heat map generation, Melissa Kendall (University of Texas Southwestern Medical Center, Dallas, TX) for providing the QseA expression vector MK208, Abhijit Bugde and the Live Cell Imaging Core (University of Texas Southwestern Medical Center, Dallas, TX) for help with FAS imaging and manipulation, and James B. Kaper (University of Maryland School of Medicine, Baltimore, MD) for the EspA, EspB, and FliC antibodies.
This work was supported by National Institutes of Health grants AI053067 and R37 AI21657, the Ellison Medical Foundation, and the Burroughs Wellcome Fund.
The contents of this article are solely the responsibility of the authors and do not represent the official views of the NIH NIAID.
Footnotes
Published ahead of print on 21 May 2010.
REFERENCES
- 1.Allen, K. J., D. Lepp, R. C. McKellar, and M. W. Griffiths. 2008. Examination of stress and virulence gene expression in Escherichia coli O157:H7 using targeted microarray analysis. Foodborne Pathog. Dis. 5:437-447. [DOI] [PubMed] [Google Scholar]
- 2.Alto, N. M., A. W. Weflen, M. J. Rardin, D. Yarar, C. S. Lazar, R. Tonikian, A. Koller, S. S. Taylor, C. Boone, S. S. Sidhu, S. L. Schmid, G. A. Hecht, and J. E. Dixon. 2007. The type III effector EspF coordinates membrane trafficking by the spatiotemporal activation of two eukaryotic signaling pathways. J. Cell Biol. 178:1265-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amberg, D. C., D. Burke, J. N. Strathern, and Cold Spring Harbor Laboratory. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual, 2005 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 4.Anderson, G. G., C. C. Goller, S. Justice, S. J. Hultgren, and P. C. Seed. 2010. Polysaccharide capsule and sialic acid-mediated regulation promote biofilm-like intracellular bacterial communities during cystitis. Infect. Immun. 78:963-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axler-Diperte, G. L., V. L. Miller, and A. J. Darwin. 2006. YtxR, a conserved LysR-like regulator that induces expression of genes encoding a putative ADP-ribosyltransferase toxin homologue in Yersinia enterocolitica. J. Bacteriol. 188:8033-8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barba, J., V. H. Bustamante, M. A. Flores-Valdez, W. Deng, B. B. Finlay, and J. L. Puente. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187:7918-7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker, C. S., B. M. Pruss, and P. Matsumura. 2004. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J. Bacteriol. 186:7529-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basineni, S. R., R. Madhugiri, T. Kolmsee, R. Hengge, and G. Klug. 2009. The influence of Hfq and ribonucleases on the stability of the small non-coding RNA OxyS and its target rpoS in E. coli is growth phase dependent. RNA Biol. 6:584-594. [DOI] [PubMed] [Google Scholar]
- 9.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 10.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke, M. B., D. T. Hughes, C. Zhu, E. C. Boedeker, and V. Sperandio. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 103:10420-10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, M. B., and V. Sperandio. 2005. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 57:1734-1749. [DOI] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deghmane, A. E., D. Giorgini, M. Larribe, J. M. Alonso, and M. K. Taha. 2002. Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol. Microbiol. 43:1555-1564. [DOI] [PubMed] [Google Scholar]
- 15.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Silva, R. S., G. Kovacikova, W. Lin, R. K. Taylor, K. Skorupski, and F. J. Kull. 2005. Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J. Biol. Chem. 280:13779-13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong, T., and H. E. Schellhorn. 2009. Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol. Genet. Genomics 281:19-33. [DOI] [PubMed] [Google Scholar]
- 18.Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Invest. 107:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 21.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 22.Gibson, K. E., and T. J. Silhavy. 1999. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator sprE. J. Bacteriol. 181:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Griffin, P. M., S. M. Ostroff, R. V. Tauxe, K. D. Greene, J. G. Wells, J. H. Lewis, and P. A. Blake. 1988. Illnesses associated with Escherichia coli O157:H7. Ann. Intern. Med. 109:705-712. [DOI] [PubMed] [Google Scholar]
- 24.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen, A. M., and J. B. Kaper. 2009. Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 73:446-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda, N., S. Iyoda, S. Yamamoto, J. Terajima, and H. Watanabe. 2009. LrhA positively controls the expression of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli by differential regulation of their master regulators PchA and PchB. Mol. Microbiol. 74:1393-1411. [DOI] [PubMed] [Google Scholar]
- 27.Hughes, D. T., M. B. Clarke, K. Yamamoto, D. A. Rasko, and V. Sperandio. 2009. The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC). PLoS Pathog. 5:e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ieva, R., C. Alaimo, I. Delany, G. Spohn, R. Rappuoli, and V. Scarlato. 2005. CrgA is an inducible LysR-type regulator of Neisseria meningitidis, acting both as a repressor and as an activator of gene transcription. J. Bacteriol. 187:3421-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyoda, S., and H. Watanabe. 2005. ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J. Bacteriol. 187:4086-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyoda, S., and H. Watanabe. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology 150:2357-2571. [DOI] [PubMed] [Google Scholar]
- 31.Janga, S. C., and E. Perez-Rueda. 2009. Plasticity of transcriptional machinery in bacteria is increased by the repertoire of regulatory families. Comput. Biol. Chem. 33:261-268. [DOI] [PubMed] [Google Scholar]
- 32.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U. S. A. 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jozic, D., J. T. Kaiser, R. Huber, W. Bode, and K. Maskos. 2003. X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases. J. Mol. Biol. 332:243-256. [DOI] [PubMed] [Google Scholar]
- 34.Kaper, J. B., and M. A. Karmali. 2008. The continuing evolution of a bacterial pathogen. Proc. Natl. Acad. Sci. U. S. A. 105:4535-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kendall, M. M., D. A. Rasko, and V. Sperandio. 2007. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 75:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 37.Kenny, B., S. Ellis, A. D. Leard, J. Warawa, H. Mellor, and M. A. Jepson. 2002. Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Mol. Microbiol. 44:1095-1107. [DOI] [PubMed] [Google Scholar]
- 38.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1988. New diagnostic test for enteropathogenic Escherichia coli. Lancet i(8598):1337. [DOI] [PubMed] [Google Scholar]
- 40.Konczy, P., K. Ziebell, M. Mascarenhas, A. Choi, C. Michaud, A. M. Kropinski, T. S. Whittam, M. Wickham, B. Finlay, and M. A. Karmali. 2008. Genomic O island 122, locus for enterocyte effacement, and the evolution of virulent verocytotoxin-producing Escherichia coli. J. Bacteriol. 190:5832-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovacikova, G., W. Lin, and K. Skorupski. 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol. Microbiol. 53:129-142. [DOI] [PubMed] [Google Scholar]
- 42.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41:393-407. [DOI] [PubMed] [Google Scholar]
- 43.Kovacikova, G., and K. Skorupski. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lahiri, A., P. Das, and D. Chakravortty. 2008. The LysR-type transcriptional regulator Hrg counteracts phagocyte oxidative burst and imparts survival advantage to Salmonella enterica serovar Typhimurium. Microbiology 154:2837-2846. [DOI] [PubMed] [Google Scholar]
- 45.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 46.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 47.Li, H., A. Granat, V. Stewart, and J. R. Gillespie. 2008. RpoS, H-NS, and DsrA influence EHEC hemolysin operon (ehxCABD) transcription in Escherichia coli O157:H7 strain EDL933. FEMS Microbiol. Lett. 285:257-262. [DOI] [PubMed] [Google Scholar]
- 48.Maddocks, S. E., and P. C. Oyston. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609-3623. [DOI] [PubMed] [Google Scholar]
- 49.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 51.Merrikh, H., A. E. Ferrazzoli, A. Bougdour, A. Olivier-Mason, and S. T. Lovett. 2009. A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc. Natl. Acad. Sci. U. S. A. 106:611-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merrikh, H., A. E. Ferrazzoli, and S. T. Lovett. 2009. Growth phase and (p)ppGpp control of IraD, a regulator of RpoS stability, in Escherichia coli. J. Bacteriol. 191:7436-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakanishi, N., H. Abe, Y. Ogura, T. Hayashi, K. Tashiro, S. Kuhara, N. Sugimoto, and T. Tobe. 2006. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61:194-205. [DOI] [PubMed] [Google Scholar]
- 53a.Orth, K., L. E. Palmer, Z. Q. Bao, S. Stewart, A. E. Rudolph, J. B. Bliska, and J. E. Dixon. 1999. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science 285:1920-1923. [DOI] [PubMed] [Google Scholar]
- 54.Peterson, C. N., V. J. Carabetta, T. Chowdhury, and T. J. Silhavy. 2006. LrhA regulates rpoS translation in response to the Rcs phosphorelay system in Escherichia coli. J. Bacteriol. 188:3175-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. U. S. A. 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rasko, D. A., M. J. Rosovitz, G. S. Myers, E. F. Mongodin, W. F. Fricke, P. Gajer, J. Crabtree, M. Sebaihia, N. R. Thomson, R. Chaudhuri, I. R. Henderson, V. Sperandio, and J. Ravel. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reading, N. C., D. A. Rasko, A. G. Torres, and V. Sperandio. 2009. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 106:5889-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 59.Rogers, M. F., L. D. Budnick, I. Kirson, E. S. Hurwitz, M. H. Hatch, C. A. Bopp, M. A. Karmali, G. W. Gary, R. Layne, and L. B. Schonberger. 1986. Hemolytic-uremic syndrome—an outbreak in Sacramento, California. West J. Med. 144:169-173. [PMC free article] [PubMed] [Google Scholar]
- 60.Russell, R. M., F. C. Sharp, D. A. Rasko, and V. Sperandio. 2007. QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli. J. Bacteriol. 189:5387-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sainsbury, D. C., G. Kessell, A. Guhan, A. Fall, V. Miller, M. R. Abouzeid, and T. Muir. 2009. Unexpected hyperpigmentation following intralesional bleomycin injection. J. Plast. Reconstr. Aesthet. Surg. 62:e497-e499. [DOI] [PubMed] [Google Scholar]
- 62.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 63.Sánchez-SanMartín, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 65.Shakhnovich, E. A., B. M. Davis, and M. K. Waldor. 2009. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol. Microbiol. 74:347-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma, V. K., and R. L. Zuerner. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186:7290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharp, F. C., and V. Sperandio. 2007. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect. Immun. 75:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781-793. [DOI] [PubMed] [Google Scholar]
- 70.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U. S. A. 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 74.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. U. S. A. 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tu, X., T. Latifi, A. Bougdour, S. Gottesman, and E. A. Groisman. 2006. The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc. Natl. Acad. Sci. U. S. A. 103:13503-13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tu, X., I. Nisan, C. Yona, E. Hanski, and I. Rosenshine. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol. Microbiol. 47:595-606. [DOI] [PubMed] [Google Scholar]
- 78.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735-2744. [DOI] [PubMed] [Google Scholar]
- 79.Vojtek, A. B., and S. M. Hollenberg. 1995. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 255:331-342. [DOI] [PubMed] [Google Scholar]
- 80.Wales, A. D., M. J. Woodward, and G. R. Pearson. 2005. Attaching-effacing bacteria in animals. J. Comp. Pathol. 132:1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walters, M., M. P. Sircili, and V. Sperandio. 2006. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188:5668-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walters, M., and V. Sperandio. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 74:5445-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang, Y., M. Rawlings, D. T. Gibson, D. Labbe, H. Bergeron, R. Brousseau, and P. C. Lau. 1995. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol. Gen. Genet. 246:570-579. [DOI] [PubMed] [Google Scholar]
- 84.Yang, H., E. Wolff, M. Kim, A. Diep, and J. H. Miller. 2004. Identification of mutator genes and mutational pathways in Escherichia coli using a multicopy cloning approach. Mol. Microbiol. 53:283-295. [DOI] [PubMed] [Google Scholar]
- 85.Zhang, L., R. R. Chaudhuri, C. Constantinidou, J. L. Hobman, M. D. Patel, A. C. Jones, D. Sarti, A. J. Roe, I. Vlisidou, R. K. Shaw, F. Falciani, M. P. Stevens, D. L. Gally, S. Knutton, G. Frankel, C. W. Penn, and M. J. Pallen. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72:7282-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]