Abstract
Faithful DNA replication involves the removal of RNA residues from genomic DNA prior to the ligation of nascent DNA fragments in all living organisms. Because the physiological roles of archaeal type 2 RNase H are not fully understood, the substrate structure requirements for the detection of RNase H activity need further clarification. Biochemical characterization of a single RNase H detected within the genome of Pyrococcus abyssi showed that this type 2 RNase H is an Mg- and alkaline pH-dependent enzyme. PabRNase HII showed RNase activity and acted as a specific endonuclease on RNA-DNA/DNA duplexes. This specific cleavage, 1 nucleotide upstream of the RNA-DNA junction, occurred on a substrate in which RNA initiators had to be fully annealed to the cDNA template. On the other hand, a 5′ RNA flap Okazaki fragment intermediate impaired PabRNase HII endonuclease activity. Furthermore, introduction of mismatches into the RNA portion near the RNA-DNA junction decreased both the specificity and the efficiency of cleavage by PabRNase HII. Additionally, PabRNase HII could cleave a single ribonucleotide embedded in a double-stranded DNA. Our data revealed PabRNase HII as a dual-function enzyme likely required for the completion of DNA replication and DNA repair.
DNA replication in all living organisms takes place concurrently on two separate strands. The lagging strand consists of multiple discontinuous segments called Okazaki fragments, whereas the leading strand consists of one large continuous segment. The production of each individual lagging strand by DNA polymerase is primed by a short stretch of RNA. Later on, these RNA primers are eliminated and the resulting gap is filled with deoxyribonucleotides prior to ligation. Priming and DNA elongation at the replication fork involve a set of specialized polymerizing enzymes which differ from replicative DNA polymerases, and one another, to correct erroneously inserted nucleotides. In archaeal cells, the priming complex lacks proofreading 3′-5′ exonuclease activity (13, 14) present in replicative DNA polymerases B and D (1, 8). Consequently, mismatches in the vicinity of the RNA-DNA junction could arise in replicating cells, as already observed in eukaryotes (28, 31). Similarly, single ribonucleotides incorporated during DNA replication (20, 26) or by external agents (32) would represent another source of erroneous nucleotides. Persistence of residual RNA during DNA replication would be detrimental for the cells, suggesting that a combination of specific and efficient nucleolytic processes is absolutely required to preserve DNA integrity.
RNases H are enzymes which degrade the RNA portion of RNA/DNA or RNA-DNA/DNA duplexes (29). RNases H are classified into two major families, type 1 and type 2, based on amino acid sequence (21). The type 1 family includes bacterial RNase HI, mammalian RNase HII, the RNase H domain of reverse transcriptase, and archaeal RNase HI, and the type 2 family contains bacterial RNase HII and RNase HIII, mammalian RNase HI, and archaeal RNase HII (21). While type 2 RNase H enzymes are universally conserved in the three domains of life, their physiological role remains elusive. Much less is known about the type 2 family than the type 1 RNase H enzymes. The multiplicity of RNases H within a single cell complicates the situation, although presumable roles in DNA replication, DNA repair, and transcription have been assigned, as recently reviewed (2, 30). In archaea, structural and biochemical characterization of type 2 RNases H (3, 4, 10, 12, 16, 19) suggested that they can initiate RNA removal from DNA duplexes, based on their ability to specifically cleave 5′ to the junctional ribonucleotide. However, despite this information, the physiological role of type 2 RNases H remains elusive. They could be involved in the completion of either leading or lagging strands or both. Additional biochemical experiments with catalytic intermediates should provide valuable knowledge on the participation of these archaeal cellular enzymes at the replication fork and in DNA repair.
To investigate this question, we have designed an RNA/DNA duplex, a cognate RNA-DNA/DNA duplex (15), and a single ribonucleotide embedded in a DNA duplex (DNA-1RNA-DNA/DNA) for use as substrates and employed the type 2 RNase H from the hyperthermophilic deep-sea euryarchaeon Pyrococcus abyssi (PabRNase HII). Since a single rnh gene exists in the genome of P. abyssi (6), RNase HII is likely the key enzyme involved in RNA elimination in this organism. Thus, PabRNase HII can be considered the representative type 2 RNase H.
Here, we analyzed the cleavage specificity of PabRNase HII for substrates with Okazaki fragment-like structure. We also tested PabRNase HII activity on Okazaki fragment-like substrates in the presence of mismatched base pairs in order to assess the molecular mechanism of recognition of the RNA-DNA junction and the subsequent cleavage specificity. In addition, we examined whether PabRNase HII can incise the DNA backbone on the 5′ side of a single ribonucleotide embedded in a DNA duplex. Our data provide substantial evidence that the single RNase H in P. abyssi has a dual role in the maintenance of genome integrity. The results from this study are further discussed to define potent roles of type 2 RNase H from P. abyssi in the resolution of RNA fragments at the replication fork and in the repair of single embedded ribonucleotides.
MATERIALS AND METHODS
Nucleic acid substrates.
Gel-purified oligonucleotides for preparing the substrates for RNase HII assays were purchased from Eurogentec (Belgium), and their sequences are listed in Table 1. Fluorescent labeling at the 5′ end was performed with the 5′ EndTag Nucleic Acid End Labeling System from Vector Laboratories (Burlingame, CA). Free fluorescent dyes were removed on MicroSpin G-25 columns. For some experiments, 5′-end or 3′-end fluorescently labeled oligonucleotides were chemically synthesized and high-performance liquid chromatography purified by Eurogentec (Belgium). To generate the substrates for the RNase HII assays, the appropriate oligonucleotides were mixed in a 1:1 molar ratio in 20 mM Tris-HCl (pH 7.4)-150 mM NaCl, heated to 75°C, and slowly cooled to room temperature.
TABLE 1.
Oligonucleotide sequences used to create structural duplex substratesa
| Primer or template | Length (nt) | Sequence |
|---|---|---|
| Primers | ||
| 1 | 32 | ugccaagcuugcaugccugcaggucgacucua |
| 2 | 30 | auucguaaucauGGTCATAGCTGTTTCCTG |
| 3 | 17 | TGCCAAGCTTGCATGCC |
| 4 | 37 | TGCCAAGCTTGCATGCCTGCAGGTCGACTCTAGAGGA |
| 5 | 12 | TGGGTGGGGTGG |
| 6 | 30 | ATTCGTAATCAuGGTCATAGCTGTTTCCTG |
| Templates | ||
| 7 | 87 | CAGGAAACAGCTATGACCATGATTACGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTGGCA |
| 8 | 30 | CAGGAAACAGCTATGACCATGATTACGAAT |
| 9 | 30 | CAGGAAACAGCTATGACCCCACCCCACCCA |
| 10 | 30 | CAGGAAACAGCTATGACCAGGATTACGAAT |
| 11 | 30 | CAGGAAACAGCTATGACCGTGATTACGAAT |
| 12 | 30 | CAGGAAACAGCTATGACTATGATTACGAAT |
The S1 substrate comprises primer 2 and template 8, the S2 substrate is primer 2, the S3 substrate comprises primers 2 and template 8, the S4 substrate consists of primers 2 and 3 and template 7, the S5 substrate contains primers 2 and 4 and template 7, the S6 substrate includes primers 2 and 5 and template 9, the S7 substrate consists of primer 2 and template 12, the S8 substrate is composed of primer 2 and template 11, the S9 substrate consists of primer 2 and template 10, the S10 substrate comprises primer 6 and template 8, and the S11 substrate contains primer 1 and template 7. Deoxyribonucleotides and ribonucleotides are shown by uppercase and lowercase letters, respectively.
Cloning, production, and purification of PabRNase HII.
The gene encoding PabRNase HII (PAB0352) was cloned into the pQE-80L expression vector (Qiagen). PabRNase HII was overexpressed in Escherichia coli BL21-CodonPlus-RIL (Stratagene) as a histidine-tagged protein and purified to near homogeneity via Ni-nitrilotriacetic acid beads (Qiagen) and S200 gel filtration using fast protein liquid chromatography (GE Healthcare) as previously described (16). Protein integrity was analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry analyses (Innova Proteomics, Rennes, France). PabRNase HII purity was controlled by 4 to 20% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Thermo Scientific).
Amino acid sequence alignments and secondary structure.
Amino acid sequence alignments have been constructed by ClustalW2 (available at www.ebi.ac.uk/clustalW2/). Secondary structure elements calculated with the program ESPript 2.2 (available at http://espript.ibcp.fr/ESPript/ESPript/) refer to the structure of TkoKOD1RNase HII (Protein Data Bank code 1IO2).
Assays for RNase HII activity.
Assays to monitor cleavage by PabRNase HII were performed in RNase HII buffer (10 μl) containing 50 mM Tris-HCl (pH 8), 5 mM dithiothreitol, 5 mM MgCl2, and 50 nM DNA substrates. Enzymes were diluted from concentrated stocks in 20 mM Tris-HCl (pH 7.5)-20% glycerol prior to use. The enzyme concentrations in a typical reaction mixture ranged from 4 to 400 nM, unless otherwise specified. After the addition of PabRNase HII, reaction mixtures were incubated at 60°C for 30 min and stopped on ice with 15 μl of stop buffer (98% formamide, 10 mM EDTA). Samples were heated at 95°C for 5 min. A base hydrolysis ladder was prepared by incubation of the labeled RNA-DNA strand (10 μM) with snake venom phosphodiesterase I (0.018 U) for 10 min at 37°C. Product analysis was carried out by 15% denaturing PAGE. After visualization with a Mode Imager Typhoon 9400 (GE Healthcare), quantification of the results was performed using ImageQuant 5.2 software. In all cases, the percentage of substrate hydrolysis was determined by the ratio of products to products plus substrate, allowing a correction for loading errors and a comparison of cleavage efficiency, irrespective of the different products generated.
To analyze divalent cations or pH dependence, RNase HII assays were carried out with 50 nM PabRNase HII and the S1 substrate at 50 nM at 60°C for 30 min. The data shown are averages of triplicate measurements and standard deviations.
To determine the kinetic parameters, steady-state kinetic reactions were carried out under the same conditions as described above by using substrate 1 at concentrations ranging from 0.03 to 3 μM. Initial velocity experiments were monitored as a function of time with 60 nM PabRNase HII at 60°C such that the rate of converted substrate did not exceed 20% of the total. Velocity measurements were reported as the concentration (micromolar) of hydrolyzed substrate over time (minutes). The observed rates of converted substrates with PabRNase HII were firstly determined from Lineweaver-Burk plots. The data were fitted by nonlinear regression using the Marquardt-Levenberg algorithm (EnzFitter 2.0; BioSoft) to the Michaelis-Menten equation. Km and Vmax values were obtained from the fit and used to calculate the catalytic efficiency (kcat/Km) of PabRNase HII. The kinetic values shown are averages of at least triplicate determinations and include standard deviations. Any adjustments to the above are noted in the figure legends.
RESULTS
Archaeal RNase HII homologues.
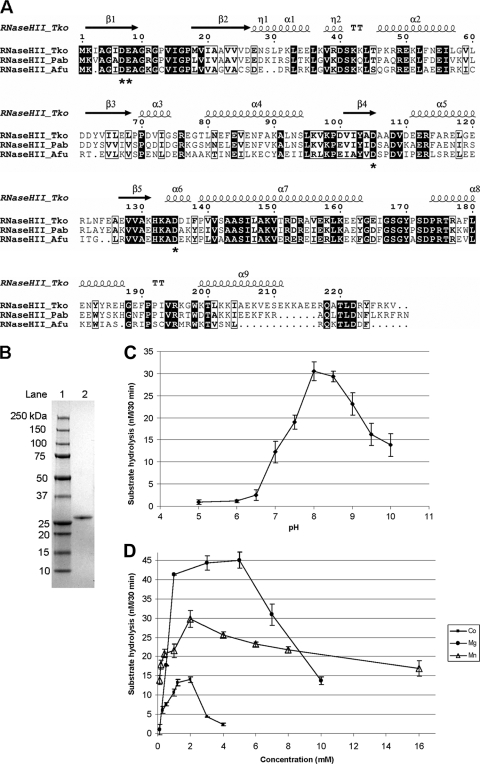
PabRNase HII showed amino acid sequence similarities of 75.2% to Thermococcus kodakaraensis RNase HII (TkoRNase HII) and 65% to Archaeoglobus fulgidus RNase HII (AfuRNase HII). Analysis of protein primary structures and related secondary structures outlined subtle differences among the three proteins (Fig. 1 A). On the one hand, PabRNase HII and AfuRNase HII are isoelectric at basic pH (isoelectric points of 9 and 7.6, respectively), whereas TkoRNase HII is isoelectric at acidic pH (isoelectric point of 5.5). On the other hand, PabRNase HII, TkoRNase HII, and AfuRNase HII exhibit conserved secondary structure elements, with the exception that the α9 helix is incomplete in AfuRNase. This secondary structure element is important for TkoRNase HII binding to the substrate (19). Structural alignments resulted in the identification of conserved active site residues (Asp7, Glu8, Asp105, and Asp135) in PabRNase HII, suggesting that the three enzymes have similar catalytic mechanisms.
FIG. 1.
Enzymatic properties of PabRNase HII. (A) Alignment of the amino acid sequences of archaeal RNase HII homologues. Sequences are from the Euryarchaeota species P. abyssi (Pab; accession number gi 14520734), T. kodakaraensis (Tko; accession number gi 57640740), and A. fulgidus (Afu; accession number gi 11498229). Conserved amino acid residues are shaded in black. Similar amino acid residues are boxed. Proposed active-site residues are indicated by asterisks. Secondary structure is shown above the sequences, denoting β sheets (arrows) and α helices (ribbons). (B) Gradient (4 to 20%) SDS-PAGE of purified, recombinant, His6-tagged PabRNase HII (0.5 μg; lane 2) and molecular mass markers (lane 1) stained with Coomassie blue. (C) pH dependence. Enzymatic activities were determined at 60°C for 30 min in reaction buffer containing 50 mM Tris-HCl, 5 mM dithiothreitol, 5 mM MgCl2, 50 nM PabRNase HII, and 50 nM RNA-DNA/DNA substrate (S1) with pH values ranging from 5 to 10. Data are the averages of triplicate measurements. (D) Divalent cation dependence. Enzymatic activities were determined at 60°C for 30 min in reaction buffer containing 50 mM Tris-HCl (pH 8), 5 mM dithiothreitol, 50 nM PabRNase HII, and 50 nM RNA-DNA/DNA substrate (S1) at the indicated concentrations of MgCl2 (•), MnCl2 (▵), and CoCl2 (▪). Data are the averages of triplicate measurements.
Enzymatic properties of P. abyssi RNase HII.
His-tagged PabRNase HII was overproduced in E. coli and purified to give a single band on SDS-PAGE (Fig. 1B, lane 2). Reaction mixtures were incubated at 60°C, the optimum temperature for PabRNase HII activity (16). PabRNase HII was assayed under different pH and ionic conditions, varying both the nature and the concentration of divalent cations, according to the general procedure described in Materials and Methods using the RNA-DNA/DNA substrate (S1). The optimum pH for its activity was found between pHs 8.0 and 8.5 (Fig. 1C). However, at pHs 7.5 and 9, the enzyme retained about 60% of the activity measured at pH 8.
PabRNase HII exhibited enzymatic activity in the presence of MgCl2, MnCl2, and CoCl2 (Fig. 1D). While PabRNase HII activity was entirely dependent on the presence of a divalent cation, the enzyme was not active in the presence of NiCl2, FeCl2, ZnCl2, CaCl2, or CuSO4. The most preferred metal ion for PabRNase HII was MgCl2, but MnCl2 and CoCl2 could substitute for MgCl2 with reduced cleavage activity. As shown in Fig. 1D, the metal concentrations which gave the highest enzymatic activity were 5 mM for MgCl2 and 2 mM for MnCl2 and CoCl2. The substrate hydrolysis levels in the presence of 5 mM MgCl2 were 1.5- and 3.2-fold higher than those determined at 2 mM for MnCl2 and CoCl2, respectively.
The kinetic parameters of PabRNase HII were determined in the presence of RNA-DNA/DNA substrate S1 and 5 mM MgCl2. The results are summarized in Table 2 and compared to those of the type 2 RNase H archaeal homologue from A. fulgidus as already described (4). Interestingly, AfuRNase HII showed stronger substrate binding affinity than PabRNase HII, as attested by a 10-fold lower Km value, while the catalytic rate constants of the two enzymes were similar (Table 2). As a consequence, PabRNase HII displayed a lower catalytic efficiency on the RNA-DNA/DNA substrate than AfuRNase HII (Table 2).
TABLE 2.
Kinetic parameters of archaeal RNases HIIa
| Enzyme | Km (μM) | kcat (min−1) | kcat/Km |
|---|---|---|---|
| PabRNase HII | 0.50 ± 0.15 | 5.57 ± 0.54 | 11.14 |
| AfuRNase HII | 0.06 ± 0.15 | 8.0 ± 0.23 | 133.3 |
Hydrolyses of RNA-DNA/DNA substrates (S1) were carried out at 60°C in PabRNase HII reaction buffer as described in Materials and Methods. The data were fitted by nonlinear regression using the Marquardt-Levenberg algorithm (EnzFitter 2.0; BioSoft) to the Michaelis-Menten equation. The Km and Vmax values obtained from the fit were used to calculate the catalytic efficiency (kcat/Km) of PabRNase HII. The kinetic values shown are the averages of at least triplicate determinations and are shown with standard deviations. The values for AfuRNase HII are from a previous study (4).
RNase activity of P. abyssi RNase HII.
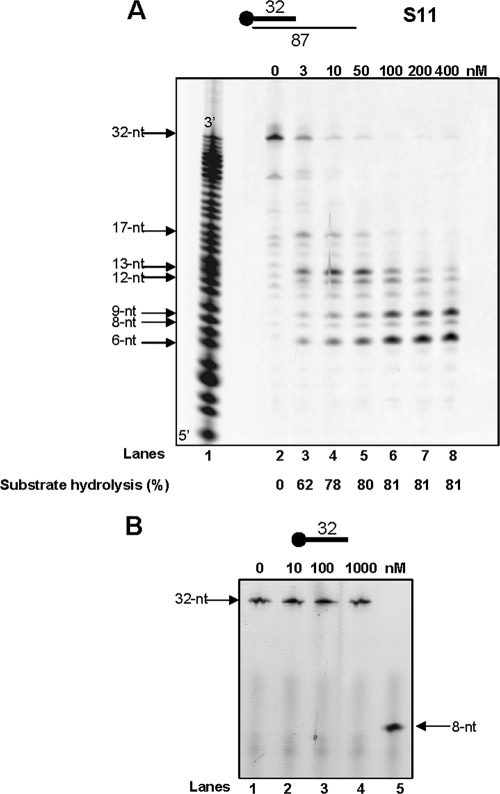
First, we examined whether PabRNase HII could cleave the RNA strand of an RNA/DNA duplex (S11). As shown in Fig. 2, different 5′-terminal RNA products accumulated, depending on the enzyme concentration, indicating that PabRNase HII exhibited endoribonuclease activity. Because control assays without enzyme showed background degradation of the RNA primer (Fig. 2A, lane 2), it was subtracted from the cleavage product signal. Less than 19% intact RNA was present upon incubation with 100 nM PabRNase HII for 30 min at 60°C (Fig. 2A, lane 6). Multiple cleavage sites were detected (Fig. 2A, lanes 2 to 8), and comparative analysis of products with those from the snake venom phosphodiesterase digest of a 5′-end-labeled 32-nucleotide (nt) RNA ladder (Fig. 2A, lane 1) pointed out main cleavage events (6, 8, 9, 12, 13, and 17 nt). Moreover, further processing of short RNA fragments could be observed by increasing enzyme concentrations. It is important to note that the PabRNase HII used in this study did not exhibit nuclease activity on single-stranded RNA (Fig. 2B). Together, these results provided evidence that PabRNase HII acts as an endoribonuclease on RNA/DNA duplexes.
FIG. 2.
RNase activity by PabRNase HII. (A) The indicated amounts of PabRNase HII were incubated with the S11 substrate (lanes 2 to 8), and a base-hydrolyzed ladder (lane 1) was prepared as described in Materials and Methods. Fluorescently 5′-end-labeled products were visualized with a Mode Imager Typhoon 9400 (GE Healthcare), and quantification was performed using ImageQuant 5.2 software. (B) PabRNase HII was incubated with the 32-base single-stranded RNA oligonucleotide at the indicated amounts (lanes 1 to 4). An 8-nt RNA oligonucleotide was used as a ladder (lane 5). Fluorescently 5′-end-labeled products were visualized with a Mode Imager Typhoon 9400 (GE Healthcare).
Structure-specific cleavage activities in PabRNase HII.
The absence of cleavage specificity of RNA/DNA duplexes prompted us to look for the digestion of other relevant physiological substrates. We hypothesize that PabRNase HII participates in the mechanism of RNA primer removal, an activity which can occur once at the leading strand or much more frequently at the lagging strand. Therefore, we explored the cleavage specificity of PabRNase HII for substrates with Okazaki fragment-like intermediates.
PabRNase HII specifically cleaves RNA-initiated DNA segments fully annealed to a DNA template.
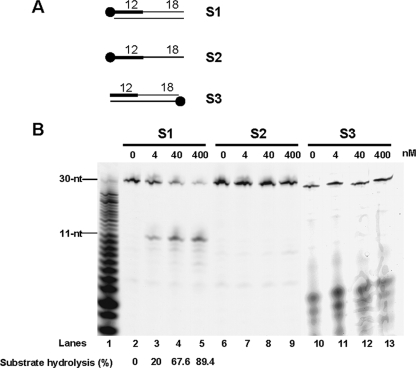
We initially began to examine whether PabRNase HII could hydrolyze a cognate double-stranded Okazaki fragment (15). A single strand composed of 12 nt of RNA (RNA12nt) followed by 18 nt of DNA (DNA18nt), fluorescently labeled at the 5′ end, was annealed to a complementary 30-nt DNA template to form the S1 substrate as shown in Fig. 3A. When this substrate was incubated in the presence of increasing PabRNase HII amounts, a major product appeared (Fig. 3B, lanes 2 to 5). This oligomer was shown to correspond to 11 nt of RNA by migration with respect to a snake venom phosphodiesterase-generated digest of RNA12ntDNA18nt (Fig. 3B, lane 1). In addition to this main cleavage site, minor cleavage sites characteristic of nonspecific nuclease activity were also found throughout the length of the RNA (Fig. 3B, lanes 2 to 5). Over time, PabRNase HII activity released the same oligomer, which was basically free of any additional shorter fragments, suggesting that this product was not transiently formed and prevailed during the reaction (data not shown). Interestingly, PabRNase HII did not hydrolyze single-stranded RNA12ntDNA18nt (Fig. 3B, lanes 6 to 9), indicating that cleavage is dependent on the heteroduplex structure. It is of note that the pale band at ∼9 nt present at relatively constant levels did not correspond to a specific cleavage product (Fig. 3B, lanes 2 to 9). Absence of specific cleavage was also observed with substrates lacking the cDNA template to either the RNA12nt or the DNA18nt sequence (data not shown). While a fully annealed RNA12ntDNA18nt/DNA is definitely required to detect cleavage specificity, PabRNase HII did not hydrolyze the cDNA template (Fig. 3B, lanes 10 to 13). Shorter bands were not due to cleavage activity since they were detectable in all lanes with equal intensities, even in the absence of enzyme. These data demonstrated that PabRNase HII can act endonucleolytically on initiator RNA and displays a specific cleavage activity dependent on the heteroduplex structure.
FIG. 3.
PabRNase HII specifically cleaves RNA-initiated DNA segments fully annealed to a DNA template. (A) Structures of substrates S1, S2, and S3. The thick line and the closed circle represent the RNA portion and the fluorescent label, respectively. (B) The indicated amounts of PabRNase HII were incubated with the S1 substrate (lanes 2 to 5), the S2 substrate (lanes 6 to 9), and the S3 substrate (lanes 10 to 13). A base-hydrolyzed ladder (lane 1) was prepared as explained in Materials and Methods. Fluorescently labeled products were visualized with a Mode Imager Typhoon 9400 (GE Healthcare), and quantification was performed using ImageQuant 5.2 software.
PabRNase HII specifically cleaves the fully annealed RNA strand of Okazaki fragment-gapped intermediates but not a 5′ RNA flap.
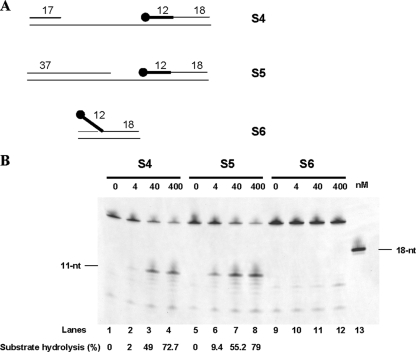
At both the leading and lagging strands, sequential enzymatic steps are thought to be part of the RNA primer elimination mechanism in P. abyssi. As a consequence, diverse structural Okazaki fragment-like substrates would arise. Therefore, we examined whether structural intermediates (S4, S5, and S6) which can be captured during the process (outlined in Fig. 4A) could direct the cleavage activity of PabRNase HII. On the 40-gapped S4 intermediate composed of an upstream DNA primer and a downstream RNA-DNA fragment fully annealed to the cDNA template, PabRNase HII specifically cleaved the RNA segment, releasing one ribonucleotide attached to the DNA segment (Fig. 4B, lanes 1 to 4). During the elongation step, the size of the gap would decrease to reach the next RNA initiator. By reconstitution of model transient substrates, we demonstrated that both the 20-nt gapped S5 intermediate (Fig. 4B, lanes 5 to 8) and a nicked intermediate (data not shown) were specifically cleaved. Collectively, the cleavage efficiencies of the gapped and nicked Okazaki fragment intermediates were comparable to those of double-stranded RNA-DNA fragments (Fig. 3B, lanes 2 to 5). However, on a 5′ RNA flap which can result from strand displacement activity by P. abyssi PolD of the next Okazaki fragment (11), PabRNase HII did not significantly release oligomers (Fig. 4B, lanes 9 to 12). It is of note that a faint intensifying band at 8 nt (Fig. 4B, lanes 9 to 12) did not correspond to a specific cleavage product. These results clearly indicated that PabRNase HII is not involved in the cleavage of a single-stranded RNA initiator despite the presence of surrounding DNA duplexes. These data are consistent with our observations from Fig. 3B that PabRNase HII exclusively cuts double-stranded RNA-DNA/DNA substrates. Importantly, we demonstrated that PabRNase HII cleaves the RNA initiator fully annealed to the cDNA template independently of the size of the gap. In addition, we provided evidence that a 5′ RNA flap is not an appropriate substrate for PabRNase HII, suggesting the requirement of additional enzymes to fully ensure the removal of Okazaki fragment intermediates at the lagging strand.
FIG. 4.
PabRNase HII specifically cleaves the fully annealed RNA strand of Okazaki fragment-gapped intermediates but not a 5′ RNA flap. (A) Structures of substrates S4, S5, and S6. The thick line and the closed circle represent the RNA portion and the fluorescent label, respectively. (B) The indicated amounts of PabRNase HII were incubated with the S4 substrate (lanes 1 to 4), the S5 substrate (lanes 5 to 8), and the S6 substrate (lanes 9 to 12). An 18-nt ladder was used (lane 13). Fluorescently labeled products were visualized with a Mode Imager Typhoon 9400 (GE Healthcare), and quantification was performed using ImageQuant 5.2 software.
PabRNase HII specifically cuts the RNA-DNA/DNA duplex when the RNA is completely annealed to the DNA template.
The above results indicated that PabRNase HII specifically cleaves the RNA in an RNA-DNA/DNA duplex 1 ribonucleotide upstream of the RNA-DNA junction. Based on this observation, we attempted to gain further information about the structure-specific recognition of the RNA-DNA junction. We predicted that mismatches located either downstream or upstream of the site of cleavage would alter the structure of the junction and prevent PabRNase HII from recognizing and cutting the substrate. Such substrates, which can be created during priming and DNA synthesis in eukaryotes (28, 31), could also be relevant in P. abyssi cells. In particular, the priming heterodimeric polymerase in P. abyssi, the P. abyssi p46/41 complex, does not possess 3′-5′ exonucleolytic activity and can consequently misincorporate nucleotides, creating mismatched base pairs at or near the RNA-DNA junction.
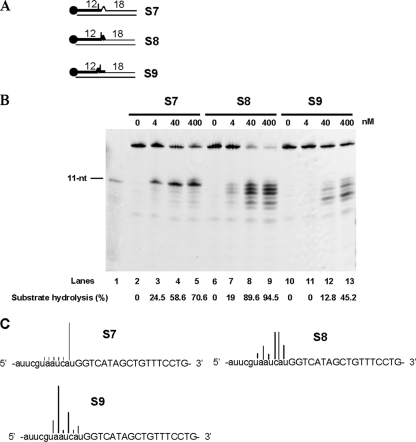
The cDNA template was designed to produce specific mismatches with the RNA12ntDNA18nt strand (Fig. 5A). When the mismatch was the deoxynucleotide downstream of the site of cleavage, PabRNase HII efficiently cleaved the S7 substrate and cut at one site into the RNA segment, leaving a monoribonucleotide attached to the DNA18nt strand (Fig. 5B, lanes 2 to 5). Cleavage efficiencies were still comparable to those of the model Okazaki fragments described above. This result seems to point out that a deoxynucleotide-mismatched Okazaki fragment does not affect recognition and specific cleavage by PabRNase HII. We next considered that ribonucleotide mismatches positioned downstream (Fig. 5A, S8 substrate) or upstream (Fig. 5A, S9 substrate) of the cutting site would be crucial for directing the cleavage specificity of PabRNase HII. Interestingly, the presence of the ribonucleotide just downstream of the cutting site induced random endonucleolytic cleavage with predominant products (Fig. 5B, lanes 6 to 9, and C, S8 substrate) and the percentage of hydrolyzed products was equivalent to that of Okazaki fragment-like substrates. When the ribonucleotide mismatch was positioned upstream of the site of cleavage, random endonucleolytic activity was enhanced but cleavage efficiencies were lowered (Fig. 5B, lanes 10 to 13). Multiple cleavage sites due to the loss of specificity appeared (Fig. 5C, S9 substrate). Taken together, these data showed for the first time that an archaeal RNase HII requires complete hybridization of the RNA segment to the DNA template in order to confer specific cleavage of RNA-DNA/DNA duplexes.
FIG. 5.
PabRNase HII specifically cuts the RNA-DNA/DNA duplex when the RNA is completely annealed to the DNA template. (A) Structures of substrates S7, S8, and S9. The thick line and the closed circle represent the RNA portion and the fluorescent label, respectively. (B) The indicated amounts of PabRNase HII were incubated with the S7 substrate (lanes 2 to 5), the S8 substrate (lanes 6 to 9), and the S9 substrate (lanes 10 to 13). An 11-nt ladder was used (lane 1). Fluorescently labeled products were visualized with a Mode Imager Typhoon 9400 (GE Healthcare), and quantification was performed using ImageQuant 5.2 software. (C) Graphical representation of sites and extents of cleavage at mismatches in RNA-DNA/DNA substrates. Cleavage sites are denoted by different bars. Deoxyribonucleotides and ribonucleotides are shown by uppercase and lowercase letters, respectively.
PabRNase HII specifically cuts a single embedded ribonucleotide in a DNA duplex.
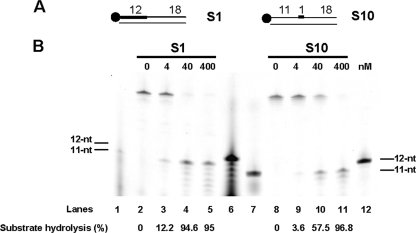
We anticipated that PabRNase HII nuclease could act on a single ribonucleotide embedded in DNA. A single ribonucleotide in a DNA duplex could arise via misincorporation of ribonucleotide during DNA synthesis or by ligation of the monoribonucleotide attached to the DNA after the cleavage of Okazaki fragments by type 2 RNase H (26). To determine whether an embedded ribonucleotide in DNA (S10 substrate) is a hydrolysable substrate, the endonuclease activity of PabRNase HII was assayed. Figure 6B, lanes 8 to 11, demonstrated that PabRNase HII was able to recognize and to cleave endonucleolytically on the 5′ side of an embedded monoribonucleotide. Additional fragments, shorter than the released 11 nt, were faintly detectable. Basically, cleavage efficiencies of a single embedded ribonucleotide were similar to those of the model Okazaki fragment S1 substrate (Fig. 6B, compare lanes 2 to 5 and 8 to 11). Overall, we showed that PabRNase HII is active on single embedded ribonucleotides in a DNA duplex and releases a major product consisting of a single ribonucleotide on the 5′ end of the downstream DNA segment.
FIG. 6.
PabRNase HII specifically cuts a single embedded ribonucleotide in a DNA duplex. (A) Structures of substrates S1 and S10. The thick line and the closed circle represent the RNA portion and the fluorescent label, respectively. (B) The indicated amounts of PabRNase HII were incubated with the S1 substrate (lanes 2 to 5) and the S10 substrate (lanes 8 to 11). Both substrates and the corresponding hydrolyzed products were manually labeled. Lanes 1 and 6 are appropriate 11- and 12-nt ladders for the hydrolyzed S1 substrates. Lanes 7 and 12 are suitable 11- and 12-nt ladders for the hydrolyzed S10 substrates. Fluorescently labeled products were visualized with a Mode Imager Typhoon 9400 (GE Healthcare), and quantification was performed using ImageQuant 5.2 software.
DISCUSSION
Two types of RNase H, 1 and 2, have been identified in a multiplicity of archaeal genomes. While most archaeal microorganisms have only one type of RNase H, a few archaea, such as Sulfolobus tokodaii and Haloferax volcanii, possess both types of RNase H. Although the physiological significance of multiple rnh genes in single archaeal genomes is not well understood, RNases H are thought to be involved in important cellular processes (3, 10, 16, 23, 24). Interestingly, archaeal type 2 RNase H appears more universal because the encoding gene is distributed in almost all archaeal genomes. Sequence comparison within archaeal type 2 RNases H revealed a high degree of sequence similarity with conserved active-site residues, suggesting that these enzymes may have biochemical properties in common (3, 9). In this report, we demonstrated that PabRNase HII, the type 2 RNase H from P. abyssi, is an alkaline enzyme. This property seems to be a hallmark of type 2 thermostable RNases H (3, 9, 22). In addition, PabRNase HII appeared to prefer the Mg2+ ion to Mn2+ or Co2+ for RNase activity. Distinct metal dependencies have been described for the A. fulgidus and T. kodakaraensis KOD1 RNases HII, with Mn2+ and Co2+ preferences, respectively (3, 9). Metal ion usage by archaeal RNases HII may be a consequence of the environmental conditions under which they thrive. It may also dictate the substrate requirement for hydrolysis and confer a specialized function on the enzyme in the maintenance of genome integrity. Determination of kinetic parameters highlighted that the homologous archaeal enzymes AfuRNase HII and PabRNase HII showed distinct catalytic efficiencies on RNA-DNA/DNA substrates. These results mainly reflected differences in substrate binding affinity. In general, biochemical discrepancies observed among the three enzymes are possibly related to variations in secondary structure elements and physicochemical parameters (e.g., isoelectric point). Despite these subtle differences, archaeal RNases HII seem to possess conserved structural features required to specifically recognize a comparable region of the substrates and to produce similar products. Like other type 2 archaeal RNases H, PabRNase HII behaved as an efficient endoribonuclease on RNA/DNA duplexes, stalling at particular sites (3, 9). Moreover, most of the biochemical features of PabRNase HII overlapped those of the eukaryotic equivalent, type 2 RNase H, described as a key enzyme in Okazaki fragment processing (17).
With diverse constructs representing replication fork intermediates, PabRNase HII made structure-specific endonucleolytic cleavage in the RNA initiator, leaving a single ribonucleotide at the 5′ end of the RNA-DNA junction. Cleavage 5′ to the junctional ribonucleotide required the presence of double-stranded substrates with the RNA segment fully annealed to the complementary strand. Gapped double-stranded substrates containing RNA-DNA junctions did not alter cleavage specificity. However, a single-stranded 5′ RNA flap was resistant to cleavage activity, indicating that PabRNase HII does not carry out this reaction at the replication fork. On the other hand, other results have demonstrated that the structure-specific nuclease Flap endonuclease I (Fen I) can cleave substrates with RNA flap structures, bypassing the need for RNase HII in Okazaki fragment processing (18, 27). Furthermore, we demonstrated that mismatches in the RNA portion, produced by erroneous priming and polymerizing activities during the initiation of DNA replication in eukaryotes (28, 31), resulted in loss of specificity by PabRNase HII. These results demonstrate, for the first time, that the RNA residues in the vicinity of the RNA-DNA junction are key structural determinants of the cleavage specificity of type 2 archaeal RNase H. Notably, archaeal type 2 RNase H seems to differ from eukaryotic type 2 (17) in that it recognizes the RNA strand rather than the RNA-DNA junction. Possibly, the RNA portion of the RNA-DNA junction annealed to a DNA template adopts an intermediate helical structure, which might target RNase HII recognition and induce specific cleavage. This hypothesis is sustained by the observation that RNA/DNA and DNA/DNA duplexes form A-type and B-type helices, respectively (5, 7).
We recently proposed a model of DNA replication in P. abyssi that involves the family B DNA polymerase P. abyssi PolB at the leading strand and the family D DNA polymerase P. abyssi PolD at the lagging strand (11). This model is reinforced by complementary studies demonstrating that P. abyssi PolB is likely the leading-strand DNA polymerase (25). Typically, P. abyssi PolD has the capacity to displace the downstream fragment, including the RNA initiator, while P. abyssi PolB is not active on this substrate. In this situation, RNA-initiated DNA segments fully annealed to a DNA template would arise only at the leading strand. Because PabRNase HII cannot cleave 5′ RNA flap templates, PabRNase HII would recognize the annealed RNA primer at the leading strand and promotes its endonucleolytic cleavage. The resulting 5′ phosphorylated junction ribonucleotide attached to the DNA would be subsequently displaced by P. abyssi PolB and cleaved by P. abyssi Fen I prior to ligation by P. abyssi DNA ligase I. Thus, the functional importance of RNase HII in the completion of leading-strand DNA replication in P. abyssi awaits the in vitro reconstitution of this multistep enzymatic process (unpublished data). Despite having biochemical properties in common with the eukaryotic type 2 RNase H, single archaeal RNases HII could be cellular enzymes involved in the removal of RNA residues at the leading strand rather than at the lagging strand. Such biological assumptions would indicate that these microorganisms have evolved differently by targeting analogous enzymes to unrelated biological functions.
Moreover, we demonstrated that PabRNase HII is able to cleave at the 5′ end of single embedded ribonucleotides with an efficiency similar to that seen with cognate Okazaki fragments (15). Since such structural substrates can appear in vivo during Okazaki fragment processing from intrinsic RNA ligation activity or erroneous nucleotide incorporation (26) and during exposure to external damaging agents (32), we suggest that PabRNase HII can participate in the removal of inappropriate ribonucleotides from the hyperthermophilic chromosome. These biochemical characteristics would imply that PabRNase HII promotes the initial step in the repair process, as already observed in eukaryotes (26). However, reconstitution of the complete enzymatic process awaits further assessment.
Acknowledgments
We are grateful to Hannu Myllykallio for providing the expression clone encoding PabRNase HII and critical reading of the manuscript.
This work was financially supported by the French Institute of Marine Research and Exploitation (Ifremer). Sébastien Le Laz thanks the University of Western Brittany for funding.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Cann, I. K., K. Komori, H. Toh, S. Kanai, and Y. Ishino. 1998. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 95:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerritelli, S. M., and R. J. Crouch. 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chai, Q., J. Qiu, B. R. Chapados, and B. Shen. 2001. Archaeoglobus fulgidus RNase HII in DNA replication: enzymological functions and activity regulation via metal cofactors. Biochem. Biophys. Res. Commun. 286:1073-1081. [DOI] [PubMed] [Google Scholar]
- 4.Chapados, B. R., Q. Chai, D. J. Hosfield, J. Qiu, B. Shen, and J. A. Tainer. 2001. Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J. Mol. Biol. 307:541-556. [DOI] [PubMed] [Google Scholar]
- 5.Chou, S. H., P. Flynn, and B. Reid. 1989. Solid-phase synthesis and high-resolution NMR studies of two synthetic double-helical RNA dodecamers: r(CGCGAAUUCGCG) and r(CGCGUAUACGCG). Biochemistry 28:2422-2435. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, G. N., V. Barbe, D. Flament, M. Galperin, R. Heilig, O. Lecompte, O. Poch, D. Prieur, J. Querellou, R. Ripp, J. C. Thierry, J. Van der Oost, J. Weissenbach, Y. Zivanovic, and P. Forterre. 2003. An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 47:1495-1512. [DOI] [PubMed] [Google Scholar]
- 7.Dickerson, R. E., H. R. Drew, B. N. Conner, R. M. Wing, A. V. Fratini, and M. L. Kopka. 1982. The anatomy of A-, B-, and Z-DNA. Science 216:475-485. [DOI] [PubMed] [Google Scholar]
- 8.Gueguen, Y., J. L. Rolland, O. Lecompte, P. Azam, G. Le Romancer, D. Flament, J. P. Raffin, and J. Dietrich. 2001. Characterization of two DNA polymerases from the hyperthermophilic euryarchaeon Pyrococcus abyssi. Eur. J. Biochem. 268:5961-5969. [DOI] [PubMed] [Google Scholar]
- 9.Haruki, M., K. Hayashi, T. Kochi, A. Muroya, Y. Koga, M. Morikawa, T. Imanaka, and S. Kanaya. 1998. Gene cloning and characterization of recombinant RNase HII from a hyperthermophilic archaeon. J. Bacteriol. 180:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haruki, M., Y. Tsunaka, M. Morikawa, and S. Kanaya. 2002. Cleavage of a DNA-RNA-DNA/DNA chimeric substrate containing a single ribonucleotide at the DNA-RNA junction with prokaryotic RNases HII. FEBS Lett. 531:204-208. [DOI] [PubMed] [Google Scholar]
- 11.Henneke, G., D. Flament, U. Hubscher, J. Querellou, and J. P. Raffin. 2005. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 350:53-64. [DOI] [PubMed] [Google Scholar]
- 12.Lai, L., H. Yokota, L. W. Hung, R. Kim, and S. H. Kim. 2000. Crystal structure of archaeal RNase HII: a homologue of human major RNase H. Structure 8:897-904. [DOI] [PubMed] [Google Scholar]
- 13.Lao-Sirieix, S. H., and S. D. Bell. 2004. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J. Mol. Biol. 344:1251-1263. [DOI] [PubMed] [Google Scholar]
- 14.Le Breton, M., G. Henneke, C. Norais, D. Flament, H. Myllykallio, J. Querellou, and J. P. Raffin. 2007. The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and repair? J. Mol. Biol. 374:1172-1185. [DOI] [PubMed] [Google Scholar]
- 15.Matsunaga, F., C. Norais, P. Forterre, and H. Myllykallio. 2003. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 4:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meslet-Cladiére, L., C. Norais, J. Kuhn, J. Briffotaux, J. W. Sloostra, E. Ferrari, U. Hubscher, D. Flament, and H. Myllykallio. 2007. A novel proteomic approach identifies new interaction partners for proliferating cell nuclear antigen. J. Mol. Biol. 372:1137-1148. [DOI] [PubMed] [Google Scholar]
- 17.Murante, R. S., L. A. Henricksen, and R. A. Bambara. 1998. Junction ribonuclease: an activity in Okazaki fragment processing. Proc. Natl. Acad. Sci. U. S. A. 95:2244-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murante, R. S., J. A. Rumbaugh, C. J. Barnes, J. R. Norton, and R. A. Bambara. 1996. Calf RTH-1 nuclease can remove the initiator RNAs of Okazaki fragments by endonuclease activity. J. Biol. Chem. 271:25888-25897. [DOI] [PubMed] [Google Scholar]
- 19.Muroya, A., D. Tsuchiya, M. Ishikawa, M. Haruki, M. Morikawa, S. Kanaya, and K. Morikawa. 2001. Catalytic center of an archaeal type 2 ribonuclease H as revealed by X-ray crystallographic and mutational analyses. Protein Sci. 10:707-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nick McElhinny, S. A., B. E. Watts, D. Kumar, D. L. Watt, E. B. Lundstrom, P. M. Burgers, E. Johansson, A. Chabes, and T. A. Kunkel. 2010. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U. S. A. 107:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtani, N., M. Haruki, M. Morikawa, and S. Kanaya. 1999. Molecular diversities of RNases H. J. Biosci. Bioeng. 88:12-19. [DOI] [PubMed] [Google Scholar]
- 22.Ohtani, N., M. Tomita, and M. Itaya. 2008. Junction ribonuclease: a ribonuclease HII orthologue from Thermus thermophilus HB8 prefers the RNA-DNA junction to the RNA/DNA heteroduplex. Biochem. J. 412:517-526. [DOI] [PubMed] [Google Scholar]
- 23.Ohtani, N., H. Yanagawa, M. Tomita, and M. Itaya. 2004. Cleavage of double-stranded RNA by RNase HI from a thermoacidophilic archaeon, Sulfolobus tokodaii 7. Nucleic Acids Res. 32:5809-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohtani, N., H. Yanagawa, M. Tomita, and M. Itaya. 2004. Identification of the first archaeal type 1 RNase H gene from Halobacterium sp. NRC-1: archaeal RNase HI can cleave an RNA-DNA junction. Biochem. J. 381:795-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouillon, C., G. Henneke, D. Flament, J. Querellou, and J. P. Raffin. 2007. DNA polymerase switching on homotrimeric PCNA at the replication fork of the euryarchaea Pyrococcus abyssi. J. Mol. Biol. 369:343-355. [DOI] [PubMed] [Google Scholar]
- 26.Rumbaugh, J. A., R. S. Murante, S. Shi, and R. A. Bambara. 1997. Creation and removal of embedded ribonucleotides in chromosomal DNA during mammalian Okazaki fragment processing. J. Biol. Chem. 272:22591-22599. [DOI] [PubMed] [Google Scholar]
- 27.Sato, A., A. Kanai, M. Itaya, and M. Tomita. 2003. Cooperative regulation for Okazaki fragment processing by RNase HII and FEN-1 purified from a hyperthermophilic archaeon, Pyrococcus furiosus. Biochem. Biophys. Res. Commun. 309:247-252. [DOI] [PubMed] [Google Scholar]
- 28.Sheaff, R. J., and R. D. Kuchta. 1994. Misincorporation of nucleotides by calf thymus DNA primase and elongation of primers containing multiple noncognate nucleotides by DNA polymerase alpha. J. Biol. Chem. 269:19225-19231. [PubMed] [Google Scholar]
- 29.Stein, H., and P. Hausen. 1969. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science 166:393-395. [DOI] [PubMed] [Google Scholar]
- 30.Tadokoro, T., and S. Kanaya. 2009. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 276:1482-1493. [DOI] [PubMed] [Google Scholar]
- 31.Thomas, D. C., J. D. Roberts, R. D. Sabatino, T. W. Myers, C. K. Tan, K. M. Downey, A. G. So, R. A. Bambara, and T. A. Kunkel. 1991. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry 30:11751-11759. [DOI] [PubMed] [Google Scholar]
- 32.Von Sonntag, C., and D. Schulte-Frohlinde. 1978. Radiation-induced degradation of the sugar in model compounds and in DNA. Mol. Biol. Biochem. Biophys. 27:204-226. [PubMed] [Google Scholar]