Abstract
Monitoring the external environment and responding to its changes are essential for the survival of all living organisms. The transmission of extracellular signals in prokaryotes is mediated mainly by two-component systems. In addition, genomic analyses have revealed that many bacteria contain eukaryotic-type Ser/Thr protein kinases. The human pathogen Streptococcus pneumoniae encodes 13 two-component systems and has a single copy of a eukaryotic-like Ser/Thr protein kinase gene designated stkP. Previous studies demonstrated the pleiotropic role of the transmembrane protein kinase StkP in pneumococcal physiology. StkP regulates virulence, competence, and stress resistance and plays a role in the regulation of gene expression. To determine the intracellular signaling pathways controlled by StkP, we used a proteomic approach for identification of its substrates. We detected six proteins phosphorylated on threonine by StkP continuously during growth. We identified three new substrates of StkP: the Mn-dependent inorganic pyrophosphatase PpaC, the hypothetical protein spr0334, and the cell division protein DivIVA. Contrary to the results of a previous study, we did not confirm that the α-subunit of RNA polymerase is a target of StkP. We showed that StkP activation and substrate recognition depend on the presence of a peptidoglycan-binding domain comprising four extracellular penicillin-binding protein- and Ser/Thr kinase-associated domain (PASTA domain) repeats. We found that StkP is regulated in a growth-dependent manner and likely senses intracellular peptidoglycan subunits present in the cell division septa. In addition, stkP inactivation results in cell division defects. Thus, the data presented here suggest that StkP plays an important role in the regulation of cell division in pneumococcus.
Protein phosphorylation is considered the universal language for inter- and intracellular communication in all living organisms. This process, catalyzed by protein kinases, enables translation of extracellular signals into cellular responses and adaptation to a constantly changing environment. Although previous work indicated that histidine kinases of two-component regulatory systems are the most prominent kinases in prokaryotes, recent data provide evidence that eukaryotic-type Ser/Thr protein kinases (ESTPKs) play an important role in prokaryotic cell signaling. The distribution of these enzymes among bacteria is broad but not universal. ESTPKs have been found in nearly two-thirds of the sequenced bacterial strains. Most of these enzymes are encoded by genes in strains belonging to the phyla Proteobacteria (Myxococcales), Actinobacteria, Cyanobacteria, Chloroflexi, Acidobacteria, and Planctomycetes (38). We surveyed sequenced bacterial genomes and found ESTPK-encoding genes also in the bacterial taxa Aquificae, Chlamydiae, Spirochaetes, Bacteroidetes, Deinococcus, Fusobacteria, and Firmicutes. Hence, ESTPKs are present in phylogenetically diverse groups of bacteria that have various life strategies.
Streptococcus pneumoniae encodes 13 two-component systems and one orphan regulator. In addition, analysis of the genome sequence of this species revealed the presence of a single gene coding for a eukaryotic-type Ser/Thr protein kinase, which was designated stkP. Gene disruption data for S. pneumoniae proved that StkP is not essential for growth in laboratory conditions but is indispensable for successful lung infection and blood invasion in mice (9). Phenotypic studies have shown that StkP plays a role in the regulation of genetic competence and autolysis (9, 45), contributes to the resistance of S. pneumoniae to environmental stresses, and plays a role in the regulation of expression of genes employed in competence, cell wall biosynthesis, pyrimidine biosynthesis, DNA repair, iron uptake, and the oxidative stress response (45). Recently, StkP was identified as an important pneumococcal antigen and was selected as one of the lead candidates for development of a new antipneumococcus protein-based vaccine (17).
StkP is homologous to the PknB protein kinase of Mycobacterium tuberculosis (3). PknB-like protein kinases make up a distinct family of transmembrane ESTPKs that are found exclusively in Gram-positive bacteria. Their structure consists of an intracellular kinase domain, a transmembrane span, and repeated extracellular penicillin-binding protein- and Ser/Thr kinase-associated domains (PASTA domains) (54). PASTA domains are found in penicillin-binding proteins (PBP) and Ser/Thr protein kinases, and their function was deduced from the crystal structure of pneumococcal PBP2X, which revealed that PASTA domains bind the beta-lactam ring of the antibiotic cefuroxime (18). The beta-lactam ring resembles the d-Ala-d-Ala motif of the peptidoglycan stem peptide; therefore, it was proposed that ESTPKs containing PASTA domains sense unlinked peptidoglycan subunits and regulate cell wall biosynthesis (54). Recently, Shah et al. (47) proved that protein kinase PrkC from Bacillus subtilis binds purified peptidoglycan or muropeptides derived from cultures of growing cells and that these molecules act as activation signals.
Inactivation of PknB-like sensor kinases showed that they are essential for growth in M. tuberculosis (13) and Corynebacterium glutamicum (15). Further study confirmed their roles in the regulation of development, cell shape, and cell wall integrity in M. tuberculosis (23), Streptococcus agalactiae (41), Streptococcus pyogenes (21), C. glutamicum (14, 15), Enterococcus faecalis (24), Staphylococcus aureus (4), and B. subtilis (47). Depletion of these signaling enzymes attenuated virulence in streptococci (9, 21, 41) and S. aureus (8) and affected biofilm formation in B. subtilis (31) and Streptococcus mutans (20).
Identification of several substrates in different bacteria has revealed in part the signaling pathways controlled by PknB-like kinases. The spectrum of substrates is broad and, except for hypothetical proteins, ranges from enzymes involved in cell wall biosynthesis (1, 14, 33, 35, 51), cell division (23, 49, 50), transcription regulation (21, 35, 43, 48), translation factors (1, 16), and chaperones (6) to enzymes involved in central metabolic pathways (41, 42). The great diversity of substrates indicates that PknB-like kinases regulate cellular processes on various levels, which may enable very flexible cell responses to the signals received.
Three substrates of pneumococcal StkP have been reported to date. In a previous study, we identified two potential substrates of StkP after in vivo S. pneumoniae cultures were labeled with 33P-orthophosphate: RpoA, the α-subunit of RNA polymerase, and GlmM, a phosphoglucosamine mutase, which was proved to be phosphorylated by StkP in vitro as well (35). GlmM catalyzes the interconversion of glucosamine-6-phosphate (GlcN-6-P) and GlcN-1-P isomers, which is the first step in the biosynthetic pathway leading to the formation of UDP-N-acetylglucosamine, an essential common precursor of cell envelope components, such as peptidoglycan, lipopolysaccharides, and teichoic acids. Ulijasz et al. (52) reported that StkP phosphorylates a DNA-binding domain of the orphan regulator RitR in vitro. This modification affects the RitR interaction with the piu promoter, suggesting a new mechanism for regulation of gene expression in S. pneumoniae.
In this study, we surveyed dynamic changes in StkP activity in vivo and assessed the role of StkP-associated PASTA domains in kinase activation. We proved that peptidoglycan-binding domains at the C terminus of StkP are indispensable for kinase activation and subsequent substrate phosphorylation. We showed that StkP is constitutively active; it phosphorylates a set of six substrates continuously throughout growth in a complex medium, and the activity of StkP decreases after growth arrest. Thus, we demonstrated for the first time that PknB-like protein kinase is activated in a growth-dependent manner. We attempted to identify multiple StkP substrates in vivo by immunodetection with specific antibody against phosphothreonine (pThr). Using a proteomic approach and mass spectrometry analysis, we identified three new StkP substrates: the cell division protein DivIVA, the hypothetical protein spr0334, and the Mn-dependent inorganic pyrophosphatase PpaC. Further, we obtained evidence that a putative substrate identified in a previous screen, the α-subunit of RNA polymerase, is not phosphorylated by StkP. Consistent with identification of the cell division protein DivIVA as a substrate of StkP, we showed that stkP mutant strains have defects in cell division. Hence, we concluded that StkP monitors intracellular levels of peptidoglycan subunits in actively growing cells and regulates the cell division cycle of S. pneumoniae by controlling the activity of DivIVA.
MATERIALS AND METHODS
Bacterial strains and growth media.
Cultures of S. pneumoniae strains (Table 1) were grown in casein tryptone medium (CAT medium) (34). Cultures of Escherichia coli were routinely propagated in Luria broth. When necessary, antibiotics were added at the following concentrations: for E. coli host strains, 50 μg/ml kanamycin and 100 μg/ml ampicillin; and for S. pneumoniae strains, 10 μg/ml chloramphenicol. E. coli JM109 was used as the recipient strain for most DNA manipulations. E. coli BL21(DE3) was used as a host for protein overexpression.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Other designation | Description | Source or reference |
|---|---|---|---|
| E. coli strains | |||
| JM109 | endA1 recA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Promega | |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen | |
| S. pneumoniae strains | |||
| Cp1015 | Sp1 | Rx derivative, str1 hexA | 34 |
| Cp1015ΔstkP | Sp10 | Cp1015 ΔstkP; Cmr | 35 |
| Cp1015stkPK42R | Sp19 | Cp1015, stkP(K42R)-His tag, Cmr, allelic exchange mutant | This study |
| Cp1015stkP-KDtm (stkPΔ372-659) | Sp25 | Cp1015, stkP::kdtm-His tag, Cmr, allelic exchange mutant | 37 |
| D39 | Sp6 | Serotype 2, wild type | 2 |
| Plasmids | |||
| pBluescript II SK+ | Ampr | Stratagene | |
| pEVP3 | Cmr | 7 | |
| pBSK-StkPK42RCm | pBluescript II SK+ derivative used for generation of strain Sp19 by allelic exchange; Ampr Cmr | This study | |
| pEXstkP-T | pET28b derivative used for expression of rStkP-KD; Kanr | 35 | |
| pETPhos | pET derivative used for expression of DivIVA, spr0334, PpaC, and RpoA; Ampr | 5 |
Preparation of protein lysates and cell fractionation.
Cultures of S. pneumoniae were grown at 37°C in 200 ml CAT medium until the desired optical density was reached. Cultures were harvested by centrifugation at 5,000 × g for 10 min at 4°C, and cell pellets were resuspended in 1 ml of precooled lysis buffer containing 25 mM Tris (pH 7.5), 100 mM NaCl, Benzonase (Merck), and protease inhibitors (Roche). Cells were disrupted with a French pressure cell (SLM Instruments), and cell debris was separated by centrifugation at 10,000 × g for 10 min. Cell lysates were subjected to ultracentrifugation (100,000 × g for 1 h), and the cytoplasmic and membrane fractions were obtained. The protein concentration was determined using a bicinchoninic acid (BCA) protein estimation kit (Pierce).
Cloning, expression, and purification of recombinant proteins DivIVA, spr0334, PpaC, RpoA, and StkP kinase domain (StkP-KD).
The sequences corresponding to the divIVA (spr1505), spr0334, ppaC (spr1389), and rpoA (spr0215) genes were PCR amplified using S. pneumoniae Rx1 genomic DNA as the template and the primers shown in Table 2. The resulting PCR fragments were digested with the appropriate restriction enzymes and cloned into a modified pETPhos vector (5). Prior to cloning, the original pETPhos vector was modified by introduction of a linker (TCGACCTCGAGCAATTG) to generate new restriction sites in the multiple cloning site. The resulting plasmids, pETPhos1505, pETPhos0334, pETPHos1389, pETPhos0215, and pEXStkP-T (35), were transformed into E. coli BL21(DE3). The expression strains were cultivated at 30°C until the optical density at 600 nm (OD600) was 0.6. Overproduction of recombinant proteins was initiated by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and the cultures were then incubated for 3 h. Recombinant proteins were purified at room temperature by using Ni-nitrilotriacetic acid (NTA) metal affinity resin (Qiagen) according to the manufacturer's instructions. Purified proteins were dialyzed against buffer containing 25 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 10% (vol/vol) glycerol.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Use |
|---|---|---|
| CAT 1 | CGCGGATCCGAAAATTTGTTTGATTTTTAA | Selection marker |
| CAT 2 | GCTCTAGAGGGTTCC GACCGTCAACGTCAA | Selection marker |
| STK-f | CGCGAATTCATGATCCAAATCGGCAA | Allelic exchange |
| STK-r | CGCGGATCCTTAGTGATGGTGATGGTGATGGTGATGGTGATGAGGAGTAGCTGAAGTTGTTTTAGGT | Allelic exchange |
| X-f | GCTCTAGAGAGTCCAGATTTGTGATA | Allelic exchange |
| X2-r | TGCCCGCGGGCGTTGTTAATTTATTTA | Allelic exchange |
| LN89/spr1505-F | CGGCCATATGCCAATTACATCATTAG | DivIVA expression |
| LN90/spr1505-R | GCCGGAATTCCTACTTCTGGTTCTTCA | DivIVA expression |
| LN72/spr0334-F | CGGCCATATGAGTAAAAAAAGACGAAATCG | spr0334 expression |
| LN73/spr0034-R | GCCGGAATTCTTAGTAGTCCAAGTCATC | spr0334 expression |
| LN1/SPR0215-f | GGCCCGCATATGATCGAGTTTGAAAAA | RpoA expression |
| LN2/SPR0215-r | CCGCTCGAGTTATTTATCTTTTAATCC | RpoA expression |
| LN70/ppi-F | GCCGCATATGTCCAAGATTCTAGTATTTGG | PpaC expression |
| LN71/ppi-R | CGGCGGATCCTTACGCATTAAAGCTTTCAG | PpaC expression |
Restriction sites are underlined.
In vitro protein phosphorylation.
The in vitro protein kinase reaction mixture contained 0.4 μg of recombinant substrate protein and 0.4 μg of purified StkP kinase domain (StkP-KD) (35) in kinase buffer (25 mM Tris-HCl [pH 7.5], 25 mM NaCl, 5 mM MnCl2, 10 μM ATP). The reaction was started by addition of ATP and was terminated after 30 min of incubation in 37°C by addition of 5× SDS sample buffer. Samples were separated on 12% acrylamide SDS-polyacrylamide gel electrophoresis (PAGE) gels and electroblotted. Proteins were detected either with anti-phosphothreonine polyclonal antibody (Cell Signaling) or with peroxidase-conjugated anti-polyhistidine monoclonal antibody (Sigma).
Construction of S. pneumoniae stkP(K42R) strain.
A mutant strain of S. pneumoniae expressing StkP(K42R) was prepared by transformation of S. pneumoniae wild-type strain Sp1 with a vectorless DNA fragment containing the oligohistidine-tagged stkP(K42R) gene, the cat cassette, and the downstream sequence of the stkP gene [5′-stkP(K42R)-cat-flanking region-3′], a procedure similar to the procedure described by Pallova et al. (37). The identity of the mutant was verified by PCR using appropriate primers and restriction analysis of the PCR products, followed by sequencing to detect the presence of the K42R mutation. To create epitope-labeled StkP(K42R), the gene was amplified using primers STK-f and STK-r from plasmid pEXstkP-K42R (35), which resulted in a 2,000-bp product. The downstream fragment was amplified with primers X-f and X2-r using chromosomal DNA as the template. Plasmid pEVP3 (7) was used as the source of the cat cassette, which was amplified with primers CAT1 and CAT2. The pBluescript II SK+ vector was used for cloning and sequencing experiments.
Two-dimensional (2D) SDS-PAGE.
Proteins were precipitated overnight using 4 volumes of acetone at −20°C. Protein pellets were dissolved in lysis buffer containing 8 M urea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.8% Pharmalyte 3-10, 65 mM dithiothreitol (DTT), and bromophenol blue. Protein pellets derived from membranes were dissolved in lysis buffer containing 8 M urea, 2 M thiourea, 4% CHAPS, 1% amido sulfobetaine-14 (ASB14), 1% Triton X-100, 0.8% Pharmalyte 3-10, 65 mM DTT, and bromophenol blue. Samples were soaked into IPG strips (Immobiline DryStrips; 18 cm; pH 3-10 NL or pH 3-5.6 NL; Amersham Biosciences) and rehydrated overnight. In the first dimension, isoelectric focusing (IEF) was performed using voltage that was linearly increased to the steady state (the voltage was limited to 150 V for 2 h, 300 V for 2 h, and 3,500 V for 5 h) and then stabilized and kept at 3,500 V for 31 h (Multiphor II; Amersham Pharmacia Biotech). After IEF, the strips were first washed in equilibration solution (50 mM Tris-HCl [pH 6.8], 6 M urea, 30% glycerol, 2% SDS) containing 0.02 g/ml DTT for 10 min, which was followed by a second 10-min wash in equilibration buffer containing 0.025 g/ml iodoacetamide and a few grains of bromophenol blue. SDS-PAGE was carried out using 12.5% polyacrylamide slab gels (Investigator; Genomic Solutions). The gels were either stained with colloidal Coomassie brilliant blue G-250 (CBB G-250) or electroblotted.
Doubled SDS-PAGE (dSDS-PAGE).
Protein extracts were prepared by mixing 30 μg of proteins from the membrane fraction with 5× SDS sample buffer (250 mM Tris-HCl [pH 6.8], 40% [vol/vol] glycerol, 5% [wt/vol] SDS, 0.005% [wt/vol] bromophenol blue) and heating the preparation for 10 min at 37°C. Proteins were resolved on 10% acrylamide SDS-PAGE gels containing 10.7% glycerol, 1 M Tris (pH 8.8), 0.1% SDS, 6% bisacrylamide cross-linker, 0.05% ammonium peroxodisulfate (APS), and 0.05% N,N,N′,N′-tetramethylethylenediamine (TEMED), as described elsewhere (53). Protein gels (138 by 130 by 1 mm) were run using a bicine-based buffer system, as described by Williams et al. (53), in an ATTO electrophoresis slab chamber. After staining with Coomassie blue R-250, gel strips were excised, equilibrated in buffer (100 mM Tris, pH 6.8), and placed on top of a 1.5-mm-thick second-dimension gel (140 by 160 mm). The gel strip was overloaded with a stacking gel mixture, using the Laemmli protocol. Second-dimension electrophoresis was run as described by Laemmli (25) using a Hoefer SE600 dual-gel electrophoresis unit. The gels were either stained with colloidal CBB G-250 or electroblotted.
Immunoblotting.
Proteins resolved on SDS-PAGE gels were electroblotted at 0.8 mA/cm2 for 2 h onto a polyvinylidene difluoride (PVDF) membrane (Millipore) using the OWL semidry electroblotting system. Phosphorylated proteins were detected with anti-phosphothreonine polyclonal antibody (Cell Signaling) or anti-phosphoserine monoclonal antibody (Sigma) used at a 1:2,000 dilution. StkP was detected using polyclonal anti-StkP serum at a dilution of 1:1,000 (35). Peroxidase-conjugated anti-polyhistidine monoclonal antibody (Sigma) at a dilution of 1:20,000 was used for detection of recombinant His-tagged proteins. Polyclonal anti-DivIVA serum (12) and polyclonal serum raised against RpoA were used at a 1:20,000 dilution. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit serum was used as a secondary antibody (1:10,000), and detection was carried out using the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
MALDI-FTMS.
The spots of interest were cut from the gel, destained by addition of 100 mM ethylmorpholine acetate buffer (Sigma-Aldrich) and acetonitrile at a 1:1 ratio, and reduced by using tris(2-carboxyethyl)phosphine hydrochloride (Sigma-Aldrich). The reduced cysteines were then alkylated with 50 mM iodoacetamide (Sigma-Aldrich). The gel pieces were washed three times by addition of acetonitrile and water. Trypsin protease was added to the gel in digestion buffer (50 mM ethylmorpholine acetate buffer, 10% acetonitrile; pH 8.3). After overnight incubation, tryptic peptides were extracted from the gel by addition of 80% acetonitrile, 0.1% trifluoroacetic acid (TFA). The extracted peptides were spotted onto a 384-position matrix-assisted laser desorption ionization (MALDI) plate (Bruker Daltonics, Billerica, MA) and overlaid with α-cyano-4-hydroxycinnamic acid as a matrix (Bruker Daltonics, Germany). Samples were ionized by MALDI using a Dual II ion source (Bruker Daltonics, Billerica, MA). Mass spectra were acquired with an APEX-Qe Fourier transform mass spectrometry (FTMS) instrument equipped with a 9.4-T superconducting magnet (Bruker Daltonics, Billerica, MA). The cell was opened for 4 ms, and the accumulation time was set at 0.2 s. One experiment consisted of the average of four spectra. The acquisition data set size was set to 512,000 points with the mass range starting at m/z 600 atomic mass units. The instrument was externally calibrated with Bruker Daltonics calibration standard II (Bruker Daltonics, Germany), resulting in a mass error greater than 1 ppm. The spectra were processed by using the Data Analysis 4.0 software (Bruker Daltonics, Billerica, MA) and were searched by using the Mascot search engine against a database created from all known bacterial proteins.
Microscopy.
For light microscopy, cells were cultured in CAT medium until the OD400 was 0.4, and living cells were observed with an Olympus BX-60 microscope equipped with an oil immersion objective (100×; 1.35 NA), using differential interference contrast (Nomarski). Photographs were obtained with a charge-coupled device Fluoview camera and were processed with AnalySIS imaging software in order to evaluate cell sizes. A total of 470 cells of each strain were measured, and the minimal, maximal, and average sizes were calculated with the program R for statistical computing (www.r-project.org).
RESULTS AND DISCUSSION
Detection of putative StkP substrates in vivo.
Previously, we used in vivo labeling with 33P-orthophosphate to detect phosphorylated proteins in S. pneumoniae (35). This method is limited by a requirement for specific growth medium and identifies proteins that are phosphorylated exclusively during the pulse-labeling event. These constraints prevent certain substrates from being detected. Therefore, we decided to use an alternative approach to identify StkP substrates: immunodetection with specific antibodies raised against phosphothreonine (anti-pThr) and phosphoserine (anti-pSer).
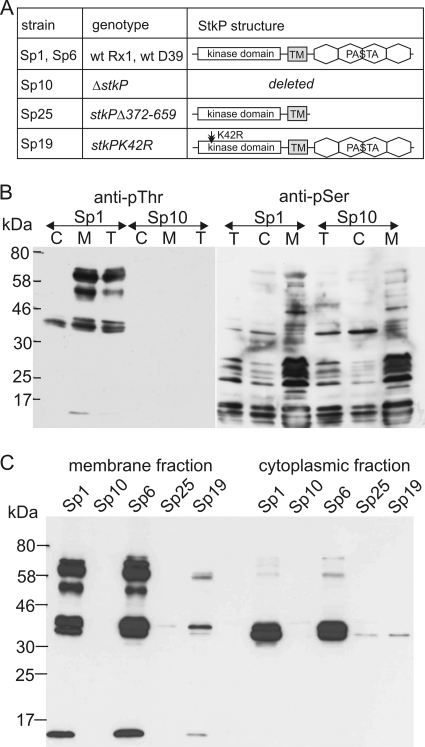
We performed immunodetection with cell lysates from cultures of a wild-type strain of S. pneumoniae (Sp1) and a ΔstkP mutant strain (Sp10) and compared the patterns of phosphorylated proteins in the cytoplasmic and membrane fractions (Fig. 1 B). The anti-pThr antibody reacted with at least six protein bands in the wild type; however, no phosphorylated proteins were detected in the Sp10 strain (Fig. 1B, anti-pThr gel). This result indicated that phosphorylation on Thr residues in S. pneumoniae is dependent on the presence of StkP. On the other hand, numerous phosphorylated protein bands were detected with anti-pSer antibody in both the wild-type strain and the ΔstkP mutant, and we did not detect major differences between these two strains (Fig. 1B, anti-pSer gel). Remarkably, our data indicate that there is overrepresentation of phosphoserine residues in S. pneumoniae, which is consistent with data for B. subtilis and E. coli, which are model bacteria that have an established Ser/Thr/Tyr phosphorylation ratio of 70:20:10 (29, 30).
FIG. 1.
Detection of in vivo phosphorylated proteins in S. pneumoniae. wt, wild type; TM, transmembrane domain. (A) Pneumococcal strains used in this study and schematic diagrams of the StkP structure. (B) Phosphorylation of proteins on Thr is StkP dependent. Proteins phosphorylated on Thr (anti-pThr) and Ser (anti-pSer) were immunodetected in fractionated protein lysates of wild-type strain Sp1 and ΔstkP mutant strain Sp10. C, cytoplasmic fraction; M, membrane fraction; T, total protein lysate; anti-pThr, immunodetection with antibody against phosphothreonine; anti-pSer, immunodetection with antibody against phosphoserine. (C) Phosphorylation of proteins on Thr in S. pneumoniae depends on StkP catalytic activity. Proteins phosphorylated on Thr were immunodetected in membrane and cytoplasmic fractions of wild-type strains Sp1 (Rx1) and Sp6 (D39) and the following three stkP mutant strains: Sp10, a ΔstkP strain; Sp25, a strain expressing an isolated kinase domain anchored in the membrane; and Sp19, a strain expressing StkPK42R with reduced catalytic activity.
In conclusion, StkP specifically phosphorylates several putative substrates in the cytoplasm, as well as in the cell membrane of S. pneumoniae, mainly on threonine residues. In contrast, phosphorylation on serine residues is predominantly independent of StkP and could therefore be attributed to either intrinsic autophosphorylation activity or protein kinases not homologous to eukaryotic enzymes. Indeed, phosphoproteomic analyses of B. subtilis and E. coli showed that there was frequent Ser/Thr/Tyr phosphorylation of proteins involved in essential metabolic pathways, particularly enzymes involved in carbon metabolism. In this context, it is noteworthy that E. coli lacks enzymes homologous to eukaryotic-type protein kinases (30).
Similar substrates are phosphorylated in avirulent and virulent strains of S. pneumoniae in an StkP-dependent manner.
To prove that there is a relationship between protein phosphorylation and StkP activity, we compared the patterns of proteins phosphorylated on Thr in wild-type strains Rx1 and D39 and in a set of stkP mutant strains (Fig. 1A and 1C). A previous study demonstrated that insertional inactivation of stkP in the virulent strain D39 resulted in significant attenuation of virulence in the pneumonia and bacteremia models of infection (9). Here we found that the patterns of phosphorylated proteins were similar for S. pneumoniae Rx1 and D39 (avirulent and virulent strains, respectively) when they were grown in complex CAT medium (Fig. 1C, Sp1 and Sp6 lanes). Thus, in both strains, StkP phosphorylates the same set of substrates that are probably important for sustaining normal growth in laboratory conditions.
To confirm that phosphorylation of proteins on Thr in S. pneumoniae depends on StkP kinase activity, we constructed strain Sp19 expressing catalytically inactive StkP(K42R). The invariant lysine K42 in the ATP-binding pocket of the conserved kinase domain is essential for kinase activity (19). Previously, we showed that although autophosphorylation of recombinant StkP(K42R) in vitro is not completely abolished, it is reduced to 5% of the wild-type level (35). In agreement with this result, we observed a significant decrease in Thr phosphorylation in Sp19 cell lysates (Fig. 1C, Sp19 lanes). In this mutant, there was still residual activity of StkP(K42R), but phosphorylation of the substrates was reduced and could be visualized only after prolonged exposure of the immunodetection reaction mixture. Hence, we confirmed that Thr phosphorylation of proteins in vivo depends on StkP enzymatic activity.
PASTA domains are essential for kinase activation.
Previously, a model for activation of the protein kinase PknB, an StkP homolog in M. tuberculosis, was proposed based on its crystal structure (55). PknB, like StkP, is composed of an intracellular kinase domain, a transmembrane helix, and an extracellular C-terminal portion containing four PASTA repeats. According to the model, substrate specificity is determined by oligomerization and/or localization of the protein kinase, as it is for several classes of eukaryotic Ser/Thr protein kinases (36, 39). It was proposed that upon ligand binding to the sensory domain the protein kinase dimerizes. Dimerization brings the two kinase domains into intimate contact, and a subsequent transphosphorylation reaction results in conformational changes and substrate recognition.
In a previous study of StkP dimerization using an in vivo genetic assay system (37), we showed that the StkP kinase domain (StkPΔ270-659) is monomeric, whereas the kinase domain associated with the transmembrane span (StkPΔ372-659) and the full-length StkP are dimeric. Thus, the dimerization is mediated by the transmembrane region and the C-terminal PASTA domains in vivo. These results were confirmed in vitro using blue native PAGE (37). The truncated StkPΔ372-659 kinase and the full-length StkP expressed in S. pneumoniae were able to autophosphorylate in an in vitro kinase assay (37).
In this study, we aimed to assess the importance of the PASTA domains in StkP activation and substrate recognition in vivo. We therefore tested the phosphorylation of proteins in pneumococcal strain Sp25 expressing the membrane-anchored StkP kinase domain StkPΔ372-659, which lacks all of the PASTA domains (37). As shown in Fig. 1C (Sp25 lanes), this deletion prevents StkP from phosphorylating its substrates in vivo. Although the membrane-spanning region of StkP is necessary for dimer formation in the λ phage genetic assay system in E. coli and the truncated kinase is able to autophosphorylate in vivo (37), it is not sufficient for activation of StkP in S. pneumoniae. In conclusion, we obtained evidence that dimerization through the transmembrane region cannot stimulate kinase activity in vivo and that the C-terminal PASTA domains are essential for StkP activation and substrate phosphorylation. We concluded that binding of a ligand to the PASTA domains is a crucial event that results in kinase activation. The PASTA domains were proposed to bind free peptidoglycan (PG) subunits (54), and recently it was shown that muropeptides in a crude preparation were indeed ligands of the B. subtilis PrkC protein kinase in vitro (47). In vivo, addition of PG subunits resulted in activation of a PrkC-dependent phosphorylation pathway, leading to germination of dormant B. subtilis spores (47).
Kinetics of StkP substrate phosphorylation during growth.
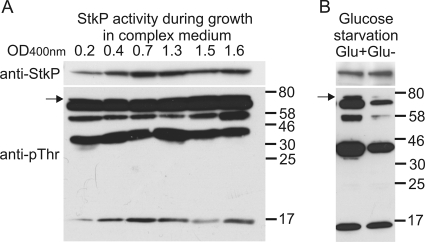
Ser/Thr phosphorylation of proteins in bacteria responds dynamically to changes in environmental conditions (27). To investigate the kinetics of protein phosphorylation, we detected phosphorylated proteins at six time points during the growth of S. pneumoniae in complex medium. As shown in Fig. 2 A, we observed Thr phosphorylation in all samples examined, and neither the number of phosphorylated proteins nor the amounts of these proteins differed significantly. We concluded that under the conditions tested StkP is constantly active and phosphorylates a set of substrates steadily during cell growth. As mentioned above, PknB-like ESTPKs are most likely activated upon binding of PG subunits. There are two potential sources of free PG subunits: (i) endogenous PG at sites of active cell wall biosynthesis (septum) and (ii) exogenous PG, represented by muropeptides released during cell lysis. We hypothesized that in our experimental setup StkP is activated by internal PG molecules present in the septum of dividing cells. To prove this, we arrested cell growth by glucose depletion and then analyzed protein phosphorylation in starving cells (Fig. 2B). We observed a significant decrease in Thr phosphorylation, indicating that StkP activity ceased upon growth arrest, probably due to a decrease in ligand concentration. On the other hand, PrkC, an StkP homolog from B. subtilis, was shown to be activated by exogenous PG subunits released into the medium. Sequence comparison revealed that multiple PASTA domains in a single protein are very diverse, and it was proposed that each domain in a protein has evolved so that it has a binding affinity for a specific stem peptide ligand (22). We hypothesize that muropeptides released during cell lysis might be also an efficient activating signal for StkP, although they could induce a different cell response.
FIG. 2.
Dynamics of StkP substrate phosphorylation during growth. (A) Activity of StkP during growth in complex medium. Proteins phosphorylated on Thr were detected with anti-pThr antibody (anti-pThr) in cell lysates of wild-type strain Sp1 prepared from logarithmic-phase (OD400, 0.2 to 1.5) and stationary-phase (OD400, 1.6) cultures. (B) Activity of StkP during growth arrest. Cultures of strain Sp1 were grown in glucose-containing CAT medium until the OD400 was 0.4 and then centrifuged, resuspended in CAT medium with glucose (Glu+) or without glucose (Glu−), and cultivated for 1 h. Phosphorylated proteins were detected with anti-pThr antibody. The total amount of StkP protein in both experiments was evaluated with anti-StkP serum (anti-StkP). The arrow indicates the position of StkP.
Identification of StkP substrates in the cytoplasm of S. pneumoniae.
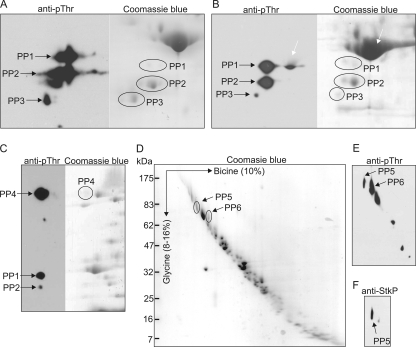
To identify proteins phosphorylated by StkP, soluble proteins were separated by two-dimensional (2D) SDS-polyacrylamide gel electrophoresis (PAGE) using isoelectric focusing (IEF) and SDS-PAGE in the first and second dimensions, respectively. In the first screen, the proteins were separated by using ready-made IPG strips (pH 3 to 10), followed by SDS-PAGE. All phosphoproteins migrated in the narrow pI range from pI 4 to 5 (data not shown). Therefore, in the following experiments, a narrow pH range, pH 3 to 5.6, was used. Three phosphorylated protein spots, PP1 to PP3, were detected by using anti-pThr antibody and were visualized by Coomassie blue staining (Fig. 3 A; see Fig. S1 in the supplemental material). These proteins were identified further by mass spectrometry (Table 3). The PP1 spot was identified as DivIVA, a cell division protein. The PP2 spot contained two comigrating proteins; the more abundant protein was RpoA, the α-subunit of RNA polymerase, and the less abundant protein was DivIVA. The PP3 spot contained PpaC, a Mn-dependent inorganic pyrophosphatase.
FIG. 3.
Identification of putative StkP substrates. (A) Identification of StkP substrates in the cytoplasmic fraction. Cytoplasmic proteins were separated by 2D SDS-PAGE using an IPG strip (pH 3 to 5.6) in the first dimension and a 12.5% acrylamide gel in the second dimension. The gels were stained with Coomassie blue or electroblotted and probed with anti-pThr antibody (anti-pThr). The PP1 to PP3 protein spots corresponding to phosphorylated proteins were excised and analyzed by MALDI-FTMS. (B) Identification of StkP substrates in the membrane fraction using 2D SDS-PAGE. Membrane proteins were solubilized in the presence of 4% CHAPS and separated by 2D SDS-PAGE using an IPG strip (pH 3 to 5.6) in the first dimension and a 12.5% acrylamide gel in the second dimension. The gels were stained with Coomassie blue or electroblotted and probed with anti-pThr antibody (anti-pThr). Phosphorylated proteins in the PP1, PP2, and PP3 protein spots were analyzed by MALDI-FTMS. The white arrow indicates a protein spot corresponding to enolase. (C) Identification of StkP substrates in the membrane fraction using 2D SDS-PAGE and a combination of detergents. Membrane proteins were solubilized in the presence of 4% CHAPS, 1% ASB14, 1% Triton X-100 and separated by 2D SDS-PAGE using an IPG strip (pH 3 to 10) in the first dimension and a 12.5% acrylamide gel in the second dimension. Gels stained with Coomassie blue were compared with immunoblots (anti-pThr), and the PP4 protein spot was analyzed by MALDI-FTMS. (D) Identification of StkP substrates in the membrane fraction using doubled SDS-PAGE. Membrane proteins derived from strain Sp1 were separated using a 10% acrylamide bicine-SDS-PAGE gel in the first dimension and a 8 to 16% gradient acrylamide glycine-SDS-PAGE gel in the second dimension. The gel was stained with Coomassie blue. (E) Membrane phosphoproteins separated by doubled SDS-PAGE were electroblotted, detected with anti-pThr antibody, and compared with a Coomassie blue-stained gel. Phosphorylated proteins in the PP5 and PP6 protein spots were analyzed by MALDI-FTMS. (F) Detection of StkP resolved by doubled SDS-PAGE with specific anti-StkP serum (anti-StkP). The protein corresponding to the PP5 protein spot was identified as StkP.
TABLE 3.
Identification of putative StkP substrates by MALDI-FTMS
| Protein spot | Locus | Gene | Protein function | Predicted molecular mass (kDa) | pI | Other study(ies)a |
|---|---|---|---|---|---|---|
| PP1 | spr1505 | divIVA | Cell division protein | 30.1 | 4.5 | 23, 49 |
| PP2a | spr0215 | rpoA | α-Subunit of RNA polymerase | 34.2 | 4.5 | |
| PP2b | spr1505 | divIVA | Cell division protein | 30.1 | 4.5 | 23, 49 |
| PP3 | spr1389 | ppaC | Mn-dependent inorganic pyrophosphatase | 33.5 | 4.4 | 41 |
| PP4 | spr0334 | Conserved hypothetical protein | 51.64 | 4.7 | 49 | |
| PP5 | spr1577 | stkP | Eukaryotic-type Ser/Thr kinase | 72.2 | 8.4 | |
| PP6 | spr0334 | Conserved hypothetical protein | 51.64 | 4.7 | 49 |
Study(ies) in which the spot was identified as an ESTPK substrate in other bacteria.
Identification of StkP substrates in the cell membrane of S. pneumoniae.
Due to their hydrophobicity, membrane proteins are very difficult to resolve in the first dimension (IEF step) of conventional 2D SDS-PAGE. They often exhibit low solubility and, once solubilized, have the tendency to precipitate at their pIs. Consequently, they are often underrepresented on 2D gels. The use of different detergents and combinations of detergents may help overcome this limitation. In our first attempt, we included 4% CHAPS in the lysis buffer used for solubilization of membrane proteins, and we detected three phosphoproteins apparently identical to those detected in the cytoplasmic fraction (Fig. 3B; see Fig. S2 in the supplemental material). Indeed, the corresponding protein spots were identified by mass spectrometry as DivIVA, RpoA, and PpaC (Table 3). Another protein phosphorylated on Thr was detected in the membrane fraction of the Sp1 strain and was identified as an enolase (spr1036) (Fig. 3B). Phosphorylated enolase was also detected in Sp10 cell lysates (data not shown), and its phosphorylation is thus not StkP dependent. Phosphorylation of enolase has been reported for many species ranging from archaea to humans (29).
All phosphoproteins detected by 2D SDS-PAGE using 4% CHAPS are membrane-associated soluble proteins. To enhance solubilization of integral membrane proteins, we used a combination of 4% CHAPS, 1% Triton X-100, and 1% ASB14, which resulted in solubilization of only one more phosphoprotein, PP4, which was identified as spr0334 (Fig. 3C; see Fig. S3 in the supplemental material).
To more effectively identify putative membrane StkP substrates, we used an alternative method for membrane protein separation, doubled SDS-PAGE (dSDS-PAGE), as described by Rais et al. (40) and optimized by Williams et al. (53). In principle, the membrane proteins are separated in the first dimension by SDS-PAGE using a bicine-based buffer system, which is followed by separation in the second dimension by SDS-PAGE using a glycine-based buffer system. As a result, proteins are dispersed around a diagonal, and the separated protein spots can be resolved better. We solubilized pneumococcal membrane proteins using SDS, separated them by dSDS-PAGE (Fig. 3D), and performed an immunodetection analysis of proteins phosphorylated on Thr (Fig. 3E). We detected six proteins that reacted with the anti-pThr antibody. Only two of these proteins (PP5 and PP6) were clearly separated and selected for further analysis. Since StkP is a membrane protein, we stripped the immunoblot and probed it with a specific anti-StkP antibody (Fig. 3F). Immunodetection revealed that spot PP5 reacted with anti-StkP serum, and the identity of StkP was further confirmed by mass spectrometry. MALDI-FTMS revealed that the PP6 spot contained the hypothetical protein spr0334, which supported the results of 2D SDS-PAGE.
Phosphorylation of putative substrates in vitro.
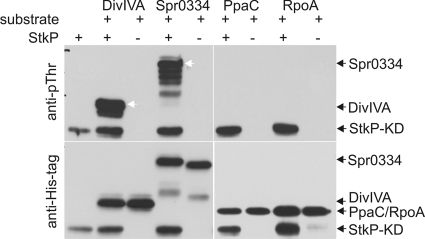
The divIVA, spr0334, rpoA, and ppaC genes, coding for putative StkP substrates, were cloned into the expression vector pETPhos, which was optimized for expression of Ser/Thr kinase substrates. The His-tagged fusion protein derived from pETPhos lacks putative phosphoacceptor Ser or Thr residues that might be targets of Ser/Thr protein kinases, as reported by Canova et al. (5). The fusion proteins were purified, and their phosphorylation was examined using an in vitro kinase assay in the presence and absence of the recombinant StkP kinase domain (rStkP-KD) and ATP (Fig. 4). Phosphorylation was monitored by immunodetection with anti-pThr antibody. The DivIVA and spr0334 proteins were phosphorylated by StkP, suggesting that they are genuine substrates of StkP. On the other hand, we did not observe phosphorylation of the RpoA or PpaC protein. The kinase reaction in the presence of [γ-32P]ATP did not result in any phosphorylation signal (data not shown).
FIG. 4.
In vitro phosphorylation of putative StkP substrates. Recombinant His-DivIVA, His-Spr0334, His-PpaC, and His-RpoA were incubated with or without StkP-KD in kinase buffer as described in Materials and Methods. Proteins were separated by SDS-PAGE, electrotransferred to a PVDF membrane, and probed with anti-pThr antibody to detect phosphorylation (anti-pThr) or with anti-His antibody to detect proteins (anti-His tag). The white arrows indicate phosphorylated DivIVA and spr0334. The black arrows on the right indicate the positions of the recombinant proteins.
The fact that RpoA was found to comigrate with DivIVA in the PP2 protein spot raised the question of whether both proteins are phosphorylated. The results of an in vitro kinase assay indicated that RpoA might be covered by the DivIVA phosphorylation signal on 2D SDS-PAGE gels. Using specific antibodies against DivIVA and RpoA, we confirmed that the DivIVA protein migrates in two protein spots (corresponding to the PP1 and PP2 spots) at different molecular masses but with similar pIs on 2D SDS-PAGE gels. The smaller DivIVA isoform migrates in the same protein spot as RpoA (PP2) (see Fig. S4 in the supplemental material). To determine the in vivo phosphorylation status of both proteins, we performed 2D SDS-PAGE with protein lysate derived from an S. pneumoniae ΔdivIVA strain (11) to locate RpoA. Immunoblotting with anti-pThr antibody revealed no signal, indicating that the protein corresponding to RpoA is not phosphorylated. Therefore, we concluded that RpoA is not a substrate of StkP and that the previously observed phosphorylation of the PP2 protein spot was due solely to phosphorylated DivIVA.
DivIVA is a cell division protein that has been studied extensively using B. subtilis. DivIVA, together with MinC and MinD, defines the midcell. MinC and MinD interact to form an inhibitor complex that prevents division at the cell poles. MinCD is controlled by DivIVA, which keeps MinCD at the cell poles after division (12, 32). Clear orthologs of DivIVA are found in a wide range of Gram-positive bacteria. Gram-negative organisms lack DivIVA and contain MinE instead (10). Disruption of divIVA in S. pneumoniae resulted in the formation of chains of unseparated, morphologically altered cells with incomplete septa that were often devoid of nucleoids (11). Immunofluorescence microscopy showed that DivIVA is localized to the division septum and the cell poles in S. pneumoniae and that it interacts with a number of pneumococcal cell division proteins. These results indicate that DivIVA has a role in controlling cell morphology, completion of cell division, and cell separation, including chromosome segregation (12). DivIVA is reported to be a substrate of homologous ESTPKs in S. agalactiae (49) and M. tuberculosis (23).
The function of spr0334, a hypothetical membrane protein phosphorylated by StkP, remains to be elucidated. spr0334 does not show significant homology to any known protein family, and homologs of it are found mainly in streptococci. Interestingly, the homolog SAK_0375 was identified as a putative substrate of Stk1 in S. agalactiae (49).
Phosphorylation of PpaC, a Mn-dependent inorganic pyrophosphatase, was extensively studied using S. agalactiae (41, 44). Pyrophosphatases (Ppases) are essential enzymes that catalyze hydrolysis of the inorganic pyrophosphates (PPi) produced during various biosynthetic reactions, including carbohydrate metabolism, ATP hydrolysis, and biosynthesis of amino acids and nucleotides. PPi regulates many enzymes without participating in their reactions, and its effect is mostly inhibitory; therefore, regulation of its concentration in the cell is crucial for sustaining normal growth (26). PpaC from S. agalactiae (PpaCSa) and PpaC from S. pneumoniae (PpaCSp) share 85% sequence identity. PpaCSa is a soluble enzyme that is phosphorylated by the Stk1 kinase on Ser residues. However, the phosphoacceptor amino acids have not been identified, and the significance of PpaC phosphorylation has not been elucidated. We inferred that PpaCSp may be phosphorylated on Thr based on its reaction with anti-pThr antibody. Even though PpaCSp phosphorylation in vivo is StkP dependent, PpaCSp is not phosphorylated by StkP in vitro. We hypothesize that StkP-dependent phosphorylation of PpaC in vitro may require specific reaction conditions or the presence of other interaction partners. A similar observation was made for in vitro phosphorylation of FtsZ from S. agalactiae (49).
Effect of stkP inactivation on bacterial morphology.
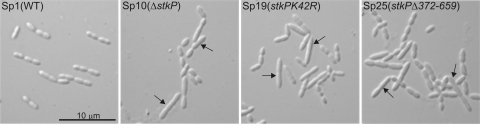
PknB-like protein kinases in several bacteria have been shown to regulate cell shape and cell division. The identification of the cell division protein DivIVA as a substrate of StkP prompted us to examine morphological changes in S. pneumoniae caused by StkP inactivation. Different stkP mutant strains were inspected by using light microscopy (Fig. 5). Mutant strain Sp10 lacking StkP differed from wild-type strain Sp1 in cell shape and size. The morphological changes were similar to those reported for the ΔstkP mutant of S. pneumoniae TIGR4 (17). A significant number of cells did not have the typical ovoid shape but appeared to be longer with rather oval poles, and they exhibited an apparent cell division defect. Subsequent image analysis and quantification confirmed these observations (see Table S1 in the supplemental material). The minimum cell size was the same for both strains, but the average cell size of ΔstkP mutant cells (2,360 μm) was greater than that of the wild-type cells (2,047 μm). The maximum cell lengths were 4,290 μm for the wild type and 8,280 μm for the mutant strain. Strain Sp25, expressing the kinase with PASTA domains deleted, and strain Sp19, expressing a kinase-dead mutant protein, had a phenotype very similar to that of the ΔstkP strain. We hypothesize that the morphological defects may be attributed to deregulation of DivIVA activity. It is noteworthy that even hypophosphorylation of DivIVA in the Sp19 strain leads to morphological defects comparable to those caused by a complete loss of phosphorylation. These results indicate that StkP plays an important role in the cell division cycle of S. pneumoniae through control of DivIVA activity.
FIG. 5.
Cell morphology of the wild-type Rx1 and stkP mutant strains. Strains were grown in complex CAT medium until mid-exponential phase and observed by differential interference contrast microscopy (Nomarski). stkP mutant strains Sp10, Sp19, and Sp25 produced elongated cells (indicated by arrows) with abnormal septation.
Conclusion.
Eukaryotic-type protein kinases possessing PASTA domains are signaling enzymes that are unique to Gram-positive bacteria belonging to the phyla Firmicutes and Actinobacteria (22). PASTA domains bind β-lactam antibiotics with low affinity and are implicated in sensing of unlinked peptidoglycan (54). The first evidence that muropeptides can activate ESTPK signaling in vivo was described by Shah et al. (47). Protein kinase PrkC in B. subtilis was shown to be activated upon binding of external muropeptides released from a cell culture and to signal bacteria to exit dormancy. In contrast to this finding, we showed that pneumococcal StkP is activated continuously during growth and that its activity is inhibited upon growth arrest. Therefore, we assume that the signal molecules that trigger autophosphorylation of StkP are intracellular peptidoglycan subunits rather than muropeptides released into the medium during cell lysis. The model of PknB-like kinase activation presumes that oligomerization and/or localization of the protein kinase is required for kinase activation. We showed that dimerization of StkP through the transmembrane region is not sufficient for substrate recruitment, whereas the presence of PASTA domains restores the kinase activity. Therefore, we hypothesize that affinity of PASTA domains to unlinked PG subunits may be important for kinase localization and may target StkP to cell division septa. In response to intracellular unlinked peptidoglycan, StkP phosphorylates a set of substrates, including cell division protein DivIVA, and thus regulates the cell division cycle.
Several substrates of ESTPKs in different species have been characterized. So far, the most complex study of the ESTPK-dependent phosphoproteome was conducted with S. agalactiae by Silvestroni et al. (49). These workers identified nine putative substrates of the Stk1 protein kinase in vivo: DivIVA, DivIVA domain protein, FtsZ, and six hypothetical proteins without known functions. PpaC, PurA, and CovR have previously been reported to be phosphorylated by Stk1 (28, 41-43). Interestingly, the S. pneumoniae StkP substrates identified in our screen overlap significantly with the substrates of Stk1 from S. agalactiae. Evidently, PknB-like protein kinases regulate vital processes in all Gram-positive bacteria, and, therefore, it is not surprising that the signaling pathways under the control of Stk1 and StkP are very similar, despite the different lifestyles of the two microorganisms. However, we believe that they are not identical. At least one Stk1 substrate, the hypothetical protein SAK_0856 of S. agalactiae, does not have a homolog in S. pneumoniae. Moreover, the phosphoglucosamine mutase GlmM of S. pneumoniae was proven to be a substrate of StkP, but phosphorylation of S. agalactiae GlmM is not Stk1 dependent. Thus, these two microorganisms seem to customize their ESTPKs to fine-tune their cellular responses. Such an adaptation of ESTPK is well-illustrated by mycobacterial PknF, which phosphorylates the chaperone GroEL in a species-dependent manner in M. tuberculosis and Mycobacterium smegmatis (6).
Interestingly, there are two proteins that are substrates of PknB-like protein kinases in more bacterial species, the cell division proteins FtsZ and DivIVA. FtsZ was found to be phosphorylated in M. tuberculosis (50), S. agalactiae (49), and C. glutamicum (46). DivIVA homologues are phosphorylated in M. tuberculosis (23) and S. agalactiae (49). In this study we identified DivIVA as an in vivo substrate of the StkP protein kinase of S. pneumoniae. These findings suggest that phosphorylation of DivIVA and FtsZ is common in Firmicutes and Actinobacteria and that corresponding protein kinases control similar signaling pathways, indicating their universal role in regulation of the cell division cycle in Gram-positive bacteria.
Supplementary Material
Acknowledgments
This work was supported by the Czech Science Foundation (grant 204/07/082 to L.N. and grant 204/08/0783 to P.B.), by the Grant Agency of the Academy of Sciences of the Czech Republic (project IAA600200801 to P.B.), and by Institutional Research Concept grant AV0Z50200510.
We gratefully acknowledge the gift of the ΔdivIVA mutant strain and anti-DivIVA antibody from Orietta Massidda. We thank Virginie Molle for the kind gift of the pETPhos vector. We thank Oldrich Benada for assistance with analysis of the microscopy images.
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Absalon, C., M. Obuchowski, E. Madec, D. Delattre, I. B. Holland, and S. J. Seror. 2009. CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis. Microbiology 155:932-943. [DOI] [PubMed] [Google Scholar]
- 2.Avery, O. T., C. M. McLeod, and M. McCarthy. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Av-Gay, Y., S. Jamil, and S. J. Drews. 1999. Expression and characterization of the Mycobacterium tuberculosis serine/threonine protein kinase PknB. Infect. Immun. 67:5676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltramini, A. M., C. D. Mukhopadhyay, and V. Pancholi. 2009. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect. Immun. 77:1406-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canova, M. J., L. Kremer, and V. Molle. 2008. pETPhos: a customized expression vector designed for further characterization of Ser/Thr/Tyr protein kinases and their substrates. Plasmid 60:149-153. [DOI] [PubMed] [Google Scholar]
- 6.Canova, M. J., L. Kremer, and V. Molle. 2009. The Mycobacterium tuberculosis GroEL1 chaperone is a substrate of Ser/Thr protein kinases. J. Bacteriol. 191:2876-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 8.Debarbouille, M., S. Dramsi, O. Dussurget, M. A. Nahori, E. Vaganay, G. Jouvion, A. Cozzone, T. Msadek, and B. Duclos. 2009. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J. Bacteriol. 191:4070-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echenique, J., A. Kadioglu, S. Romao, P. W. Andrew, and M. C. Trombe. 2004. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72:2434-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65, table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fadda, D., C. Pischedda, F. Caldara, M. B. Whalen, D. Anderluzzi, E. Domenici, and O. Massidda. 2003. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J. Bacteriol. 185:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fadda, D., A. Santona, V. D'Ulisse, P. Ghelardini, M. G. Ennas, M. B. Whalen, and O. Massidda. 2007. Streptococcus pneumoniae DivIVA: localization and interactions in a MinCD-free context. J. Bacteriol. 189:1288-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez, P., B. Saint-Joanis, N. Barilone, M. Jackson, B. Gicquel, S. T. Cole, and P. M. Alzari. 2006. The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 188:7778-7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiuza, M., M. J. Canova, D. Patin, M. Letek, I. Zanella-Cleon, M. Becchi, L. M. Mateos, D. Mengin-Lecreulx, V. Molle, and J. A. Gil. 2008. The MurC ligase essential for peptidoglycan biosynthesis is regulated by the serine/threonine protein kinase PknA in Corynebacterium glutamicum. J. Biol. Chem. 283:36553-36563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiuza, M., M. J. Canova, I. Zanella-Cleon, M. Becchi, A. J. Cozzone, L. M. Mateos, L. Kremer, J. A. Gil, and V. Molle. 2008. From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. J. Biol. Chem. 283:18099-18112. [DOI] [PubMed] [Google Scholar]
- 16.Gaidenko, T. A., T. J. Kim, and C. W. Price. 2002. The PrpC serine-threonine phosphatase and PrkC kinase have opposing physiological roles in stationary-phase Bacillus subtilis cells. J. Bacteriol. 184:6109-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giefing, C., A. L. Meinke, M. Hanner, T. Henics, M. D. Bui, D. Gelbmann, U. Lundberg, B. M. Senn, M. Schunn, A. Habel, B. Henriques-Normark, A. Ortqvist, M. Kalin, A. von Gabain, and E. Nagy. 2008. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 205:117-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon, E., N. Mouz, E. Duee, and O. Dideberg. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477-485. [DOI] [PubMed] [Google Scholar]
- 19.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42-52. [DOI] [PubMed] [Google Scholar]
- 20.Hussain, H., P. Branny, and E. Allan. 2006. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 188:1628-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, H., and V. Pancholi. 2006. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J. Mol. Biol. 357:1351-1372. [DOI] [PubMed] [Google Scholar]
- 22.Jones, G., and P. Dyson. 2006. Evolution of transmembrane protein kinases implicated in coordinating remodeling of gram-positive peptidoglycan: inside versus outside. J. Bacteriol. 188:7470-7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang, C. M., D. W. Abbott, S. T. Park, C. C. Dascher, L. C. Cantley, and R. N. Husson. 2005. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19:1692-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristich, C. J., C. L. Wells, and G. M. Dunny. 2007. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. U. S. A. 104:3508-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lahti, R. 1983. Microbial inorganic pyrophosphatases. Microbiol. Rev. 47:169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine, A., F. Vannier, C. Absalon, L. Kuhn, P. Jackson, E. Scrivener, V. Labas, J. Vinh, P. Courtney, J. Garin, and S. J. Seror. 2006. Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics 6:2157-2173. [DOI] [PubMed] [Google Scholar]
- 28.Lin, W. J., D. Walthers, J. E. Connelly, K. Burnside, K. A. Jewell, L. J. Kenney, and L. Rajagopal. 2009. Threonine phosphorylation prevents promoter DNA binding of the group B Streptococcus response regulator CovR. Mol. Microbiol. 71:1477-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macek, B., F. Gnad, B. Soufi, C. Kumar, J. V. Olsen, I. Mijakovic, and M. Mann. 2008. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7:299-307. [DOI] [PubMed] [Google Scholar]
- 30.Macek, B., I. Mijakovic, J. V. Olsen, F. Gnad, C. Kumar, P. R. Jensen, and M. Mann. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6:697-707. [DOI] [PubMed] [Google Scholar]
- 31.Madec, E., A. Laszkiewicz, A. Iwanicki, M. Obuchowski, and S. Seror. 2002. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46:571-586. [DOI] [PubMed] [Google Scholar]
- 32.Marston, A. L., and J. Errington. 1999. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33:84-96. [DOI] [PubMed] [Google Scholar]
- 33.Molle, V., A. K. Brown, G. S. Besra, A. J. Cozzone, and L. Kremer. 2006. The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J. Biol. Chem. 281:30094-30103. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, D. A., S. A. Lacks, W. R. Guild, and J. M. Hageman. 1983. Isolation and characterization of three new classes of transformation-deficient mutants of Streptococcus pneumoniae that are defective in DNA transport and genetic recombination. J. Bacteriol. 156:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novakova, L., L. Saskova, P. Pallova, J. Janecek, J. Novotna, A. Ulrych, J. Echenique, M. C. Trombe, and P. Branny. 2005. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 272:1243-1254. [DOI] [PubMed] [Google Scholar]
- 36.Oliver, A. W., S. Knapp, and L. H. Pearl. 2007. Activation segment exchange: a common mechanism of kinase autophosphorylation? Trends Biochem. Sci. 32:351-356. [DOI] [PubMed] [Google Scholar]
- 37.Pallova, P., K. Hercik, L. Saskova, L. Novakova, and P. Branny. 2007. A eukaryotic-type serine/threonine protein kinase StkP of Streptococcus pneumoniae acts as a dimer in vivo. Biochem. Biophys. Res. Commun. 355:526-530. [DOI] [PubMed] [Google Scholar]
- 38.Perez, J., A. Castaneda-Garcia, H. Jenke-Kodama, R. Muller, and J. Munoz-Dorado. 2008. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc. Natl. Acad. Sci. U. S. A. 105:15950-15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pike, A. C., P. Rellos, F. H. Niesen, A. Turnbull, A. W. Oliver, S. A. Parker, B. E. Turk, L. H. Pearl, and S. Knapp. 2008. Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites. EMBO J. 27:704-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rais, I., M. Karas, and H. Schagger. 2004. Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification. Proteomics 4:2567-2571. [DOI] [PubMed] [Google Scholar]
- 41.Rajagopal, L., A. Clancy, and C. E. Rubens. 2003. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 278:14429-14441. [DOI] [PubMed] [Google Scholar]
- 42.Rajagopal, L., A. Vo, A. Silvestroni, and C. E. Rubens. 2005. Regulation of purine biosynthesis by a eukaryotic-type kinase in Streptococcus agalactiae. Mol. Microbiol. 56:1329-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajagopal, L., A. Vo, A. Silvestroni, and C. E. Rubens. 2006. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol. Microbiol. 62:941-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rantanen, M. K., L. Lehtio, L. Rajagopal, C. E. Rubens, and A. Goldman. 2007. Structure of the Streptococcus agalactiae family II inorganic pyrophosphatase at 2.80 A resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 63:738-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saskova, L., L. Novakova, M. Basler, and P. Branny. 2007. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 189:4168-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz, C., A. Niebisch, A. Schwaiger, U. Viets, S. Metzger, M. Bramkamp, and M. Bott. 2009. Genetic and biochemical analysis of the serine/threonine protein kinases PknA, PknB, PknG and PknL of Corynebacterium glutamicum: evidence for non-essentiality and for phosphorylation of OdhI and FtsZ by multiple kinases. Mol. Microbiol. 74:724-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah, I. M., M. H. Laaberki, D. L. Popham, and J. Dworkin. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma, K., M. Gupta, A. Krupa, N. Srinivasan, and Y. Singh. 2006. EmbR, a regulatory protein with ATPase activity, is a substrate of multiple serine/threonine kinases and phosphatase in Mycobacterium tuberculosis. FEBS J. 273:2711-2721. [DOI] [PubMed] [Google Scholar]
- 49.Silvestroni, A., K. A. Jewell, W. J. Lin, J. E. Connelly, M. M. Ivancic, W. A. Tao, and L. Rajagopal. 2009. Identification of serine/threonine kinase substrates in the human pathogen group B streptococcus. J. Proteome Res. 8:2563-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thakur, M., and P. K. Chakraborti. 2006. GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J. Biol. Chem. 281:40107-40113. [DOI] [PubMed] [Google Scholar]
- 51.Thakur, M., and P. K. Chakraborti. 2008. Ability of PknA, a mycobacterial eukaryotic-type serine/threonine kinase, to transphosphorylate MurD, a ligase involved in the process of peptidoglycan biosynthesis. Biochem. J. 415:27-33. [DOI] [PubMed] [Google Scholar]
- 52.Ulijasz, A. T., S. P. Falk, and B. Weisblum. 2009. Phosphorylation of the RitR DNA-binding domain by a Ser-Thr phosphokinase: implications for global gene regulation in the streptococci. Mol. Microbiol. 71:382-390. [DOI] [PubMed] [Google Scholar]
- 53.Williams, T. I., J. C. Combs, A. P. Thakur, H. J. Strobel, and B. C. Lynn. 2006. A novel Bicine running buffer system for doubled sodium dodecyl sulfate-polyacrylamide gel electrophoresis of membrane proteins. Electrophoresis 27:2984-2995. [DOI] [PubMed] [Google Scholar]
- 54.Yeats, C., R. D. Finn, and A. Bateman. 2002. The PASTA domain: a beta-lactam-binding domain. Trends Biochem. Sci. 27:438. [DOI] [PubMed] [Google Scholar]
- 55.Young, T. A., B. Delagoutte, J. A. Endrizzi, A. M. Falick, and T. Alber. 2003. Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat. Struct. Biol. 10:168-174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.