Abstract
FNR-dependent activation of the Escherichia coli K-12 nrf promoter is downregulated by the nitric oxide-sensitive NsrR protein together with the nucleoid-associated protein IHF, which bind to overlapping targets adjacent to the DNA site for FNR. The NsrR target is inactivated by mutation at the Salmonella enterica serovar Typhimurium nrf promoter.
The Escherichia coli K-12 nrf operon encodes the NrfA periplasmic nitrite reductase (9), and its expression is controlled by a single promoter (pnrf) whose activity is completely dependent on FNR, the principal global transcription regulator responsible for anaerobic adaptation (26). In previous work (3-6), we showed that the DNA target for FNR at pnrf is centered at position −41.5 (i.e., between 41 and 42 bp upstream from the transcript start point), that anaerobically induced FNR-dependent activation of pnrf is downregulated by the binding of the nucleoid-associated protein IHF to a target (IHF I) centered at position −54, and that this repression is relieved by the binding of either NarL or NarP to a DNA target centered at position −74.5 (Fig. 1). Recall that NarL and NarP are homologous response regulators whose activity is triggered by nitrite or nitrate ions (10), and this provides a simple mechanism for the induction of nrf operon expression in response to environmental nitrite or nitrate (26, 28). As well as reducing nitrite ions, the NrfA nitrite reductase can also reduce nitric oxide (19, 27), and two recent studies (11, 18) have suggested that pnrf is regulated by NsrR, a global transcription repressor whose activity is modulated by nitric oxide (1, 25). Chromatin immunoprecipitation experiments showed that NsrR binds to a site in the nrf operon regulatory region (18), while transcriptome analysis suggested that NsrR represses nrf operon expression (11). Since neither study was able to identify the DNA site for NsrR unambiguously, here we present direct experimental evidence for NsrR binding at the E. coli nrf promoter and for its location.
FIG. 1.
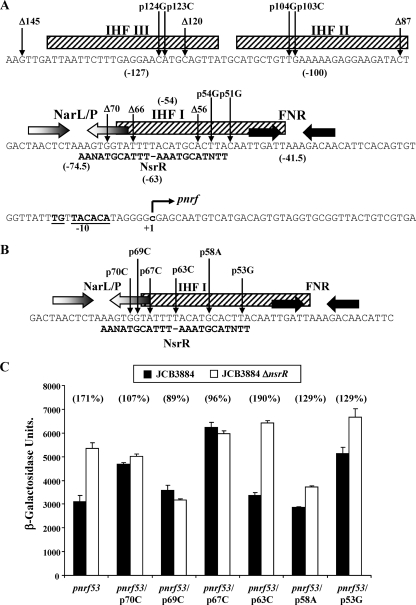
Organization of the nrf promoter and mutational analysis. (A) Shown are pnrf sequences from position −147 to position +41 and elements involved in pnrf regulation. FNR and NarL/NarP binding sites are represented by inverted arrows, while IHF sites are depicted by boxes, with the positions of binding sites shown in parentheses. The extended −10 promoter element is in bold and underlined, and the transcription start (+1) is in lowercase. The NsrR inverted repeat sequence centered at position −63 is shown. The upstream limits of the pnrf53 fragment deletions and the substitutions that inactivate each DNA site for IHF are indicated. (B) Alignment of the pnrf sequence surrounding position −63 with the NsrR consensus inverted repeat sequence. FNR and NarL/NarP binding sites are represented by inverted arrows, and the IHF I binding site is depicted by a box. The single base pair substitutions within the NsrR binding site are shown. (C) Measured β-galactosidase activities of JCB3884 (narL narP) and JCB3884 ΔnsrR cells carrying pRW50, containing wild-type and mutant pnrf53 fragments as in panel B. Assays were performed using the Miller protocol (16) as described in the table footnotes.
Deletion analysis of pnrf.
The pnrf53 promoter fragment encodes pnrf sequences from position −209 to position +131 and contains all of the cis-acting elements necessary for regulation (26). Previously, using this fragment cloned into low-copy-number lac expression vector plasmid pRW50 (15), we showed that expression from pnrf is downregulated by NsrR by comparing the expression of the resulting pnrf::lac fusion in the wild-type and ΔnsrR mutant strains (11). To locate the DNA site for NsrR, we have exploited a set of nested deletions in the pnrf53 promoter fragment that removed sequences upstream from positions −145, −120, −87, −70, −66, and −56 (Fig. 1A). Each truncated fragment was cloned into pRW50, and NsrR-dependent repression was measured. Data presented in Table 1 show that NsrR-dependent repression is lost with the deletions to positions −70, −66, and −56, and thus, we conclude that the functional DNA site for NsrR must be downstream of position −87. A further point from these data concerns the role of two secondary upstream DNA sites for IHF, IHF II at position −100 and IHF III at position −127 (Fig. 1A). We previously showed that pnrf activity is stimulated by IHF binding to IHF III at position −127, while IHF binding to IHF II at position −100 has little or no effect (6). Consistent with this, data in Table 1 show that the deletion of sequences between positions −145 and −120 causes an overall decrease in pnrf activity, while further deletion to position −87 has little effect.
TABLE 1.
Measurement of pnrf expression in JCB3884 and JCB3884 ΔnsrR cells and effects of different upstream deletions
| Promotera | β-Galactosidase activity |
Differenceb | |
|---|---|---|---|
| JCB3884 | JCB3884 ΔnsrR | ||
| pnrf53 | 3,480 | 6,144 | 177 |
| pnrf53/Δ145 | 3,393 | 5,775 | 170 |
| pnrf53/Δ120 | 1,732 | 2,955 | 171 |
| pnrf53/Δ87 | 1,691 | 2,334 | 138 |
| pnrf53/Δ70 | 5,358 | 4,693 | 88 |
| pnrf53/Δ66 | 6,992 | 5,817 | 83 |
| pnrf53/Δ56 | 12,110 | 10,098 | 83 |
The first column lists the different pnrf53 deletion fragments, and the extent of each deletion is depicted in Fig. 1A. β-Galactosidase activities were measured in narL narP Δlac mutant host strain JCB3884 (26) or in its ΔnsrR mutant derivative carrying the lac expression vector plasmid pRW50 containing the different nrf promoter fragments. Assays were performed using the Miller protocol (16), as in our previous work (13). Cells were grown anaerobically at 37°C in minimal salts medium (20). β-Galactosidase activities are expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of dry cell mass. Each value is the average of three independent determinations that varied by less than 10%. The promoter activity of all pnrf53 derivatives was negligible when cells were grown aerobically and completely dependent on FNR, since expression was undetectable in a Δfnr mutant strain (not shown).
Shown is the percent difference in anaerobic expression due to the deletion of nsrR.
Mutational analysis of the NsrR binding site.
The consensus DNA target for NsrR binding consists of two 11-bp motifs, organized as an inverted repeat, separated by 1 bp, though there is evidence that a single 11-bp motif may be sufficient for binding in some cases (18). Inspection of the pnrf DNA sequence downstream of position −87 identified two 11-bp elements that resemble the consensus (Fig. 1B). Since we had previously generated several different single base substitutions in this region of pnrf (3-6, 26), we measured their effects on NsrR-dependent downregulation of pnrf activity using the pnrf53 fragment cloned in pRW50. Results illustrated in Fig. 1C show that NsrR-dependent repression is abolished or decreased by base substitutions at positions −70, −69, −67, −58, and −53 but unchanged by the substitution at position −63, which is between the two 11-bp elements. These data are consistent with the location of our proposed DNA site for NsrR and show that both halves of the inverted repeat are required for optimal repression. Note that the differences in the overall levels of expression from pnrf seen with different base substitutions are likely due to changes in the IHF I target, which overlaps the NsrR target (Fig. 1B).
Gel retardation assays.
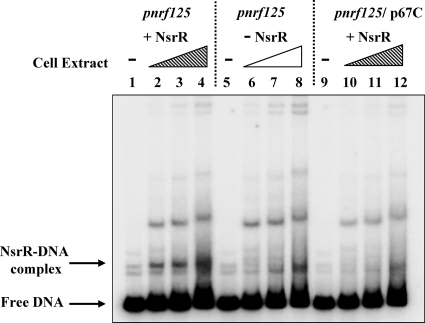
To measure the binding of NsrR to pnrf directly, we incubated an end-labeled DNA fragment that carries pnrf sequences from position −125 to position −20 with cell extracts from cells lacking NsrR and IHF that carried a plasmid encoding NsrR or the empty vector. DNA-protein complexes were separated by polyacrylamide gel electrophoresis (PAGE). Results in Fig. 2 show that a unique NsrR-DNA complex was observed with extracts containing NsrR but absent with the control. This complex was not present when the DNA fragment carried the p67C substitution that abolishes NsrR repression in vivo (Fig. 1C).
FIG. 2.
Gel retardation assays. Shown are gel retardation assays using a DNA fragment (pnrf125) carrying pnrf sequences from position −125 to position −20 that was derived from the pnrf53 fragment by PCR. The end-labeled fragment was incubated with cell extracts from a ΔnsrR derivative of ihfA mutant strain JCB38849 (2) that had been made Tets (7) and then transformed with either pGIT9, which expresses NsrR (1), or the empty vector pSTBlue-1. Cells were grown anaerobically at 37°C in 50 ml of minimum medium (20) to an optical density at 650 nm of 0.6. Cells were harvested by centrifugation, washed with 5 ml of cold wash buffer (20 mM Tris-HCl [pH 8.0], 5% glycerol, 1 mM dithiothreitol, 200 μg ml−1 phenylmethylsulfonyl fluoride, 4 μg ml−1 pepstatin), resuspended in 2 ml of wash buffer, and disrupted by sonication, and then cell debris was removed by centrifugation. The purified DNA fragment was end labeled with [γ-32P]ATP, and 0.5 ng of each fragment was incubated with cell extracts in a mixture of l0 mM potassium phosphate (pH 7.5), 100 mM potassium glutamate, 1 mM EDTA, 50 μM dithiothreitol, 5% glycerol, and 25 μg ml−1 herring sperm DNA. Samples were run in 0.25× Tris-borate-EDTA on a 6% polyacrylamide gel containing 2% glycerol at 12 V cm−1 and analyzed using a Bio-Rad Molecular Imager FX and Quantity One software (Bio-Rad). Reaction mixtures contained extracts from JCB38849S ΔnsrR cells carrying either pGIT9 (lanes 1 to 4 and 9 to 12) or pSTBlue-1 (lanes 5 to 8). The fragments used were as follows: lanes 1 to 8, wild-type fragment; lanes 9 to 12, fragment carrying the p67C substitution. The amount of total protein in each reaction mixture was as follows: lanes 1, 5, and 9, no protein; lanes 2, 6, and 10, 1.5 μg of protein; lanes 3, 7, and 11, 3 μg of protein; lanes 4, 8, and 12, 6 μg of protein. The NsrR-DNA complex is indicated.
NsrR and IHF corepress at pnrf.
To investigate whether repression of pnrf by NsrR is affected by IHF binding at its three targets, we exploited derivatives of the pnrf53 fragment carrying substitutions that disrupt IHF I (p54Gp51G), IHF II (p104Gp103C), or IHF III (p124Gp123C) (Fig. 1A) (3, 6). NsrR-dependent effects were measured by using the pnrf53 fragment cloned in pRW50. Data in Table 2 show that, as expected, expression from pnrf is increased when IHF I is inactivated, decreased when IHF III is inactivated, but largely unaltered when IHF II is inactivated. However, similar repression by NsrR was observed in each instance, suggesting that downregulation of pnrf by NsrR is independent of IHF-dependent regulation.
TABLE 2.
Measurements of pnrf expression in JCB3884 and JCB3884 ΔnsrR cells and effects of mutations in upstream DNA sites for IHFa
| Promoter | Mutant site | β-Galactosidase activity |
|
|---|---|---|---|
| JCB3884 | JCB3884 ΔnsrR | ||
| pnrf53 | 3,480 | 6,144 | |
| pnrf53/p54Gp51G | IHF I | 7,526 | 9,331 |
| pnrf53/p104Gp103C | IHF II | 3,901 | 6,789 |
| pnrf53/p124Gp123C | IHF III | 2,067 | 3,495 |
The first column lists the different pnrf53 fragments used. The p54Gp51G, p104Gp103C, and p124Gp123C substitutions disrupt the IHF I, IHF II, and IHF III sites, respectively (see Fig. 1A) and were constructed in our previous work (3, 6). Measured β-galactosidase activities in JCB3884 and JCB3884 ΔnsrR cells carrying pRW50 containing the different nrf promoter fragments are shown. Assays were performed using the Miller protocol (16) as previously described (13). Cells were grown anaerobically at 37°C in minimal salts medium (20). β-Galactosidase activities are expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of dry cell mass. Each value is the average of three independent determinations that varied by less than 10%. The promoter activity of all pnrf53 derivatives was negligible when cells were grown aerobically, and expression was undetectable in a Δfnr mutant strain (not shown).
DNA sampling of the nrf promoter.
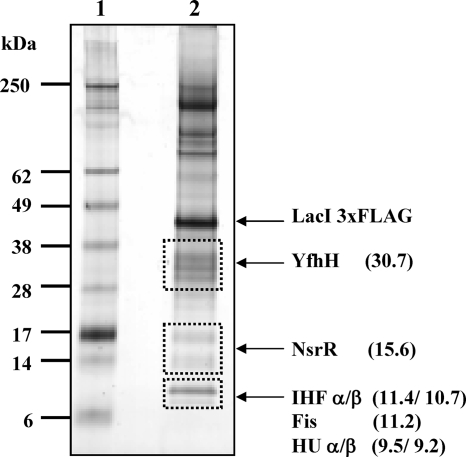
To investigate the binding of NsrR to pnrf in vivo directly, we used the recently developed DNA sampling method, which enables the rapid isolation of specific DNA fragments from E. coli, together with associated proteins (8). Cells were grown anaerobically in nitrate-free minimal medium in order to “sample” bound proteins in the absence of NarL or NarP binding. The pnrf DNA, together with associated protein, was purified as previously described (8), and Fig. 3 shows a silver-stained sodium dodecyl sulfate (SDS)-PAGE gel of the isolated proteins. Three sections of this gel were analyzed by mass spectrometry, and peptide fragments from NsrR were detected, together with the nucleoid-associated proteins IHF, Fis, and HU. In addition, we unexpectedly found YfhH, a member of the RpiR/AslR family of transcription factors that has yet to be characterized (23). Interestingly, there is evidence for an NsrR binding site in the yfhH promoter region (18).
FIG. 3.
DNA sampling at the nrf operon regulatory region. Shown is a silver-stained SDS-PAGE gel of proteins bound to pnrf in vivo. DNA sampling was carried out as described by Butala et al. (8). A derivative of E. coli K-12 MG1655 encoding lacI-3×FLAG was cotransformed with pRW901, carrying the pnrf53 fragment adjacent to five lac operators, and pACBSR-DL1, which encodes the I-SceI meganuclease and bacteriophage lambda Gam protein. Cells were grown anaerobically in 500 ml of minimum medium (20) to an optical density at 600 nm of 0.6, and l-arabinose was added to a final concentration of 0.5% for 20 min of incubation to induce I-SceI and Gam protein expression. Cells were harvested by centrifugation, and the DNA-protein complexes bound to pnrf were isolated as described in reference 8. Proteins were resolved by SDS-PAGE in 4 to 12% gradient gels (Invitrogen) and visualized using a SilverQuest silver staining kit (Invitrogen). Three gel slices (shown as dashed boxes) were excised and destained. The proteins in each slice were then reduced, alkylated, and digested with trypsin, and the resulting peptides were analyzed using a Thermo-Finnigan FT-ICR mass spectrometer using a NanoMate chip-based electrospray (8). The gel was loaded as follows: lane 1, molecular weight markers; lane 2, affinity-isolated proteins. The molecular masses of the identified proteins are indicated.
NsrR does not regulate the Salmonella nrf promoter.
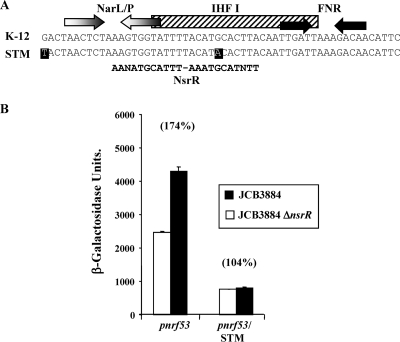
Alignment of the pnrf sequence from Salmonella enterica serovar Typhimurium with that of E. coli K-12 revealed a base difference in the DNA site for NsrR that would be expected to decrease NsrR binding (Fig. 4 A). To investigate NsrR-dependent effects at the Salmonella nrf promoter, the fragment corresponding to pnrf53 was cloned into pRW50. Data illustrated in Fig. 4B show that anaerobic expression from the Salmonella nrf promoter is unaffected by the disruption of nsrR, suggesting that it lacks a functional DNA site for NsrR.
FIG. 4.
NsrR action at a Salmonella nrf promoter. (A) Shown is an alignment of the E. coli K-12 pnrf sequence from position −85 to position −30 with the corresponding sequence from S. enterica serovar Typhimurium (STM) (NC003197). The locations of FNR and NarL/NarP binding sites are represented by inverted arrows, and the IHF binding site is shown by a box. Differences between the two promoters are highlighted by black boxes, and the NsrR consensus inverted repeat binding sequence is shown. (B) Measured β-galactosidase activities in JCB3884 or JCB3884 ΔnsrR cells carrying pRW50 containing the pnrf53 fragment or the corresponding fragment from S. enterica serovar Typhimurium (6). Assays were performed using the Miller protocol (16) as described in the table footnotes. Activities were negligible when cells were grown aerobically and undetectable in a Δfnr mutant strain (not shown). Note that NsrR from Salmonella shows 96% identity with the E. coli K-12 protein.
Discussion.
The E. coli NsrR protein is a nitric oxide-sensitive transcription repressor that downregulates the expression of many genes involved in nitric oxide detoxification or the repair of damage caused by reactive nitrogen species (1, 11, 18). The consensus DNA binding sequence for NsrR consists of an inverted repeat, and at all of the target promoters characterized to date, the DNA site for NsrR overlaps the −35 or −10 element (1, 14, 18, 22). Results from mutational analysis (Fig. 1), a gel retardation assay (Fig. 2), and DNA sampling (Fig. 3) all argue that, at the E. coli nrf promoter, NsrR binds further upstream, recognizing a target centered at position −63. To confirm this, we exploited the observation that the E. coli hcp-hcr promoter (phcp) is repressed by NsrR but is derepressed by the presence of a high-copy-number plasmid carrying a DNA target for NsrR (11). Thus, the presence of a pBR322-based plasmid carrying the pnrf125 fragment with pnrf sequences from position −125 to position −20 (Fig. 2) induced the expression of a phcp::lac fusion by up to 2-fold, while this induction was suppressed by the presence of the p67C substitution in the pnrf125 fragment (D.F.B., unpublished data). Note that this argues against the existence of a second significant DNA site for NsrR at pnrf.
Although the primary role of the nrf operon appears to be in formate-dependent reduction of nitrite to ammonia, it can also protect cells from nitrosative stress by reducing nitric oxide (19, 27), and hence, the observed downregulation by NsrR makes biological sense. The upstream position of the pnrf NsrR binding site is novel, and a likely consequence is that NsrR modulates pnrf activity rather than switching it off completely. In fact, measured effects of NsrR at pnrf are small, and this suggests that NsrR here acts as a fine-tuner rather than as an on-off switch. The E. coli nrf operon regulatory region is extremely complex, with binding targets for at least six transcription factors: FNR, NarL, NarP, IHF, Fis, and NsrR (3-6). The nrf promoter is dependent on FNR for activation and can be shut off by Fis or high levels of NarL that appear to override all other positive regulatory inputs (5, 10, 26). The primary role of IHF, mediated by binding at the IHF I target, is also to downregulate FNR-dependent activation, but this can be reversed by NarP or NarL in response to nitrite or nitrate ions in the environment (6). NsrR binds to a target that overlaps IHF I, and the effects of NsrR and IHF appear to be additive. Since IHF binds to the DNA via the minor groove (21) and NsrR is likely to bind through the major groove, the simplest model suggests that both proteins bind simultaneously and repress expression from the nrf promoter. Removal of either protein leads to some induction, but removal of both is needed for maximum induction (Table 2). This is reminiscent of the situation at the galactose operon promoter, where multiple repressors are involved and maximum induction, termed “ultrainduction,” is seen only when each of the repressors can be removed from their target (24). Interestingly, the Salmonella nrf promoter appears to have become “blind” to repression by NsrR, though it remains to be seen if this has any biological significance.
Finally, we used the newly developed DNA sampling method to show directly that NsrR binds to the E. coli nrf operon regulatory region. As well as identifying IHF and Fis, as expected, the sampling experiment (Fig. 3) suggested that HU and YfhH also bind to the pnrf53 fragment. Concerning HU, a recent transcriptome analysis of its regulon showed that HU activates nrf operon expression during exponential growth (17). Concerning YfhH, although its role has yet to be established, it may be noteworthy that YfhH is a member of the RpiR-AslR family of transcription factors and the activity of many family members is modulated by the binding of sugars (12, 23). Hence, nrf operon expression may be modulated by additional signals and we clearly still have more to learn about its congested regulatory region.
Acknowledgments
We are grateful to Jeff Cole for helpful discussions and for providing different bacterial strains and to Diane Bodenmiller for helping to develop the gel retardation assay for NsrR binding.
This work was supported by a Wellcome Trust Programme Grant. S.S. was supported by a grant (MCB-0702858) from the National Science Foundation.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188:874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browning, D. F., J. A. Cole, and S. J. W. Busby. 2000. Suppression of FNR-dependent transcription activation at the Escherichia coli nir promoter by Fis, IHF and H-NS: modulation of transcription by a complex nucleo-protein assembly. Mol. Microbiol. 37:1258-1269. [DOI] [PubMed] [Google Scholar]
- 3.Browning, D. F., C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. W. Busby. 2002. Independent regulation of the divergent Escherichia coli nrfA and acsP1 promoters by a nucleoprotein assembly at a shared regulatory region. Mol. Microbiol. 43:687-701. [DOI] [PubMed] [Google Scholar]
- 4.Browning, D. F., C. M. Beatty, E. A. Sanstad, K. A. Gunn, S. J. W. Busby, and A. J. Wolfe. 2004. Modulation of CRP-dependent transcription at the Escherichia coli acsP2 promoter by nucleoprotein complexes: anti-activation by the nucleoid proteins FIS and IHF. Mol. Microbiol. 51:241-254. [DOI] [PubMed] [Google Scholar]
- 5.Browning, D. F., D. C. Grainger, C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. W. Busby. 2005. Integration of three signals at the Escherichia coli nrf promoter: a role for Fis in catabolite repression. Mol. Microbiol. 57:496-510. [DOI] [PubMed] [Google Scholar]
- 6.Browning, D. F., D. J. Lee, A. J. Wolfe, J. A. Cole, and S. J. Busby. 2006. The Escherichia coli K-12 NarL and NarP proteins insulate the nrf promoter from the effects of integration host factor. J. Bacteriol. 188:7449-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browning, D. F., J. A. Cole, and S. J. Busby. 2008. Regulation by nucleoid-associated proteins at the Escherichia coli nir operon promoter. J. Bacteriol. 190:7258-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butala, M., S. J. Busby, and D. J. Lee. 2009. DNA sampling: a method for probing protein binding at specific loci on bacterial chromosomes. Nucleic Acids Res. 37:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darwin, A., H. Hussain, L. Griffiths, J. Grove, Y. Sambongi, S. Busby, and J. Cole. 1993. Regulation and sequence of the structural gene for cytochrome c552 from Escherichia coli: not a hexahaem but a 50 kDa tetrahaem nitrite reductase. Mol. Microbiol. 9:1255-1265. [DOI] [PubMed] [Google Scholar]
- 10.Darwin, A. J., and V. Stewart. 1996. The NAR modulon systems: nitrate and nitrite regulation of anaerobic gene expression, p. 343-359. In E. Lin and A. Lynch (ed.), Regulation of gene expression in Escherichia coli. R. G. Landes Company, Austin, TX.
- 11.Filenko, N., S. Spiro, D. F. Browning, D. Squire, T. W. Overton, J. Cole, and C. Constantinidou. 2007. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 189:4410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaeger, T., and C. Mayer. 2008. The transcriptional factors MurR and catabolite activator protein regulate N-acetylmuramic acid catabolism in Escherichia coli. J. Bacteriol. 190:6598-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayaraman, P. S., J. Cole, and S. Busby. 1989. Mutational analysis of the nucleotide sequence at the FNR-dependent nirB promoter in Escherichia coli. Nucleic Acids Res. 17:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, H., P. J. Bledsoe, and V. Stewart. 2007. Activation of yeaR-yoaG operon transcription by nitrate-responsive regulator NarL is independent of oxygen-responsive regulator FNR in Escherichia coli K-12. J. Bacteriol. 189:7539-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N. R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74:271-276. [DOI] [PubMed] [Google Scholar]
- 16.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 17.Oberto, J., S. Nabti, V. Jooste, H. Mignot, and J. Rouviere-Yaniv. 2009. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One 4:e4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partridge, J. D., D. M. Bodenmiller, M. S. Humphrys, and S. Spiro. 2009. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 73:680-694. [DOI] [PubMed] [Google Scholar]
- 19.Poock, S. R., E. R. Leach, J. W. Moir, J. A. Cole, and D. J. Richardson. 2002. Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J. Biol. Chem. 277:23664-23669. [DOI] [PubMed] [Google Scholar]
- 20.Pope, N. R., and J. Cole. 1982. Generation of a membrane potential by one of two independent pathways of nitrite reduction by Escherichia coli. J. Gen. Microbiol. 128:219-222. [DOI] [PubMed] [Google Scholar]
- 21.Rice, P. A., S. Yang, K. Mizuuchi, and H. A. Nash. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87:1295-1306. [DOI] [PubMed] [Google Scholar]
- 22.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 1:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sørensen, K. I., and B. Hove-Jensen. 1996. Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J. Bacteriol. 178:1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokeson, J. P., S. Garges, and S. Adhya. 1991. Further inducibility of a constitutive system: ultrainduction of the gal operon. J. Bacteriol. 173:2319-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tucker, N., P. N. E. Le Brun, R. Dixon, and M. I. Hutchings. 2010. There's NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol. 18:149-156. [DOI] [PubMed] [Google Scholar]
- 26.Tyson, K. L., J. A. Cole, and S. J. W. Busby. 1994. Nitrite and nitrate regulation at the promoters of two Escherichia coli operons encoding nitrite reductase: identification of common target heptamers for NarP- and NarL-dependent regulation. Mol. Microbiol. 13:1045-1055. [DOI] [PubMed] [Google Scholar]
- 27.van Wonderen, J. H., B. Burlat, D. J. Richardson, M. R. Cheesman, and J. N. Butt. 2008. The nitric oxide reductase activity of cytochrome c nitrite reductase from Escherichia coli. J. Biol. Chem. 283:9587-9594. [DOI] [PubMed] [Google Scholar]
- 28.Wang, H., and R. P. Gunsalus. 2000. The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than nitrite. J. Bacteriol. 182:5813-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]