Abstract
The dimorphic bacterium Caulobacter crescentus has evolved marked phenotypic changes during its 50-year history of culture in the laboratory environment, providing an excellent system for the study of natural selection and phenotypic microevolution in prokaryotes. Combining whole-genome sequencing with classical molecular genetic tools, we have comprehensively mapped a set of polymorphisms underlying multiple derived phenotypes, several of which arose independently in separate strain lineages. The genetic basis of phenotypic differences in growth rate, mucoidy, adhesion, sedimentation, phage susceptibility, and stationary-phase survival between C. crescentus strain CB15 and its derivative NA1000 is determined by coding, regulatory, and insertion/deletion polymorphisms at five chromosomal loci. This study evidences multiple genetic mechanisms of bacterial evolution as driven by selection for growth and survival in a new selective environment and identifies a common polymorphic locus, zwf, between lab-adapted C. crescentus and clinical isolates of Pseudomonas aeruginosa that have adapted to a human host during chronic infection.
Colonization of new environments or changes in resource availability, predatory regime, or climate can drive adaptive evolution. Determining the genetic basis of these changes informs our understanding of the evolution of diversity and the nature of selection. Domestication of crop plants, adaptive radiations, and in-host evolution during chronic microbial infection are characterized by the evolution of a suite of phenotypes that are advantageous in the new environment. Recent work has successfully identified several of the polymorphisms responsible for this type of adaptive evolution in a variety of species (3, 7, 11, 12, 15, 22, 25, 35-37). With comparative genome sequencing emerging as a powerful tool for identifying genetic polymorphism (5, 14, 23), these studies are becoming faster and easier. Still, large genome sizes and countless sequence differences between individuals, isolates, strains, and species have made comprehensive analyses intractable.
Upon isolation and introduction into the laboratory, model research organisms experience extreme environmental changes, with associated selection pressures. Indeed, adaptation to life in captivity has been observed in a wide range of domesticated and model research organisms (2) and in zoo populations of endangered species (31). Many phenotypes that evolve in these nonnative environments do so repeatedly and become common features of human-cultured, -raised, or -cultivated organisms (2), providing evidence of positive selection. Likewise, the aquatic bacterium Caulobacter crescentus has evolved marked phenotypic changes during the 50 years it has been cultured in the laboratory environment. At least six phenotypic differences (Fig. 1) between two closely related strains (NA1000 and CB15) derived from the same common ancestor have evolved over decades of laboratory cultivation. It is presumed that these phenotypes evolved in response to the dynamic culture conditions and associated selection pressures experienced by bacteria in the laboratory environment. However, the extent of genetic divergence between these strains was uncharacterized, and it was not known whether the phenotypes could be explained by a few single nucleotide polymorphisms (SNPs), insertions/deletions, or genome rearrangements or by the accumulation of many mutations, each with a small contribution to particular phenotypes. In an effort to comprehensively characterize their divergence, we identified the genetic basis of all known phenotypic differences between two laboratory strains (NA1000 and CB15) of C. crescentus.
FIG. 1.
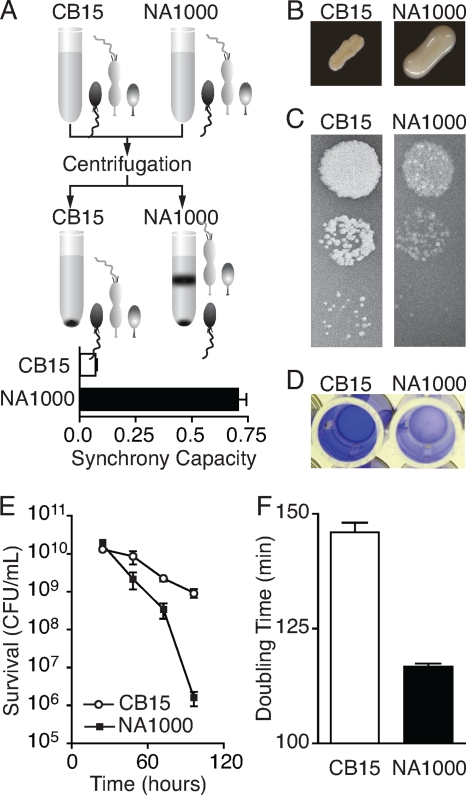
Evolved phenotypic differences between CB15 (Crosson2) and NA1000 (Crosson1). (A) Caulobacter cells divide asymmetrically to yield a swarmer and a stalked cell, which are mixed in culture. NA1000 stalked and predivisional cells (light gray) pellet less efficiently than swarmer cells (dark gray), allowing them to be physically separated. Synchrony capacity is quantified by calculating the proportion of cultured cells remaining in suspension. Error bars are ±standard errors of the mean (SEM). (B) When patched and grown on high-sugar media, NA1000 colonies develop a mucoid morphology, while CB15 colonies do not. (C) The transducing phage φCR30 efficiently infects and lyses CB15 cells, resulting in clear plaques, while infection of NA1000 with the same phage lysate results in fewer plaques that are visually turbid. (D) Holdfast-mediated attachment to a surface can be measured using a crystal violet assay. CB15 cells attach, resulting in robust staining, while NA1000 exhibits negligible adherence. (E) Upon continued aeration and incubation of stationary-phase Caulobacter cultures, NA1000 (▪) loses viability more rapidly than CB15 (○). Error bars are ±SEM. (F) In glucose minimal medium, NA1000 generation time is 20% shorter than that of CB15. Error bars are ±SEM.
Our study revealed 11 coding, noncoding, and insertion/deletion polymorphisms between these two strains, five of which completely account for the evolved differences between the strains. The results presented herein provide insight into prokaryotic evolution driven by selection for growth and survival in a research laboratory and demonstrate the utility of combining whole-genome sequencing and alignment with molecular genetic tools to reveal the genetic basis of multiple derived phenotypes. Our work demonstrates that rapid adaptation of C. crescentus to the laboratory environment occurred in both strain lineages and is characterized by relatively few genetic changes, including nonsynonymous mutation, noncoding regulatory changes, acquisition of new genes, and inactivation of existing genes, each with a large phenotypic effect.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Strains and plasmids are listed in Table 1. Rich medium was PYE (0.2% Bacto peptone, 0.1% yeast extract, 1 mM MgSO4, 0.5 mM CaCl2). Solid medium was PYE with 1.5% Bacto agar (16). Top agar was PYE with 0.3% Bacto agar and 0.3% glucose. High-sugar PYE medium was PYE with 1.5% glucose or 3% sucrose. Minimal medium was M2G (20 mM Na2HPO4, 20 mM KH2PO4, 9.3 mM NH4Cl, 0.5 mM MgSO4, 0.5 mM CaCl2, 1 μM FeSO4, 1 μM EDTA, 0.2% glucose). All cultures were grown at 30°C, unless otherwise noted.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description, genotype, relevant characteristics, and/or date frozen (mo/day/yr) | Source; reference |
|---|---|---|
| Caulobacter strains | ||
| FC19 | CB15; wild-type, sequenced strain; Crosson2 | Lucy Shapiro; 30 |
| FC20 | NA1000; synchronizable derivative of CB15; Crosson1 | Lucy Shapiro; 18 |
| FC746 | CB15 with NA1000 allele at SNP1 | This study |
| FC747 | CB15 with NA1000 allele at SNP2 | This study |
| FC748 | CB15 with NA1000 allele at SNP3 | This study |
| FC749 | CB15 with NA1000 allele at SNP4 | This study |
| FC750 | CB15 with NA1000 allele at SNP5 | This study |
| FC751 | CB15 with NA1000 allele at SNP6 | This study |
| FC752 | CB15 with NA1000 allele at SNP9 | This study |
| FC753 | CB15 with NA1000 allele at SNP10 | This study |
| FC754 | CB15 with NA1000 allele at SNP7 | This study |
| FC755 | CB15 with NA1000 allele at SNP8 | This study |
| FC756 | NA1000 with CB15 allele at SNP1 | This study |
| FC757 | NA1000 with CB15 allele at SNP2 | This study |
| FC758 | NA1000 with CB15 allele at SNP3 | This study |
| FC759 | NA1000 with CB15 allele at SNP4 | This study |
| FC760 | NA1000 with CB15 allele at SNP5 | This study |
| FC761 | NA1000 with CB15 allele at SNP6 | This study |
| FC762 | NA1000 with CB15 allele at SNP9 | This study |
| FC763 | NA1000 with CB15 allele at SNP10 | This study |
| FC764 | NA1000 with CB15 allele at SNP7 | This study |
| FC765 | NA1000 with CB15 allele at SNP8 | This study |
| FC766 | NA1000 with a deletion of the large indel | This study |
| FC1049 | CB15 with NA1000 alleles at SNP8 and SNP10 | This study |
| FC1050 | NA1000 with CB15 alleles at SNP8 and SNP10 | This study |
| FC846 | CB15 (ATCC 19089, ML28); frozen in 2001 | Mike Laub |
| FC858 | NA1000 (Laub); frozen in 2001 | Mike Laub |
| FC859 | NA1000 (Smit); frozen in 1983 | John Smit |
| FC860 | NA1000 (Ely); frozen in 1979 | Bert Ely |
| FC847 | CB15 (Brun4); frozen 1/11/94 | Yves Brun |
| FC855 | NA1000 (Brun1); frozen 6/11/90 | Yves Brun |
| FC856 | NA1000 (Brun2); frozen 2/2/97 | Yves Brun |
| FC857 | NA1000 (Brun3); frozen 11/8/01 | Yves Brun |
| FC854 | CB15 (Poindexter, YB1358); frozen 10/15/97 | Yves Brun (originally from Jeanne Poindexter) |
| FC1054 | CB15/pRKlac290-PzwfCB15 | This study |
| FC1055 | CB15/pRKlac290-PzwfNA1000 | This study |
| FC1056 | CB15/pRKlac290-PzwfNA1000JS | This study |
| Escherichia coli strains | ||
| FC728 | Top10/pNPTS138-SNP1CB15 | This study |
| FC730 | Top10/pNPTS138-SNP2CB15 | This study |
| FC732 | Top10/pNPTS138-SNP3CB15 | This study |
| FC734 | Top10/pNPTS138-SNP4CB15 | This study |
| FC736 | Top10/pNPTS138-SNP5CB15 | This study |
| FC738 | Top10/pNPTS138-SNP6CB15 | This study |
| FC740 | Top10/pNPTS138-SNP9CB15 | This study |
| FC742 | Top10/pNPTS138-SNP10CB15 | This study |
| FC727 | Top10/pNPTS138-SNP7CB15 | This study |
| FC744 | Top10/pNPTS138-SNP8CB15 | This study |
| FC729 | Top10/pNPTS138-SNP1NA1000 | This study |
| FC731 | Top10/pNPTS138-SNP2NA1000 | This study |
| FC733 | Top10/pNPTS138-SNP3NA1000 | This study |
| FC735 | Top10/pNPTS138-SNP4NA1000 | This study |
| FC737 | Top10/pNPTS138-SNP5NA1000 | This study |
| FC739 | Top10/pNPTS138-SNP6NA1000 | This study |
| FC741 | Top10/pNPTS138-SNP9NA1000 | This study |
| FC743 | Top10/pNPTS138-SNP10NA1000 | This study |
| FC725 | Top10/pNPTS138-SNP7NA1000 | This study |
| FC745 | Top10/pNPTS138-SNP8NA1000 | This study |
| FC1051 | Top10/pRKlac290-PzwfCB15 | This study |
| FC1052 | Top10/pRKlac290-PzwfNA1000(Crosson1) | This study |
| FC1053 | Top10/pRKlac290-PzwfNA1000(Smit) | This study |
| Plasmids | ||
| pNPTS138 | Allele replacement vector | M. R. K. Alley |
| pRKlac290 | β-Galactosidase vector | M. R. K. Alley and James Gober |
DNA sequencing.
C. crescentus NA1000 cells (10 ml) were grown in PYE and pelleted by centrifugation. DNA was extracted and purified using standard guanidium thiocyanate extraction and isopropanol/ethanol precipitation and sheared for library preparation, and quality was assayed using an Agilent 2100 bioanalyzer (Santa Clara, CA). The genome was sequenced using a GS FLX (Roche, Branford, CT) sequencer to a depth of 20-fold and assembled de novo with the Roche-454 Life Sciences Newbler assembler (see Table S1 in the supplemental material).
Polymorphism confirmation and genome finishing.
The 151 contigs resulting from assembly were aligned against the CB15 genome sequence (GenBank accession number AE005673) using Mauve 2.0.0 (10) and Sequencher 4.7 (Gene Codes, Ann Arbor, MI). Gaps between contigs were generally small (850 bp, average size) and occurred mostly at sites of low sequence complexity or repeated elements. PCR primers flanking the gap were used to amplify the region from NA1000 and directly sequenced (primer sequences are available upon request). We designed primers flanking each position at which the fully assembled genomes disagreed (Table 2 and see Tables S2 and S3 in the supplemental material), amplified them from NA1000 by PCR, and directly sequenced the PCR product using an ABI3730xl sequencer (Applied Biosystems, Foster City, CA). Polymorphisms that were confirmed in strain NA1000 were resequenced in CB15 to determine whether the differences were real polymorphisms (Table 2) or errors in the published CB15 genome sequence (see Table S3 in the supplemental material).
TABLE 2.
Confirmed differences between Caulobacter crescentus CB15 and NA1000 genomesa
| SNP name | CB15 position | NA1000 position | Gene annotation | CB15 gene | NA1000 gene | Erroneous allele (changed aa or base) | Corrected allele (changed aa or base) | Type of mutation |
|---|---|---|---|---|---|---|---|---|
| Mobile element | 473068.5 | 473069-499098 | Several genes involved in polysaccharide biosynthesis and metabolism | NA | NA | Not present | Present | I |
| SNP1 | 525889 | 551919 | Intergenic | CC_0502-0503 | CCNA_00536-00537 | NA | NA | NC |
| SNP2 | 1499722 | 1525749 | Transcriptional regulator, TetR family | CC_1345 | CCNA_01407 | GCC (Ala) | GTC (Val) | NS |
| SNP3 | 1548826 | 1548820 | Cytochrome cbb3 oxidase subunit I, ccoN | CC_1401 | CCNA_02097-02098 | CTC (Leu) | TTC (Phe) | NS |
| SNP4 | 2221557 | 2247533 | Intergenic | CC_2018-2019 | CCNA_02070 | NA | NA | NC |
| SNP5 | 2268285 | 2294261 | Intergenic | CC_2057-2058 | CCNA_02136-02137 | NA | NA | NC |
| SNP6 | 2604988 | 2630969 | 4-Coumarate—CoA ligase | CC_2400 | CCNA_02483 | CGG (Ile) | CGA (Ile) | S |
| SNP7 | 2638118.5 | 2664099 | O-antigen chain length regulator, hfsA | CC_2431 | CCNA_02513 | Intact ORF (ΔG) | Frameshifted ORF (G) | I |
| SNP8 | 3042436.5 | 3068416 | TonB-dependent receptor | CC_2820 | CCNA_02910 | Frameshifted ORF (ΔT) | Intact ORF (T) | I |
| SNP9 | 3197735 | 3223720 | Hypothetical protein | CC_2978 | CCNA_03073 | GAG (Ala) | ACG (Thr) | NS |
| SNP10 | 3234417 | 3260402 | TonB-dependent receptor | CC_3013 | CCNA_03108 | GAC (Asp) | GGC (Glc) | NS |
Positions and gene numbers for CB15 are consistent with the published genome sequence, GenBank accession number AE005673. Positions and gene numbers for NA1000 are consistent with the newly assembled and annotated genome, GenBank accession number CP001340. Gene annotations were determined using the Integrated Genomics gene annotation algorithm and comparison with protein sequences in GenBank. Erroneous and corrected alleles are represented in the CB15 (AE005673) and NA1000 (CP001340) genome sequences, respectively. aa, amino acid; NA, not applicable; I, insertion/deletion; NC, noncoding; S, synonymous; NS, nonsynonymous; CoA, coenzyme A.
PCR conditions.
PCR for direct sequencing or diagnostic purposes was carried out using GoTaq green (Promega, Madison, WI) 2× reaction mix with 0.5 to 1.0 μM primer, 5% dimethyl sulfoxide (DMSO), and a template (0.5 to 1.0 μl fresh Caulobacter culture). We used the thermal cycling protocol 95°C for 3 min; 30 cycles of 94°C for 15 s, 55°C for 15 s, and 72°C for 1 min/kb; 72°C for 5 min; and maintenance at 4°C. When used as a template for Sanger sequencing, 2.5 μl of the fresh PCR product was mixed with 2.0 μl water and 0.5 μl ExoSAP-IT (USB Corporation, Cleveland, OH), incubated at 37°C for 30 min and at 80°C for 15 min, and stored at −20°C. PCR for cloning was carried out using KOD Hot Start polymerase (Novagen, Madison, WI) with 0.5 to 1.0 μM primer in 1× KOD buffer, 5% DMSO, 0.16 mM deoxynucleoside triphosphates (dNTPs), 2.5 mM MgSO4, and the template (0.5 to 1.0 μl fresh Caulobacter culture). We used the following thermal cycling protocol: 95°C for 3 min; 30 cycles of 94°C for 15 s, 55°C for 15 s, and 68°C for 1 min/kb; 68°C for 5 min; and maintenance at 4°C. Primer sequences are available upon request.
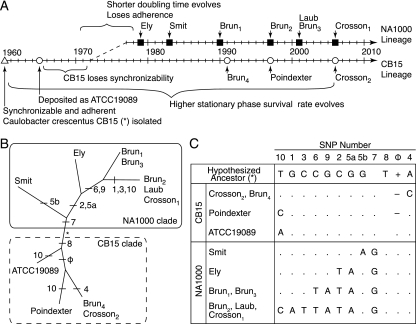
Caulobacter strain history.
The history of these Caulobacter strains was pieced together from information gathered from personal communications with longstanding members in the Caulobacter field (John Smit, Bert Ely, and Ellen Quardokus) and through careful reading of the Caulobacter literature (Fig. 2) (13, 17, 18, 32, 33).
FIG. 2.
History of laboratory-cultivated Caulobacter crescentus strains CB15 and NA1000. (A) The synchronizable CB15/NA1000 ancestor was originally isolated in 1960 (▵) and deposited with the ATCC (○). Following spontaneous loss of synchronizability, a new synchronizable derivative was isolated (NA1000). The source and freeze date for each CB15 (○) and NA1000 (▪) isolate, as well as the approximate timing of phenotypic evolution in both lineages, are indicated. The Brun, Laub, and Crosson strains are all endpoints originally derived from stock strains in the laboratory of Lucy Shapiro (Stanford University). (B) A DNA parsimony tree based on the 11 polymorphic sites forms two clades representing the NA1000 and CB15 lineages, and strain positions within the tree are consistent with their freeze dates (Table 1). The branches where SNPs evolved are indicated. Branch lengths are not drawn to scale. (C) Genotype data for the archived strains and the inferred CB15/NA1000 ancestral genotype. SNPs are numbered according to genomic position (Fig. 3).
Phylogenetic analysis.
We obtained CB15 and NA1000 strains that were archived at various points during the 50 years that Caulobacter has been in culture in the lab (Fig. 2). We genotyped each of the 11 polymorphisms between CB15 (Crosson2) and NA1000 (Crosson1) that we identified in all of these strains using PCR amplification and direct sequencing. From these data, we constructed an unrooted phylogenetic tree using DNA parsimony with 100 bootstrap replicates with PHYLIP (19) and Drawtree (28).
Genome annotation.
Open reading frames (ORFs) for the complete NA1000 genome sequence were called and annotated using ERGO (Integrated Genomics, Chicago, IL). We manually curated the annotation to verify that experimentally characterized genes were annotated correctly. Furthermore, as there is a tendency of automated gene-calling algorithms to misannotate start sites in GC-rich genomes, we made manual adjustments to the translation start positions for approximately 10% of ERGO-predicted genes. The adjustments were based on alignments to homologous sequences in GenBank.
Strain construction.
For each polymorphism that was confirmed by resequencing in both CB15 and NA1000, we built a pair of allele substitution strains using a standard double-crossover allele replacement strategy (34). One-kilobase fragments surrounding each polymorphism were PCR amplified (reaction conditions are described above), cloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA), subcloned into the suicide knockout plasmid pNTPS138 (Table 1), and electroporated into either CB15 or NA1000. Counterselection for the second chromosomal crossover event resulting in allele replacement was selected for by plating cells that had been outgrown in nonselective liquid medium for 12 h on PYE-sucrose plates. Allele substitutions were verified by PCR amplification and direct sequencing (reaction conditions are described above).
Synchrony assay.
C. crescentus cells divide asymmetrically to yield a flagellated swarmer cell and a stalked cell; exponentially growing C. crescentus cultures contain a mixture of these cell types. In strain NA1000, the stalked and predivisional cells can be physically separated from newly born swarmer cells by centrifugation in Ludox or Percoll or by repeated centrifugation and washing. Cell types from strain CB15, on the other hand, cannot be physically separated by these methods. To quantify the capacity of C. crescentus strains to be synchronized, liquid cultures were grown to an optical density at 660 nm (OD660) of 0.1 to 0.3. Percoll (50 μl; Sigma Chemical, St. Louis, MO) was added to 500 μl of culture and spun at 14,000 rpm in a 260D microcentrifuge (Denville Scientific, Metuchen, NJ) for 1 min. Under these conditions, all cells of strain CB15 collect in the pellet, while only swarmer cells pellet in strain NA1000, leaving the stalked and predivisional cells in a suspension as part of the supernatant (Fig. 1). The supernatant and cell suspension were collected, and the pellet was resuspended in 500 μl fresh M2G. We measured the OD660 of the supernatant (ODs) containing the cell suspension and the OD660 of the resuspended cell pellet (ODp) separately using a GeneSys 20 spectrophotometer (Thermo Fisher Scientific, Asheville, NC). To control for differences in total culture density, we defined synchrony capacity as ODs/(ODs + ODp).
Mucoidy assay.
We assessed colony mucoidy by patching each strain on high-sugar medium, incubating at 30°C for 48 h, and storing at 4°C for 24 h before scoring. CB15 colonies and streaks appear dry and dull, while NA1000 colonies and streaks appear moist and shiny when grown under these conditions (Fig. 1).
Phage susceptibility assay.
Phage susceptibility was determined by examining φCR30 plaque morphology. CB15 plaques have sharp edges and clear centers, while NA1000 plaques have indistinct edges and turbid centers (Fig. 1). Phage susceptibility was assessed by plating 100 μl of culture in 2 ml of PYE-glucose top agar on 60-mm PYE-glucose plates. Dilutions (10 μl) of φCR30 (17) lysate were spotted on the cooled top agar lawns and incubated overnight at 30°C. Plaques were photographed with illumination from below and appear lighter than the surrounding lawn of C. crescentus cells.
Surface attachment assay.
Measuring cellular attachment to a polystyrene culture dish assessed the development of a functional holdfast. Liquid PYE (1 ml) was inoculated with 2 μl fresh culture in a 96-well polystyrene culture dish (Corning, Corning, NY). Cultures were incubated at 30°C overnight, with shaking. The OD660 for an aliquot from each culture was measured using a GeneSys 20 spectrophotometer (Thermo Fisher Scientific, Asheville, NC) before the wells were washed thoroughly with deionized water, stained for 15 min with 0.1% crystal violet, washed thoroughly with deionized water, and solubilized in 1 ml 100% ethanol, and the OD540 was measured using a GeneSys 20 spectrophotometer (Thermo Fisher Scientific, Asheville, NC). Quantitative attachment is reported relative to CB15 levels.
Stationary-phase survival assay.
Liquid PYE (5 ml) was inoculated with freshly grown overnight culture and incubated at 30°C, with shaking, for 96 h. Dilutions were plated on PYE plates and incubated at 30°C for 48 h, and colonies were counted.
Measurement of population doubling time.
Doubling times were measured in 2-ml cultures in M2G (16) at 30°C with shaking in an Infors Multitron shaker (Infors, Bottmingen, Switzerland). Optical density measurements were taken every 20 to 40 min between an OD660 of 0.05 and an OD660 of 0.250 using a GeneSys 20 spectrophotometer (Thermo Fisher Scientific, Asheville, NC) and fit to a single exponential-growth function using Prism 4.0c (GraphPad Software, Inc., La Jolla, CA). To account for slight variations in growth rate due to different batches of media, all doubling times were normalized to the average CB15 doubling time for a given experiment and measured independently at least five times.
Expression analysis.
The zwf promoter region (285 bp) from each strain (Crosson1, Crosson2, Smit) was PCR amplified using KOD polymerase, cloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA), subcloned into the lacZ transcriptional fusion plasmid pRKLac290, and mated into CB15 and NA1000 using Escherichia coli strain FC03, carrying the RK600 helper plasmid (20). Reporter strains were grown in M2G plus 1 μg/ml tetracycline. All β-galactosidase assays were conducted with 100 μl exponential-phase culture (with an OD660 between 0.01 and 0.20), solubilized in chloroform, mixed with Z buffer (700 μl; 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7), and incubated in the presence of an o-nitrophenyl-β-d-galactopyranoside (ONPG) substrate (4 mg/ml ONPG in 0.1 M KPO4, pH 7). Reactions were stopped by adding 1 M Na2CO3 (1 ml). OD420 was measured using a GeneSys 20 spectrophotometer (Thermo Fisher Scientific, Asheville, NC), and Miller units (U) were calculated as U = (OD420·1,000)/(OD660·tv), where t is the reaction time (minutes) and v is the volume of culture used in the assay (ml) (27).
Genomic GC content analysis.
The C. crescentus NA1000 genome (GenBank accession number NC_011916) was downloaded into MATLAB R2008a (the MathWorks, Natick, MA) using the getgenbank command. The GC-AT composition of the entire chromosome was calculated using a 1,000-bp sliding window with the ntdensity command. This analysis requires the MATLAB Bioinformatics toolbox.
Statistical analyses.
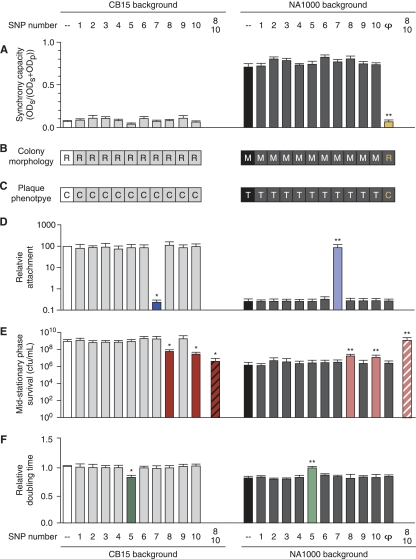
Quantitative phenotypes were compared between an allele replacement strain and its parental-background strain using pairwise two-tailed t tests calculated in Prism 4.0c (GraphPad Software, Inc., La Jolla, CA). Strains with a phenotype significantly different from the CB15 or NA1000 phenotype of the parental background are indicated with an asterisk or double asterisk, respectively, in Fig. 5D to F. Sample sizes, phenotype means, and P values are listed in Table 4.
FIG. 5.
Simultaneous mapping of laboratory-evolved Caulobacter phenotypes by comparison of allele replacement strains with their parental backgrounds (Fig. 1 and Table 1). Strains are organized by background (CB15, left; NA1000, right), CB15 (white) and NA1000 (black) phenotypes are shown for reference, and strains that differ significantly from their parent are indicated (*, CB15; **, NA1000) (Table 4). The SNP numbers at the top and bottom apply to all graphs and tables. The capacity of cultures to be physically synchronized (A), their development of mucoid colony morphology on high-sugar media (M, mucoid; R, rough nonmucoid) (B), and their φCR30 susceptibility (C, clear plaques; T, turbid plaques) (C) map to the presence of the large indel (φ and the yellow bar in panel A and the yellow letters in panels B and C). (D) The difference in adhesion between NA1000 and CB15, as measured by a crystal violet assay, maps to SNP7, a frameshift mutation in the holdfast synthesis gene hfsA. (E) Increased survival in stationary phase is conferred by the presence of CB15 alleles at SNP8 and SNP10, a frameshift mutation and a single amino acid change, respectively, in two different TBDRs. Single-allele-replacement strains have intermediate survival rates (red and pink bars), while the double-allele-replacement strains show parental levels of survival (striped red and pink bars). (F) The faster generation time in NA1000 maps to SNP5 (green bars). All error bars are ±SEM.
TABLE 4.
Quantitative phenotypic analysis of Caulobacter crescentus CB15 and NA1000 allele substitution strainsa
| Strain | SNP(s) | Strain background | Synchrony capacity |
Relative attachment |
Mid-stationary-phase survival |
Relative doubling time |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean | SEM | P value (versus CB15) | P value (versus NA1000) | n | Mean | SEM | P value (versus CB15) | P value (versus NA1000) | n | Mean | SEM | P value (versus CB15) | P value (versus NA1000) | n | Mean | SEM | P value (versus CB15) | P value (versus NA1000) | |||
| FC019 | CB15 | 10 | 0.07 | 0.01 | <0.0001 | 6 | 1 | 0 | <0.0001 | 19 | 9.63 × 108 | 2.4 × 108 | 0.001 | 19 | 1.0 | 0.01 | <0.0001 | |||||
| FC020 | NA1000 | 11 | 0.71 | 0.04 | <0.0001 | 6 | 2.65 × 10−3 | 2.8 × 10−4 | <0.0001 | ND | 16 | 1.65 × 106 | 6.8 × 105 | 0.001 | 16 | 0.8 | 0.01 | <0.0001 | ||||
| FC746 | 1 | CB15 | 10 | 0.09 | 0.01 | 0.341 | ND | 6 | 0.813 | 0.143 | 0.25 | ND | 10 | 1.30 × 109 | 3.5 × 108 | 0.424 | ND | 6 | 1.0 | 0.02 | 0.411 | ND |
| FC747 | 2 | CB15 | 11 | 0.11 | 0.03 | 0.264 | ND | 6 | 0.878 | 0.061 | 0.1 | ND | 10 | 7.27 × 108 | 1.5 × 108 | 0.512 | ND | 6 | 1.0 | 0.02 | 0.255 | ND |
| FC748 | 3 | CB15 | 11 | 0.11 | 0.02 | 0.104 | ND | 6 | 0.929 | 0.167 | 0.69 | ND | 10 | 7.92 × 108 | 1.6 × 108 | 0.635 | ND | 6 | 1.0 | 0.01 | 0.107 | ND |
| FC749 | 4 | CB15 | 10 | 0.08 | 0.01 | 0.593 | ND | 6 | 0.745 | 0.105 | 0.06 | ND | 10 | 7.87 × 108 | 1.4 × 108 | 0.622 | ND | 5 | 1.0 | 0.01 | 0.126 | ND |
| FC750 | 5 | CB15 | 10 | 0.04 | 0.01 | 0.04 | ND | 6 | 0.869 | 0.133 | 0.37 | ND | 8 | 9.56 × 108 | 2.2 × 108 | 0.985 | ND | 11 | 0.8 | 0.02 | <0.0001 | 0.676 |
| FC751 | 6 | CB15 | 11 | 0.11 | 0.02 | 0.169 | ND | 6 | 0.863 | 0.11 | 0.27 | ND | 10 | 2.06 × 109 | 5.5 × 108 | 0.045 | ND | 9 | 1.0 | 0.01 | 0.051 | ND |
| FC752 | 7 | CB15 | 11 | 0.11 | 0.03 | 0.222 | ND | 6 | 0.871 | 0.108 | 0.28 | ND | 8 | 2.02 × 109 | 8.3 × 108 | 0.114 | ND | 6 | 1.0 | 0.01 | 1 | ND |
| FC753 | 8 | CB15 | 11 | 0.07 | 0.01 | 0.6 | ND | 6 | 0.989 | 0.113 | 0.93 | ND | 17 | 3.10 × 107 | 8.3 × 106 | 0.001 | 0.002 | 9 | 1.0 | 0.01 | 0.554 | ND |
| FC754 | 9 | CB15 | 11 | 0.07 | 0.01 | 0.966 | ND | 6 | 2.39 × 10−3 | 2.4 × 10−4 | <0.0001 | 0.503 | 8 | 1.93 × 109 | 6.7 × 108 | 0.102 | ND | 6 | 1.0 | 0.02 | 0.025 | ND |
| FC755 | 10 | CB15 | 11 | 0.08 | 0.01 | 0.52 | ND | 6 | 1.14 | 0.175 | 0.46 | ND | 17 | 6.58 × 107 | 1.3 × 107 | 0.001 | <0.0001 | 7 | 1.0 | 0.01 | 0.065 | ND |
| FC756 | 1 | NA1000 | 11 | 0.72 | 0.03 | ND | 0.77 | 6 | 2.63 × 10−3 | 1.6 × 10−4 | ND | 0.97 | 9 | 1.48 × 106 | 3.0 × 105 | ND | 0.854 | 15 | 0.8 | 0.01 | ND | 0.271 |
| FC757 | 2 | NA1000 | 11 | 0.81 | 0.02 | ND | 0.04 | 6 | 2.72 × 10−3 | 2.7 × 10−4 | ND | 0.86 | 11 | 5.62 × 106 | 2.3 × 106 | ND | 0.064 | 13 | 0.8 | 0.01 | ND | 0.240 |
| FC758 | 3 | NA1000 | 11 | 0.79 | 0.02 | ND | 0.09 | 6 | 2.77 × 10−3 | 2.1 × 10−4 | ND | 0.727 | 8 | 4.13 × 106 | 2.0 × 106 | ND | 0.160 | 6 | 0.8 | 0.01 | ND | 0.438 |
| FC759 | 4 | NA1000 | 11 | 0.74 | 0.02 | ND | 0.53 | 6 | 2.66 × 10−3 | 2.0 × 10−4 | ND | 0.964 | 8 | 2.43 × 106 | 8.4 × 105 | ND | 0.500 | 6 | 0.8 | 0.01 | ND | 0.674 |
| FC760 | 5 | NA1000 | 10 | 0.75 | 0.03 | ND | 0.48 | 6 | 2.69 × 10−3 | 2.6 × 10−4 | ND | 0.902 | 8 | 3.1 × 106 | 1.1 × 106 | ND | 0.257 | 12 | 1.0 | 0.01 | 0.065 | <0.0001 |
| FC761 | 6 | NA1000 | 11 | 0.83 | 0.03 | ND | 0.02 | 6 | 3.37 × 10−3 | 3.6 × 10−4 | ND | 0.144 | 8 | 3.27 × 106 | 1.2 × 106 | ND | 0.210 | 6 | 0.9 | 0.01 | ND | 0.024 |
| FC762 | 7 | NA1000 | 11 | 0.75 | 0.02 | ND | 0.38 | 6 | 2.87 × 10−3 | 2.8 × 10−4 | ND | 0.589 | 8 | 2.78 × 106 | 1.3 × 106 | ND | 0.413 | 6 | 0.8 | 0.01 | ND | 0.337 |
| FC763 | 8 | NA1000 | 11 | 0.74 | 0.02 | ND | 0.49 | 6 | 2.93 × 10−3 | 2.8 × 10−4 | ND | 0.492 | 17 | 1.39 × 107 | 4.6 × 106 | 0.001 | 0.016 | 7 | 0.8 | 0.02 | ND | 0.593 |
| FC764 | 9 | NA1000 | 11 | 0.78 | 0.02 | ND | 0.16 | 6 | 0.901 | 0.125 | 0.46 | <0.0001 | 8 | 3.96 × 106 | 1.1 × 106 | ND | 0.079 | 6 | 0.8 | 0.01 | ND | 0.165 |
| FC765 | 10 | NA1000 | 10 | 0.81 | 0.03 | ND | 0.06 | 6 | 2.92 × 10−3 | 3.1 × 10−4 | ND | 0.53 | 17 | 2.09 × 107 | 4.3 × 106 | 0.001 | 0 | 7 | 0.8 | 0.03 | ND | 0.952 |
| FC766 | NA (large indel) | NA1000 | 10 | 0.07 | 0.02 | 0.793 | <0.0001 | 6 | 2.77 × 10−3 | 2.2 × 10−4 | ND | 0.731 | 8 | 2.65 × 106 | 8.3 × 105 | ND | 0.390 | 6 | 0.8 | 0.01 | ND | 0.243 |
| FC1049 | 8 and 10 | CB15 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 8 | 4.93 × 106 | 2.2 × 106 | 0.018 | 0.081 | ND | ND | ND | ND | ND |
| FC1050 | 8 and 10 | NA1000 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 8 | 1.58 × 109 | 6.3 × 108 | 0.273 | 0.002 | ND | ND | ND | ND | ND |
For graphical representation of these data, see Fig. 5. Synchrony capacity is defined as ODs/(ODs + ODp). Attachment is measured relative to that of CB15. Mid-stationary-phase survival is measured as the number of CFU/ml after 96 h of growth in liquid culture. Doubling times are measured relative to that of CB15. P values were calculated using unpaired two-tailed t tests to compare the phenotype of a strain with that of its parent and, only if significantly different, with that of the other parent. n, number of replicates; NA, not applicable; ND, not done.
Nucleotide sequence accession numbers.
The complete, annotated NA1000 genome was submitted to GenBank under accession numbers CP001340 and NC_011916.
RESULTS
The dimorphic aquatic bacterium Caulobacter crescentus has evolved marked phenotypic changes during the 50 years that it has been in culture in the laboratory environment. The history of the experimental strains has been documented in the literature and scientists' memories, making C. crescentus an excellent system for studying the nature of selection and phenotypic microevolution in prokaryotes (Fig. 2 and Table 1). Upon isolation, the CB15/NA1000 ancestor, called CB15 in the literature, was adherent and slow growing, and the two cell types (the motile swarmer cell and the nonmotile stalked cell) could be physically separated (i.e., synchronized) by centrifugation (32). It was maintained in slant culture before deposition as strain ATCC 19089, and after additional years of culture, CB15 lost the capacity to be synchronized in at least one laboratory. A synchronizable derivative was reisolated from this stock, maintained in serial liquid and slant culture for several years, and subsequently named NA1000. As both strains were serially passaged throughout their history, the opportunity for evolution and divergence existed in both lineages. An increased survival rate during stationary phase is likely strongly selected under this type of strain maintenance regime. During the years after the NA1000 derivative was isolated, it lost adherence, acquired a faster generation time, and was disseminated to numerous labs across North America (18). Spontaneous mutants for both of these phenotypes are positively selected in a serially propagated culture, as faster-growing cells rapidly increase in frequency, while cells that adhere to the side of a flask or culture tube are culled from the population each time the culture is diluted. CB15 (Crosson2) exhibits higher survival in stationary phase, has greater susceptibility to phage φCR30, and is less mucoid than NA1000 (Crosson1) (Fig. 1) (13). Due to its inherent capacity to be physically synchronized by centrifugation, strain NA1000 has become the predominant experimental C. crescentus strain throughout the world.
The sequencing and annotation of C. crescentus NA1000.
To define the genetic basis of phenotypic divergence in C. crescentus, we used a combination of whole-genome sequencing and classical molecular genetic tools to simultaneously map the mutations responsible for all known phenotypic differences between strain CB15 (Crosson2) and its derivative NA1000 (Crosson1). Chromosomal DNA of C. crescentus NA1000 (Crosson1) was sequenced using a model 454-GS FLX instrument and assembled de novo into 151 contigs (see Table S1 in the supplemental material). Direct sequencing of PCR products spanning gap regions completed the assembly to produce the 4.04-Mb NA1000 genome (GenBank accession number CP001340). Enhanced sequence quality, improved prokaryotic gene prediction algorithms, and more extensive protein sequence representation in GenBank resulted in an improved C. crescentus CB15 genome annotation relative to that of the previously published C. crescentus genome (30).
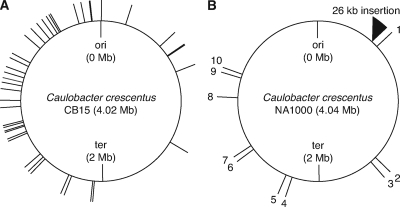
Comparison of the completed NA1000 genome (GenBank accession number CP001340) with the published CB15 sequence (accession number AE005673) (30) revealed 76 polymorphisms and no genomic rearrangements. Upon resequencing the polymorphic sites in both strains, we identified 20 errors in the 454 sequence data and 45 of the differences as errors in the published CB15 sequence (Fig. 3A; see also Tables S2 and S3 in the supplemental material) (30). The remaining 11 differences represent the entire set of real polymorphisms between these strains and include eight SNPs and two single-base and one large (26-kb) insertion-deletion site (indel) (Fig. 3B and Table 2).
FIG. 3.
Schematic representation of errors in the CB15 genome sequence and polymorphism between CB15 and NA1000. Caulobacter crescentus CB15 genome sequence (GenBank accession number AE005673) was aligned with the NA1000 sequence (accession number CP001340), and all differences identified were resequenced in both CB15 and NA1000. (A) Of the 76 differences identified, 20 were errors in the 454 sequence data (see Table S2 in the supplemental material) and 45 were errors in the published CB15 genome sequence, as both NA1000 and CB15 carry alleles identical to the newly generated NA1000 sequence (see Table S3 in the supplemental material). (B) The remaining 11 polymorphisms represent the true genotypic differences between CB15 and NA1000. They are numbered sequentially starting at the origin of replication (ori) and proceeding clockwise through the terminus (ter) and back to the origin. Figures were drawn with assistance from the Genome Tools Project software package (24). Sequencing statistics and details about sequence errors and polymorphisms are listed in Table 2 and Tables S1, S2, and S3 in the supplemental material.
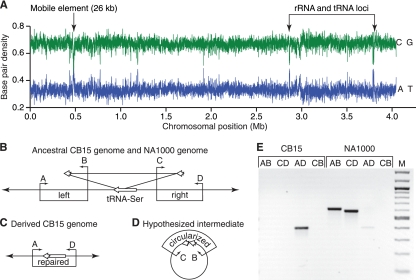
The chromosomal location and predicted ORFs within the large indel suggest that it is a mobile element. We measured the GC composition of the NA1000 genome and identified this indel as the region most different from average, indicating that it was likely acquired from a foreign source (Fig. 4). Within the indel are 22 predicted ORFs, including a P4 family integrase, an excisionase, conjugation and DNA transfer proteins, and several predicted membrane polysaccharide biosynthesis genes (Table 3). Sequence analysis of the ORFs in the indel reveals homology across a diverse group of bacterial taxa, precluding identification of its original source. This indel is inserted into a serine tRNA gene (KEGG database accession number CCNA_R0007), causing a small duplication that leaves the tRNA intact and functional (Fig. 4). Using PCR, we detected spontaneous deletion of the large indel from the NA1000 genome at a low frequency in exponentially growing culture (Fig. 4E), demonstrating that this mobile element is a cause of instability in the CB15/NA1000 ancestral and NA1000 genetic backgrounds.
FIG. 4.
The large indel in the NA1000 genome is a mobile element. (A) Analysis of the base composition of the NA1000 genome using a 1,000-bp sliding window reveals a substantially reduced GC content in the large indel, suggesting that it was acquired by horizontal transfer. Two other regions with known differences from average NA1000 GC content correspond to rRNA and tRNA loci. (B) The large indel is inserted into the 3′ end of serine tRNA gene CCNA_R0007 (long white arrow), causing a short duplication (short white arrows) but preserving the integrity of the gene. Four primers (A, B, C, and D; small black arrows) were used to determine if a strain carries the mobile element. Primer sets AB and CD amplify fragments spanning the left and right junctions, respectively. (C) Primer set AD fails to amplify a product in strains carrying the insertion but readily amplifies a product across the repaired junction. (D) Many mobile elements excise through a circular intermediate (8); the presence of a circular intermediate in NA1000 can be assayed with primer set BC. (E) Spontaneous deletion of this element occurs at low levels in exponentially growing NA1000 cultures. The only product amplified from a CB15 culture is the repaired junction (AD), whereas both the left and right junctions (AB and CD, respectively) are amplified from NA1000. A faint repaired junction (AD) product is detected in NA1000 cultures, suggesting that a small fraction of the cells in the population carry spontaneous deletions of the large indel. The AD products are identical in size and sequence in CB15, NA1000, and NA1000Δφ, indicating that deletion of this region is highly reproducible. We were unable to identify conditions that induced or enhanced excision and did not detect a circular intermediate with primers CB, suggesting that spontaneous excision events are rare, the intermediates are unstable, or excision of this element occurs via an alternate mechanism.
TABLE 3.
Annotated genes within the large indel (mobile) element present in the Caulobacter crescentus NA1000 genome
| Gene | Annotationa | Strand |
|---|---|---|
| CCNA_00460 | Hypothetical protein | + |
| CCNA_00461 | UDP-glucose 6-dehydrogenase (pseudogene) | + |
| CCNA_00463 | Hypothetical protein | − |
| CCNA_00464 | Hypothetical protein | + |
| CCNA_00465 | UDP-galactopyranose mutase | + |
| CCNA_00466 | Glycosyltransferase | + |
| CCNA_00467 | Oligosaccharide translocase/flippase | + |
| CCNA_00468 | Hypothetical protein | − |
| CCNA_00469 | Glycosyltransferase | − |
| CCNA_00470 | O-antigen polymerase | − |
| CCNA_00471 | GDP-l-fucose synthase | − |
| CCNA_00472 | GDP-mannose 4,6 dehydratase | − |
| CCNA_00473 | Hypothetical protein | − |
| CCNA_00474 | DNA relaxase/conjugal transfer nickase-helicase, trwC | − |
| CCNA_00475 | Bacterial conjugation protein, ATP binding domain, trwB | − |
| CCNA_00476 | Hypothetical protein | − |
| CCNA_00477 | Excisionase | − |
| CCNA_00478 | Hypothetical protein | − |
| CCNA_00479 | Transcriptional regulatory protein | + |
| CCNA_00480 | Bacteriophage P4 integrase | − |
| CCNA_00481 | Transcriptional regulatory protein | + |
| CCNA_00482 | Protein HipA | + |
Gene annotations were determined using the Integrated Genomics gene annotation algorithm and by comparison with protein sequences in GenBank.
Evidence of parallel adaptive evolution in C. crescentus laboratory strains.
During adaptation to the laboratory environment, C. crescentus diverged phenotypically into the contemporary CB15 and NA1000 strains. To better understand their evolutionary trajectory, we genotyped the 11 polymorphic sites in strains that had been frozen and stored at different times since their original isolation (Fig. 2A and Table 1) and constructed an unrooted parsimony tree. Comparing the tree architecture with historical information, we demonstrate that both the CB15 and NA1000 lineages have undergone evolution in the lab since they diverged; the genotyped strains cluster into two distinct monophyletic clades (Fig. 2B). Over time, three independent mutations arose in the same TonB-dependent receptor (CCNA_03108, SNP10) and two independent regulatory alleles arose upstream of the gene encoding glucose-6-phosphate dehydrogenase (G6PD) (zwf; SNP5a and SNP5b) (Fig. 2B and C). The repeated, parallel evolution of these genomic loci provides evidence that strains of C. crescentus cultivated in labs across North America are subject to similar selective pressures and that these mutations confer an adaptive advantage on C. crescentus cultured in the laboratory (22).
Simultaneous mapping of positively selected phenotypes.
New phenotypes can evolve through positive selection or genetic drift acting on newly acquired genes, regulatory or coding changes of major effect, or accumulation of many mutations, each with a small phenotypic contribution (5). To assess the genetic mechanisms in the evolution of C. crescentus CB15 and NA1000, we simultaneously mapped the causative mutation(s) for each phenotypic difference between the strains (Crosson2 and Crosson1, respectively). This was accomplished by constructing a series of reciprocal allele replacement strains for each of the 8 SNPs and 2 single-base indels, isolating an NA1000 strain with a spontaneous deletion of the large indel (NA1000Δ26kbφ), and measuring all phenotypes in each of these and the two parental strains (Fig. 5 and Tables 1 and 4).
Synchrony, mucoidy, and phage susceptibility phenotypes are determined by a putative mobile element.
Using our simultaneous mapping strategy, we determined that the ability to physically separate swarmer and stalked cells (i.e., synchronizability), mucoid colony morphology, and reduced φCR30 susceptibility require the presence of the large indel sequence. NA1000 exhibits all of these phenotypes, but CB15 and NA1000Δ26kbφ do not (Fig. 1A to C and 5A to C and Table 4). Many of the genes contained within the large indel have predicted functions in carbohydrate metabolism and biosynthesis that may alter the C. crescentus capsular phenotype (Table 3) (41). Indeed, the capsule likely plays significant roles in all three of the phenotypes that map to this region. Alteration in the abundance or complement of polysaccharides in the capsule can affect cellular sedimentation, and thus synchronizability, by differentially altering the density and hydrodynamic properties of the cell (6). Furthermore, φCR30 infection is dependent on attachment to the bacterial S-layer (13), and the presence of a mucoid capsule likely interferes with binding of the phage. As bacteriophage present a significant source of bacterial mortality in oligotrophic environments (4), the presence of a gene or genes that contribute even a modest protective effect can provide a significant selective advantage in the natural environment. As discussed above, we have shown that this sequence is inherently unstable and spontaneously excises in a subpopulation of cells in culture (Fig. 4), providing a mechanism for the host population to toggle between two disparate capsular states, depending on selective conditions.
Loss of holdfast-mediated adhesion in NA1000 is due to a frameshift mutation in the holdfast synthesis gene, hfsA.
Caulobacter species are well known for their ability to attach to inorganic and organic substrates through the holdfast organelle at the end of the stalk (26, 32, 40). Across the years of laboratory cultivation, CB15 (Crosson2) retained the ability to attach to surfaces and to other cells, while NA1000 (Crosson1) did not. Cell attachment is easily assayed by quantifying adhesion to a polystyrene surface using crystal violet staining (Fig. 1D). Analysis of the allele substitution strains reveals that the attachment defect in NA1000 is due to a single base insertion (SNP7) causing a frameshift in the middle of the known holdfast synthesis gene, hfsA (Table 2) (39). CB15 cells with the NA1000 SNP7 allele lose their ability to attach to surfaces, while the CB15 allele completely restores attachment in an NA1000 background (Fig. 5D and Table 4).
Altered survival in stationary phase is determined by mutations in two related TonB-dependent outer membrane receptors.
In liquid culture, C. crescentus reaches stationary phase within 36 h. After 96 h of culture time, 6% of CB15 (Crosson2) and 0.01% of NA1000 (Crosson1) cells remain culturable on solid agar (Fig. 1E). This difference in survival during stationary phase involves two TonB-dependent outer membrane receptor (TBDR) genes (encoding SNP8 [CCNA_02910] and SNP10 [CCNA_03108]; see Table S5 in the supplemental material). All single-allele-replacement strains involving these TBDRs have survival rates that are intermediate between those of their parental strains, and no single mutation can fully explain the dramatic difference in stationary-phase survival between CB15 (Crosson2) and NA1000 (Crosson1) (Fig. 5E and Table 4). Replacement of both TBDR alleles, however, results in recapitulation of the parental phenotypes (Fig. 5E and Table 4). Thus, cell survival during stationary phase is the result of an additive effect of distinct mutations in two genes encoding related TBDRs. SNP10 has mutated independently in at least three different branches of the C. crescentus strain phylogeny (Fig. 2B), suggesting that it is under strong positive selection in the laboratory environment.
A single base pair regulatory mutation in the zwf promoter increases growth rate.
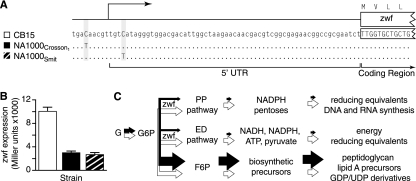
Like increased survival under stress, increased growth rate is a strongly selected phenotype that often evolves during laboratory cultivation (1, 2). Cell growth and division are complex and highly regulated processes, and there are theoretically many ways to elicit a fast-growth phenotype. We find that NA1000 (Crosson1) carries a single nucleotide polymorphism (SNP5) in the promoter of zwf, which encodes the metabolic enzyme glucose-6-phosphate dehydrogenase (Fig. 6 and Table 1). We observe an inverse relationship between zwf expression level and growth rate: the NA1000 allele results in a 70% reduction in zwf expression (Fig. 6) and is entirely responsible for the 20%-faster generation time in NA1000 (Fig. 1F and 5F and Table 4).
FIG. 6.
Regulatory mutations that reduce zwf expression increase the growth rate of Caulobacter. (A) SNP5 is located 7 bp upstream of the zwf transcriptional start site in isolate NA1000 (Crosson1). An independently evolved allele (SNP5b) in NA1000 (Smit) is located 3 bp downstream of the zwf transcriptional start. (B) Both NA1000 promoters show reduced activity relative to those of CB15, as measured using transcriptional fusions to lacZ (one-way analysis of variance; P < 0.0001). (C) Model of carbon flux in CB15 cells (white arrows) and NA1000 cells (black arrows). Glucose (G) is phosphorylated to glucose-6-phosphate (G6P), which serves as the primary substrate for three metabolic pathways in Caulobacter. G6P can enter either the pentose phosphate (PP) or the Entner-Doudoroff (ED) pathway following dehydrogenation by the gene product of zwf, glucose-6-phosphate 1-dehydrogenase. Alternatively, G6P can be isomerized to fructose-6-phosphate (F6P) by glucose-6-phosphate isomerase. As dehydrogenation of G6P is the rate-limiting step in both the PP and ED pathways, reduced zwf expression is known to increase the amount of G6P available for isomerization into F6P and, consequently, increase the concentration of substrates used for cell membrane and cell wall biogenesis (29).
G6PD catalyzes the first step in both the Entner-Doudoroff (ED) and pentose phosphate (PP) pathways (Fig. 6C). Together, these pathways generate ATP, NADH, NADPH, and pyruvate, as well as five carbon sugars important for nucleic acid biosynthesis (9). We propose a model in which reduced G6PD expression limits carbon flux through both the ED and PP pathways, resulting in a concomitant increase of G6P isomerization to fructose-6-phosphate (F6P) (Fig. 6C). F6P is a precursor for a range of biomolecules, including peptidoglycan and lipid A, and cannot be used as a substrate for glycolysis because the Caulobacter genome does not encode a phosphofructokinase (9). It has been shown experimentally that decreased zwf expression in Escherichia coli modulates carbon flux through central metabolic pathways, resulting in increased concentrations of F6P (29). Similarly, synthesis of the exopolysaccharide alginate in Pseudomonas aeruginosa requires F6P as a precursor (38). Our data provide evidence that C. crescentus growth rate is not limited by energy production (e.g., ATP, NADH, NADPH) when cultivated under laboratory conditions but rather by the availability of the cellular building block F6P.
Clearly, regulatory changes in the expression of key metabolic genes can lead to the rapid evolution of adaptive traits (11, 12). We have identified an independent mutation in the NA1000 (Smit) strain that also results in fast growth and reduced zwf expression (Fig. 6). The parallel evolution of fast growth in these NA1000 strains via two independent zwf promoter mutations suggests that this simple regulatory change is the most facile means of producing a faster-growing cell. Notably, the evolved deregulation of zwf expression in clonal isolates of Pseudomonas aeruginosa from chronically infected cystic fibrosis patients (38) suggests that changes in carbon metabolism via zwf are advantageous during the establishment of chronic infection and may be more broadly involved in the adaptive evolution of bacteria to new environments.
DISCUSSION
Distinguishing between positive selection and neutral evolution is challenging and typically requires large data sets and thorough statistical analyses (3, 21, 35, 36). While the small number of differences between C. crescentus CB15 and NA1000 makes it impossible to rigorously test these hypotheses, it is notable that repeated, parallel evolution of the same phenotype via similar genetic changes is compelling evidence that the phenotypes that we characterized are under positive selection (22). The documented parallel evolution of a TonB-dependent receptor gene and the regulatory region of zwf are linked to increased survival during stationary phase and a faster generation time, respectively. We note that both rapid growth and increased stress resistance characterize adaptation to life in captivity across a broad taxonomic section of domesticated research organisms (1, 2) and endangered zoo populations derived from long-term breeding programs (31). In short, human cultivation of prokaryotic and eukaryotic species selects for fast growth and increased stress resistance. We have deciphered the genetic bases of these, and other, adaptive phenotypes in lab-adapted C. crescentus.
All together, our data provide evidence that positive selection is responsible for the evolution of C. crescentus during its domestication as a laboratory organism. Moreover, adaptive evolution in bacteria proceeds through multiple genetic mechanisms, including nonsynonymous mutation, noncoding regulatory changes, acquisition of new sequences, and inactivation of existing genes. Future work will provide further insight into the prevalence of each of these mechanisms in the evolution of bacterial populations during domestication, infection, or colonization of other new environments.
Supplementary Material
Acknowledgments
We thank Ellen Quardokus, Yves V. Brun, Mike Laub, John Smit, Bert Ely, and Jeanne Poindexter for helpful discussions and Gwendolyn R. Marks for help with figures. We also thank Vince Magrini for helping to establish the collaboration between 454 Life Sciences, the entire Caulobacter community for suggestions on genome annotation, and the Crosson Lab.
C.M.C.-R. was supported by a Postbaccalaureate Research Education Program (PREP) grant to the University of Chicago from the NIH-NIGMS. M.E.M. is supported by NIH-NRSA fellowship 1F32-GM083424. S.C. acknowledges support for this project from the Arnold and Mabel Beckman Foundation (BYI) and the Mallinckrodt Foundation.
Footnotes
Published ahead of print on 14 May 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ackermann, M., A. Schauerte, S. C. Stearns, and U. Jenal. 2007. Experimental evolution of aging in a bacterium. BMC Evol. Biol. 7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artamonova, V. S., and A. A. Makhrov. 2006. Unintentional genetic processes in artificially maintained populations: proving the leading role of selection in evolution. Russ. J. Genet. 42:234-246. [PubMed] [Google Scholar]
- 3.Barrick, J. E., D. S. Yu, S. H. Yoon, H. Jeong, T. K. Oh, D. Schneider, R. E. Lenski, and J. F. Kim. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243-1247. [DOI] [PubMed] [Google Scholar]
- 4.Boras, J. A., M. M. Sala, E. Vazquez-Dominguez, M. G. Weinbauer, and D. Vaque. 2009. Annual changes of bacterial mortality due to viruses and protists in an oligotrophic coastal environment (NW Mediterranean). Environ. Microbiol. 11:1181-1193. [DOI] [PubMed] [Google Scholar]
- 5.Buckling, A., R. C. Maclean, M. A. Brockhurst, and N. Colegrave. 2009. The beagle in a bottle. Nature 457:824-829. [DOI] [PubMed] [Google Scholar]
- 6.Buer, C. S., K. T. Gahagan, G. A. Swartzlander, and P. J. Weathers. 1998. Differences in optical trapping prompt investigations of Agrobacterium surface characteristics. J. Ind. Microbiol. Biotechnol. 21:233-236. [Google Scholar]
- 7.Bull, J. J., M. R. Badgett, D. Rokyta, and I. J. Molineux. 2003. Experimental evolution yields hundreds of mutations in a functional viral genome. J. Mol. Evol. 57:241-248. [DOI] [PubMed] [Google Scholar]
- 8.Calendar, R. 2006. The bacteriophages, 2nd ed. Oxford University Press, New York, NY.
- 9.Caspi, R., H. Foerster, C. A. Fulcher, P. Kaipa, M. Krummenacker, M. Latendresse, S. Paley, S. Y. Rhee, A. G. Shearer, C. Tissier, T. C. Walk, P. Zhang, and P. D. Karp. 2008. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 36:D623-D631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darling, A. C., B. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean, A. M., and J. W. Thornton. 2007. Mechanistic approaches to the study of evolution: the functional synthesis. Nat. Rev. Genet. 8:675-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doebley, J. F., B. S. Gaut, and B. D. Smith. 2006. The molecular genetics of crop domestication. Cell 127:1309-1321. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, P., and J. Smit. 1991. A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface layer protein as a receptor. J. Bacteriol. 173:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellegren, H. 2008. Comparative genomics and the study of evolution by natural selection. Mol. Ecol. 17:4586-4596. [DOI] [PubMed] [Google Scholar]
- 15.Ellegren, H., and B. C. Sheldon. 2008. Genetic basis of fitness differences in natural populations. Nature 452:169-175. [DOI] [PubMed] [Google Scholar]
- 16.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 17.Ely, B., and R. C. Johnson. 1977. Generalized transduction in Caulobacter crescentus. Genetics 87:391-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA.
- 20.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillespie, J. H. 2004. Population genetics: a concise guide, 2nd ed. Johns Hopkins University Press, Baltimore, MD.
- 22.Gompel, N., and B. Prud'homme. 2009. The causes of repeated genetic evolution. Dev. Biol. 332:36-47. [DOI] [PubMed] [Google Scholar]
- 23.Koonin, E. V., and Y. I. Wolf. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36:6688-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, W., and S. L. Chen. 2002. Genome-tools: a flexible package for genome sequence analysis. Biotechniques 33:1334-1341. [DOI] [PubMed] [Google Scholar]
- 25.MacLean, R. C. 2005. Adaptive radiation in microbial microcosms. J. Evol. Biol. 18:1376-1386. [DOI] [PubMed] [Google Scholar]
- 26.Merker, R. I., and J. Smit. 1988. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl. Environ. Microbiol. 54:2078-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Neron, B., H. Menager, C. Maufrais, N. Joly, J. Maupetit, S. Letort, S. Carrere, P. Tuffery, and C. Letondal. 2009. Mobyle: a new full web bioinformatics framework. Bioinformatics 25:3005-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolas, C., P. Kiefer, F. Letisse, J. Kromer, S. Massou, P. Soucaille, C. Wittmann, N. D. Lindley, and J. C. Portais. 2007. Response of the central metabolism of Escherichia coli to modified expression of the gene encoding the glucose-6-phosphate dehydrogenase. FEBS Lett. 581:3771-3776. [DOI] [PubMed] [Google Scholar]
- 30.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelletier, F., D. Reale, J. Watters, E. H. Boakes, and D. Garant. 2009. Value of captive populations for quantitative genetics research. Trends Ecol. Evol. 24:263-270. [DOI] [PubMed] [Google Scholar]
- 32.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poindexter, J. S. 1981. The Caulobacters: ubiquitous unusual bacteria. Microbiol. Rev. 45:123-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ried, J. L., and A. Collmer. 1987. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 35.Rokyta, D. R., C. J. Beisel, P. Joyce, M. T. Ferris, C. L. Burch, and H. A. Wichman. 2008. Beneficial fitness effects are not exponential for two viruses. J. Mol. Evol. 67:368-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rokyta, D. R., P. Joyce, S. B. Caudle, and H. A. Wichman. 2005. An empirical test of the mutational landscape model of adaptation using a single-stranded DNA virus. Nat. Genet. 37:441-444. [DOI] [PubMed] [Google Scholar]
- 37.Ross-Ibarra, J., P. L. Morrell, and B. S. Gaut. 2007. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. U. S. A. 104(Suppl. 1):8641-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silo-Suh, L., S.-J. Suh, P. V. Phibbs, and D. E. Ohman. 2005. Adaptations of Pseudomonas aeruginosa to the cystic fibrosis lung environment can include deregulation of zwf, encoding glucose-6-phosphate dehydrogenase. J. Bacteriol. 187:7561-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, C. S., A. Hinz, D. Bodenmiller, D. E. Larson, and Y. V. Brun. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J. Bacteriol. 185:1432-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toh, E., H. D. Kurtz, Jr., and Y. V. Brun. 2008. Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J. Bacteriol. 190:7219-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitfield, C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39-68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.