Abstract
Although arsenic is highly toxic to most organisms, certain prokaryotes are known to grow on and respire toxic metalloids of arsenic (i.e., arsenate and arsenite). Two enzymes are known to be required for this arsenic-based metabolism: (i) the arsenate respiratory reductase (ArrA) and (ii) arsenite oxidase (AoxB). Both catalytic enzymes contain molybdopterin cofactors and form distinct phylogenetic clades (ArrA and AoxB) within the dimethyl sulfoxide (DMSO) reductase family of enzymes. Here we report on the genetic identification of a “new” type of arsenite oxidase that fills a phylogenetic gap between the ArrA and AoxB clades of arsenic metabolic enzymes. This “new” arsenite oxidase is referred to as ArxA and was identified in the genome sequence of the Mono Lake isolate Alkalilimnicola ehrlichii MLHE-1, a chemolithoautotroph that can couple arsenite oxidation to nitrate reduction. A genetic system was developed for MLHE-1 and used to show that arxA (gene locus ID mlg_0216) was required for chemoautotrophic arsenite oxidation. Transcription analysis also showed that mlg_0216 was only expressed under anaerobic conditions in the presence of arsenite. The mlg_0216 gene is referred to as arxA because of its greater homology to arrA relative to aoxB and previous reports that implicated Mlg_0216 (ArxA) of MLHE-1 in reversible arsenite oxidation and arsenate reduction in vitro. Our results and past observations support the position that ArxA is a distinct clade within the DMSO reductase family of proteins. These results raise further questions about the evolutionary relationships between arsenite oxidases (AoxB) and arsenate respiratory reductases (ArrA).
Arsenic is toxic to most organisms and is known to cause cancer in humans. However, bacteria have adapted several biotransformation pathways that function to either couple the reduction or oxidation of arsenicals to energy conservation and growth (1). The enzymologies of these two pathways have several features in common. The arsenate respiratory reductase (ArrAB) and arsenite oxidase (AoxAB) enzymes are usually composed of at least two subunits, a small iron-sulfur cluster-containing subunit (ArrB and AoxA) and a larger molybdopterin-containing catalytic subunit (ArrA and AoxB). Although they catalyze arsenic redox chemistry, ArrA and AoxB form distinct phylogenetic clades within the dimethyl sulfoxide (DMSO) reductase family of molybdenum-containing enzymes (16, 24).
Culture-dependent approaches have resulted in the isolation of a variety of diverse bacteria that metabolize arsenic (reviewed in reference 26). Many of these isolates have had their genomes sequenced, which has been insightful for understanding the composition and diversity of arr and aox gene clusters. In the arsenite-oxidizing nitrate reducer Alkalilimnicola ehrlichii strain MLHE-1 (a haloalkaliphile isolated from Mono Lake [CA]) (10, 15), bioinformatic analysis of its genome revealed the absence of genes homologous to the arsenite oxidase genes of the aoxB type. Instead, two genes (mlg_0216 and mlg_2426) were identified that better resembled the catalytic subunit of the arsenate respiratory reductase (20); however, MLHE-1 has not been shown to respire (or reduce) arsenate (15). Recent work by Richey et al. (20) showed that the Mlg_0216 protein (and not Mlg_2426) was expressed under chemolithoautotrophic (10 mM arsenite and 10 mM nitrate) growth conditions. Moreover, it was shown that Mlg_0216 exhibits both arsenate reductase and arsenite oxidase activities in vitro. These observations raised the question, is the mlg_0216 gene required for arsenite oxidation in vivo? In this report, we addressed this question by developing a genetic system in MLHE-1, generating strains with mutations in mlg_0216 and mlg_2426, and physiologically characterizing the resulting strains. Our results implicate mlg_0216 in chemolithoautotrophic arsenite oxidation coupled to nitrate respiration.
MATERIALS AND METHODS
Strains and plasmids.
Table 1 lists the strains of A. ehrlichii and Escherichia coli and plasmids used in this study.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or markers, characteristic(s), and use | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| WM3064 | Donor strain for conjugation; thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir(wt)] | 21 |
| DH5α λpir | F− Δ(argF-lac)169 φ80dlacZ58ΔM15 glnV44(AS) rfbD1 gyrA96 (Nalr) recA1 endA1 spoT1 thi-1 hsdR17 deoR λpir+ | 14 |
| A. ehrlichii strains | ||
| Wild-type MLHE-1 | Isolated from anoxic bottom water of Mono Lake; capable of chemoautotrophic growth on As(III) linked to nitrate; Kmr Ampr Cms at 1 μg ml−1 | 15 |
| MLHE1_0216 | A. ehrlichii MLHE-1 with pSMV20_0216 insertion into mlg_0216; incapable of chemoautotrophic growth on As(III); Kmr Ampr Cmr | This study |
| MLHE1_2426 | A. ehrlichii MLHE-1 with pSMV20_2426 insertion into mlg_2426; capable of chemoautotrophic growth on As(III); Kmr Ampr Cmr | This study |
| Plasmids | ||
| pBBR1MCS-1 | 5.1-kb broad-host-range plasmid; CmrlacZ | 9 |
| pSMV10 | 9.1-kb mobilizable suicide vector; oriR6K mobRP4 sacB Kmr Gmr | D. Lies, Caltech |
| pSMV20 | pSMV10 with cat gene from pHSG576 inserted into Gm resistance gene; oriR6K mobRP4 sacB Kmr Cmr | This study |
| pHSG576 | 3.475-kb vector, lac promoter, Cmr | Manel Camps, UCSC; 8 |
| pSMV20_0216 | pSMV20 with 500-bp insert from mlg_0216; Kmr Cmr | This study |
| pSMV20_2426 | pSMV20 with 500-bp insert from mlg_2426; Kmr Cmr | This study |
Growth conditions.
Luria-Bertani (LB) medium supplemented with diaminopimelic acid was used for routine culturing of E. coli strain WM3064. Aerobic medium for growth of MLHE-1 consisted of Mono Lake minimal (MLM) medium (15). MLM medium consists of (g liter−1) Na2CO3 (10.6), NaHCO3 (4.2), (NH4)2SO4 (0.1), MgSO4 (0.025), NaCl (90), K2HPO4(0.15), KH2PO4 (0.08), and 5 ml SL-10 trace elements (29). MLM medium was supplemented with 0.5 g liter−1 yeast extract, 0.5 g liter−1 Casamino Acids, 0.5 g liter−1 tryptone, 0.5 g liter−1 peptone, and 40 mM sodium acetate and is referred to as Mono Lake Complex (MLC) medium. All media were adjusted to pH 9.8. Autoclave sterilization of the medium was avoided due to formation of precipitates; instead, the medium was filter sterilized. For anaerobic growth on arsenite, the yeast extract, Casamino Acids, tryptone, and peptone were omitted and sodium arsenite and sodium nitrate were added at 5 mM. For anaerobic growth on nitrate, MLM medium was amended with 40 mM sodium acetate and 20 mM sodium nitrate. Filter-sterilized anaerobic medium was prepared by boiling under a stream of N2. The medium was placed in an anaerobic glove box (98% N2 and 2% H2) and dispensed into presterilized Balch tubes, which were sealed with sterile butyl rubber stoppers before being taken out of the anaerobic glove box. Chloramphenicol (Cm) was added to the medium at a final concentration of 1 μg ml−1. Wild-type and mutant MLHE-1 bacteria were grown at 37°C in aerobic MLC medium (with 40 mM sodium acetate) shaken at 250 rpm. Anaerobic growth curves of wild-type and mutant MLHE-1 bacteria were initiated from aerobically grown cultures prepared in MLC medium with 40 mM sodium acetate. The overnight aerobic cultures were diluted to an optical density at 600 nm (OD600) of 0.6 using anaerobic MLC medium and then inoculated at a 1:100 dilution into Balch tubes containing 10 ml of anaerobic MLC medium with the appropriate electron donor/acceptor. To monitor cell growth, tubes were sampled and the cells were fixed in 10% formaldehyde. Enumeration was done by bright-field microscopy using a hemocytometer. The growth of aerobic cultures with sodium acetate and anaerobic cultures with sodium acetate and sodium nitrate was monitored over time by measuring the OD600 with a Spectronic 20D+.
Analytical methods.
Prior to the speciation measurements, cells were removed from the growth curve samples (500 μl) by filtering through 0.2-μm Micro Centrifuge Filters (Costar no. 8169). Arsenic speciation and nitrate concentrations in culture experiments were analyzed by ion chromatography using a Dionex model DX 500 with an AS9HC column and a 9 mM sodium carbonate eluent. Nitrite was measured by using the Griess reagent. The microtiter dish method is briefly described as follows. The Griess reagent was made by mixing equal volumes of N-(1-naphthyl)ethylenediamine dihydrochloride (1 mg ml−1) and sulfanilic acid (prepared in 5% phosphoric acid at 10 mg ml−1). Samples or standards (150 μl) were mixed with 130 μl of deionized water and 20 μl of Griess reagent. The mixture was incubated at room temperature for 30 min, followed by measurement of the OD548.
Plasmid construction.
To construct pSMV20, the Cm acetyltransferase (cat) gene was amplified by PCR (see below for conditions) using plasmid pHSG576 (8) and cat primers cat_F1 (5′-GGAGATCTTCGCTGTCTTTTTCGTGACA-3′) and cat_R1 (5′-GGAGATCTGTAGCACCAGGCGTTTAAGG-3′). The PCR fragment was cloned into the BglII restriction site (underlined section in primers indicates BglII restriction site) in the gene encoding gentamicin (Gm) resistance on pSMV10, yielding pSMV20. To construct pSMV20_mlg0216 and pSMV20_mlg2426, primers IN_mlg_0216_F (5′-GGACTAGTAGACCCGTGTGACCAAGAAC-3′), IN_mlg_0216_R (5′-GGACTAGTTGGTGTTGCGGTAGTCGTAA-3′), IN_mlg_2426_F (5′-GGACTAGTATACCGTCGTCACCAAGAGC-3′), and IN_mlg_2426_R (5′-GGACTAGTCATTGTAAGGCCGGAAAGAC-3′) were used to PCR amplify (details described below) a 500-bp segment of the appropriate gene, which was cloned into the SpeI site (underlined section in primers indicates SpeI restriction site) of the multiple cloning site of pSMV20. The correct mutations were verified through sequencing of the plasmid inserts. The plasmids were maintained in E. coli DH5α λpir.
PCR.
PCR mixtures consisted of 25 μl containing 2.5 μl 10× Pfu-Turbo Hotstart buffer (Stratagene), 0.2 mM deoxynucleoside triphosphate mix, 400 nM primer, 30 ng pHSG576 or MLHE-1 genomic DNA, 1 μl Pfu-Turbo Hotstart DNA polymerase (Stratagene), and nuclease-free water. Samples were incubated with the following thermocycle profile: 95°C for 1 min; 29 cycles of 55°C for 30 s and 72°C for 1 min; and a final extension of 72°C for 10 min.
Mutagenesis.
E. coli strain WM3064 cells harboring pSMV20_mlg0216 and pSMV20_mlg2426 were grown overnight at 37°C in LB medium. MLHE-1 was grown in MLC medium overnight at 37°C. E. coli and MLHE-1 cultures (1 ml of each) were combined and spun down at 10,000 rpm for 2 min. The supernatant was removed, and the combined cells were resuspended in 1 ml of LB. The combined cells (100 μl) were spot plated on LB plates and incubated overnight at 37°C. The next day, the spots were recovered in 3 ml of MLC medium. The cell suspension was shaken at 250 rpm for 4 h, at 37°C. After the recovery time, the cells were plated on MLC medium plates with 1 μg ml−1 Cm. After 5 days, colonies were inoculated into MLC medium containing 1 μg ml−1 Cm. After 2 days, cells were harvested by centrifugation and used to extract DNA for analysis of plasmid integration.
To confirm that Cm-resistant colonies had successfully recombined pSMV20_0216/pSMV20_2426 into their genomes, the extracted DNA was analyzed by PCR using the cat_F1 (5′-GGAGATCTTCGCTGTCTTTTTCGTGACA-3′) and cat_R1 (5′-GGAGATCTGTAGCACCAGGCGTTTAAGG-3′) primers, which produce a 991-bp PCR fragment. In addition, M13_F (5′-GTAAAACGACGGCCAG-3′) and mlg_0216_R (5′-GGACTAGTCCAACTCGCGGTAGTAGTCC-3′) or mlg_2426_R (5′-GGACTAGTGATCGGACCCACCGTATAGA-3′) were used to confirm that pSMV20_0216/pSMV20_2426 had recombined into the appropriate genes.
Quantification of mlg_0216 (arxA) transcription.
The methods for reverse transcription-PCR (RT-PCR) has been described previously (22). MLHE-1 was grown in triplicate cultures aerobically (with 40 mM acetate) and anaerobically (with 40 mM acetate and 20 mM nitrate) in MLC medium. Cells were harvested in mid-log phase of growth. One milliliter of the cultures was centrifuged at 14,000 rpm at 4°C for 10 min. The cell pellets were flash frozen and stored at −80°C until they were analyzed. Immediately after collection of the initial cell samples, the cultures were amended with either 5 mM arsenate or 5 mM arsenite. After 6 h, 1-ml volumes of the cultures were once again centrifuged at 14,000 rpm at 4°C for 10 min and cell pellets were flash frozen and stored at −80°C until analyzed. RNA was extracted from the cell pellets using the Qiagen RNeasy miniprep kit according to the manufacturer's instructions, with a final elution volume of 30 μl. DNA contamination was removed by DNase (Promega RQ1 DNase) digestion. cDNA was synthesized as described previously (22). mlg_0216 cDNA was amplified using q_mlg_0216_F (5′-ACGGCAACGACTCCTACAAC-3′) and q_mlg_0216_R (5′-CGCACATCAATGGTGGTTAC-3′). For amplification of the 16S rRNA gene, the Univ340F (5′-GGACTACNNGGGTATCTAAT-3′) and Univ806R (5′-CCTACGGGRBGCASCAG-3′) primers were used. ImageJ64 (http://rsbweb.nih.gov/ij/) was used to quantify the agarose gel band intensities.
Phylogenetic analysis.
The predicted protein sequences of MLHE-1 genes mlg_0216 and mlg_2426, as well as other DMSO reductase family enzymes (5, 11), were aligned using ClustalW 1.82 (28). Phylogenetic analysis was performed on the alignment using the program PAUP* 4.0b10b (27). The distance criterion was used to construct an unrooted neighbor-joining tree. Gaps in the amino acid sequence alignment were ignored. The final tree was visualized using Dendroscope (6) and edited using Adobe Illustrator.
RESULTS
Antibiotic profile of MLHE-1.
Antibiotic resistance profiling of MLHE-1 was done in order to identify a suitable selectable marker for generating mutations. Growth of MLHE-1 on agar plates was measured in the presence of kanamycin (Km), ampicillin (Amp), Gm, tetracycline (Tc), and Cm. However, high resistance to the first three antibiotics at up to 500 μg ml−1 eliminated their usefulness as selectable markers in MLHE-1. The genome sequence of MLHE-1 contains homologs of the β-lactamase gene (confers Amp resistance) and the 2-aminoglycoside phosphotransferase gene (confers Km or Gm resistance). The use of Tc was disregarded because addition of the antibiotic to the Mono Lake medium caused it to turn a deep dark red. MLHE-1 showed sensitivity to Cm at a MIC of 1 μg ml−1. Moreover, no cat genes were identified in the MLHE-1 genome. A Cm concentration of 1 μg ml−1 was used for all further experiments.
Genetic system for MLHE-1.
After establishing that MLHE-1 was sensitive to Cm, we tested if the broad-host-range plasmid pBBR1MCS-1 (9) could be conjugated into MLHE-1. This plasmid contains the cat gene, which should confer Cm resistance on the host organism. The E. coli WM3064 conjugating strain harboring pBBR1MCS-1 was mated with MLHE-1 and subsequently plated on MLM medium containing 1 μg ml−1 Cm. The mating reaction yielded approximately 350 Cm-resistant CFU; this represented an approximate Cm resistance frequency of 10−5 relative to nonresistant MLHE-1 colonies. Replicate Cm-resistant MLHE-1 colonies were grown in MLM medium supplemented with Cm, followed by plasmid extraction. PCR analysis using plasmid-specific primers (M13 forward and reverse primers) confirmed the presence of the pBBR1MCS-1 plasmid in the Cm-resistant MLHE-1 colonies. In contrast to the wild type, only the Cm-resistant MLHE-1 colonies produced an ∼200-bp PCR product (data not shown). This validation was important because it demonstrated that (i) E. coli can conjugate a plasmid into MLHE-1, (ii) MLHE-1 can replicate and transcribe the genes on pBBR1MCS-1, and (iii) the cat gene is sufficient for Cm resistance in MLHE-1.
To construct a vector for genetic manipulation of MLHE-1, the cat gene from a different vector, pHSG576 (8) (pBBR1MCS-1 and pHSG576 have identical cat genes), was cloned into the gene encoding Gm resistance on pSMV10. This new pSMV10-based plasmid was referred to as pSMV20 and expected to only replicate in host strains containing the λ pir gene. Because MLHE-1 does not have λ pir, pSMV20 should only be maintained by chromosomal integration.
Attempts were made to generate nonpolar null mutants similar to our past work with Shewanella sp. strain ANA-3. However, counterselection of MLHE-1 using the sucrose sensitivity trait encoded by the sacB gene was ineffective in selecting for double-recombination events where the integrated plasmid had been resolved from the chromosome. The apparent sucrose-resistant colonies of MLHE-1 were still Cm resistant, indicating that the strains still contained the genome-integrated mutation plasmid.
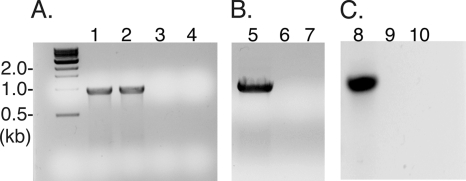
Our next approach was to develop insertion mutations by generating mutagenesis plasmids that contain 500-bp internal fragments of the target gene (mlg_0216 or mlg_2426) cloned into pSMV20 (Fig. 1). This internal region of the gene should act as a site for single recombination and produce a large insertion mutation in MLHE-1. The mutagenesis plasmids (pSMV20_0216 and pSMV20_2426) were conjugated from the E. coli mating strain (WM3064) to MLHE-1. The mating reaction typically yielded ∼100 CFU, or an estimated frequency of ∼10−4 Cm-resistant to nonresistant colonies. The resulting Cm-resistant colonies of MLHE-1 were screened by PCR for integration of pSMV20_0216 and pSMV20_2426 into the genome. Figure 2 A shows positive detection by PCR of the cat gene. In addition, recombination of pSMV20_0216 and pSMV20_2426 into the target gene was determined using PCR with a pSMV20-specific forward primer (M13F) and an MLHE-1 genome-specific reverse primer (mlg_0216 R and mlg_2426 R). The PCR should produce a 1.5-kp DNA product in strains with the integrated plasmids (Fig. 2B and C). As expected, DNA from the Cm-resistant colonies of MLHE-1, and not the wild-type strain, produced the expected 1.5-kp PCR product. These results confirmed that the Cm-resistant strains harbored pSMV20_0216 or pSMV20_2426 insertion mutations in their respective gene targets.
FIG. 1.
MLHE-1 arx operon. Genes are shown as arrows to scale, and each arrow indicates the orientation of the coding sequence. The arx operon is predicted to contain six genes (shaded in black); arxA (mlg_0216) encodes the molybdopterin oxidoreductase, arxB′ (mlg_0217) encodes a [4Fe-4S]-containing protein, arxB (mlg_0215) encodes another [4Fe-4S]-containing protein, arxC (mlg_0214) encodes a membrane protein, arxD (mlg_0213) encodes a TorD like protein, and arxE (mlg_0212) encodes a peptidylprolyl isomerase. The genes depicted in white are predicted to be a separate operon transcribed in the direction opposite to that of the arx operon and predicted to comprise the regulatory system for the arx operon. The genes in this operon include mlg_0218 (encoding an oxyanion transport protein), mlg_0219 (encoding a histidine kinase resembling AoxS), and mlg_0220 (thought to encode a response regulator resembling AoxR). Plasmid pSMV20 is shown to indicate a single homologous recombination event leading to the creation of insertion mutant strains.
FIG. 2.
Confirmation of insertion mutations in the MLHE-1 wild-type genome. Shown are an mlg_0216 (arxA) and mlg_2426 insertion mutation cat PCR (panel A), an mlg_0216 insertion PCR with M13 F/mlg_0216 R (panel B), and an mlg_2426 insertion PCR with M13 F/mlg_2426 R (panel C). Samples: MLHE_0216 DNA (lanes 1 and 5), MLHE_2426 DNA (lanes 2 and 7), MLHE-1 wild-type DNA (lanes 3, 6, and 9), and a water control (lanes 4, 7, and 10).
Growth experiments.
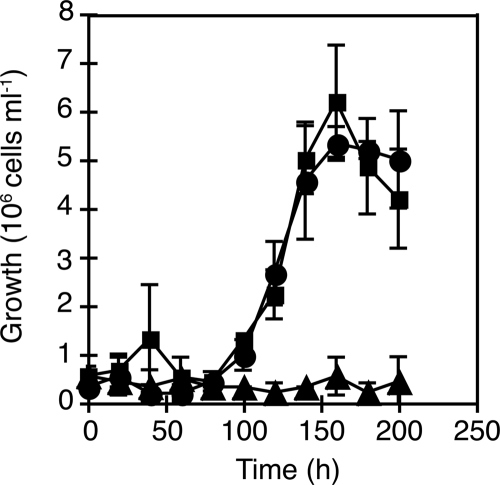
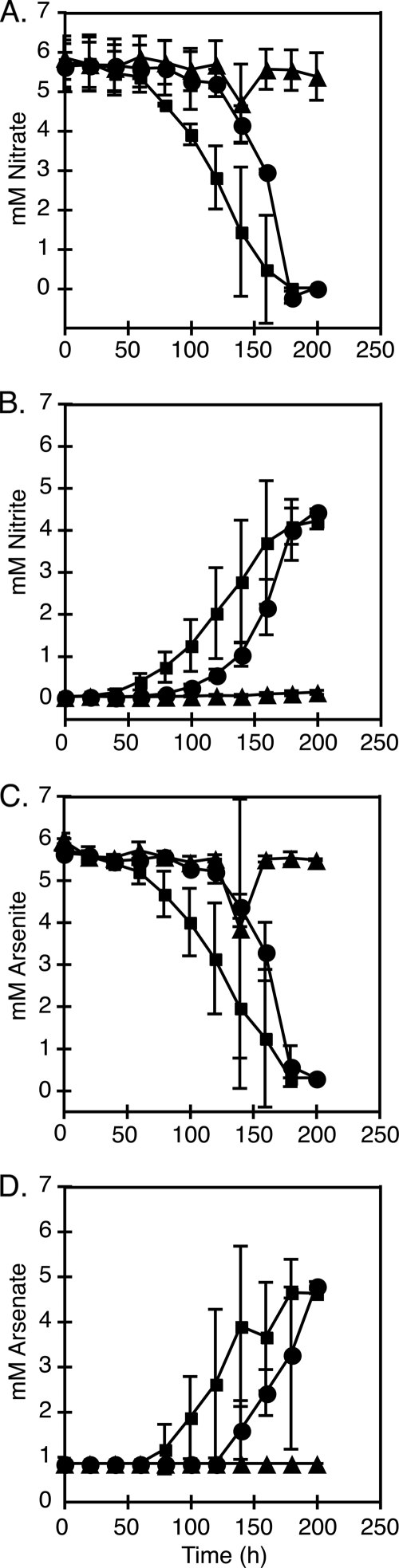
Growth curve analyses were used to test whether mlg_0216 and/or mlg_2426 is required for chemoautotrophic arsenite oxidation. The two insertion mutants, MLHE1_0216 and MLHE1_2426, and the wild-type strain were grown under anaerobic conditions with 5 mM arsenite and 5 mM nitrate in MLM medium. The wild-type strain and MLHE1_2426 reached stationary growth phase within 200 h, while MLHE1_0216 failed to grow (Fig. 3). Furthermore, the wild-type strain and MLHE1_2426 completely oxidized the arsenite to arsenate and reduced the nitrate to nitrite within 200 h (Fig. 4). In contrast, MLHE1_0216 did not reduce nitrate to nitrite (Fig. 4A and B) or oxidize arsenite to arsenate (Fig. 4C and D) during this time course.
FIG. 3.
Cell counts of MLHE-1 cultures grown anaerobically with 5 mM arsenite and 5 mM nitrate. Symbols: ▪, wild type; ▴, MLHE1_0216; •, MLHE1_2426. Data points and error bars represent averages and standard deviations of triplicate cultures, respectively.
FIG. 4.
Anaerobic growth analysis of MLHE-1 variants grown on 5 mM arsenite and 5 mM nitrate. Panels: A, mM nitrate; B, mM nitrite; C, mM arsenite; D, mM arsenate. Symbols: ▪, wild type; ▴, MLHE1_0216; •, MLHE1_2426. Data points and error bars represent averages and standard deviations of triplicate cultures, respectively.
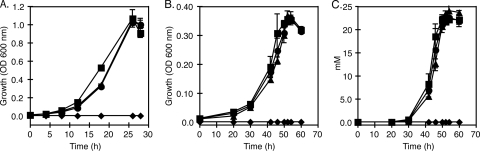
The growth specificity of the insertion mutants were determined by growing each strain chemoheterotrophically with acetate as the electron donor and nitrate or oxygen as the electron acceptor. Because of the higher cell densities achieved under these growth conditions, we chose to monitor growth by following changes in the OD600 of cultures using a spectrophotometer. Both mutant strains (MLHE1_2426 and MLHE1_0216) and the wild-type strain grew similarly on acetate and oxygen. All three strains reached stationary phase within 28 h (Fig. 5 A). Furthermore, all three strains grew comparably under anaerobic conditions with acetate and nitrate, reaching stationary phase and completely reducing nitrate to nitrite within 50 h (Fig. 5B and C).
FIG. 5.
Growth analysis of MLHE-1 variants on 40 mM acetate and (A) oxygen or (B) 20 mM nitrate and (C) nitrite analysis. Symbols: ▪, wild type; ▴, MLHE1_0216; •, MLHE1_2426; ⧫, no cells. Data points and error bars represent averages and standard deviations of triplicate cultures, respectively.
Ideally, we would like to demonstrate complementation of the mlg_0216 gene disruption in MLHE1_0216. However, difficulties were encountered in finding an appropriate broad-host-range vector that had a suitable antibiotic resistance marker (i.e., one that lacks a Cm, Km, Am, Tc, or Gm resistance cassette). Nevertheless, growth curve analyses provided sufficient evidence that mlg_0216 is required for chemoautotrophic arsenite oxidation in MLHE-1. However, mlg_2426 does not appear to be involved in arsenite oxidation. Furthermore, the growth curve experiments for acetate and other electron acceptors demonstrated that the insertion mutations did not affect oxygen and nitrate respiration pathways.
mlg_0216 transcription profile.
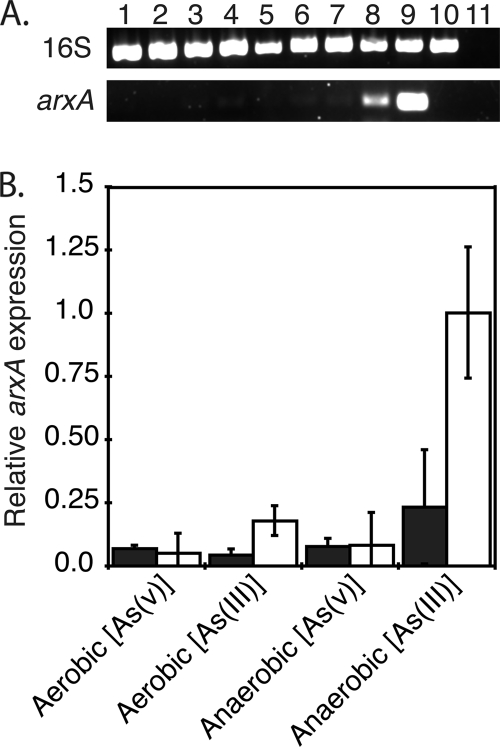
We were also interested in determining what growth conditions would influence the expression of mlg_0216. The wild-type strain was grown under aerobic and anaerobic conditions in MLC medium, and total RNA was extracted before and after the addition of 5 mM arsenite or 5 mM arsenate. Prior to the addition of arsenic under aerobic or anaerobic conditions, mlg_0216 expression was minimal (Fig. 6 A, lanes 1, 3, 5, and 7, and B). However, expression of mlg_0216 was detected within 6 h postaddition of arsenite only in anaerobically grown cells (Fig. 6 A, lane 8, and B). Slight to no mlg_0216 RT-PCR products were detected in total RNA prepared from cells grown aerobically and induced with either arsenite or arsenate (Fig. 6A, lanes 2 and 4). For all the growth conditions, similar band intensities were observed for the 16S rRNA gene RT-PCR products (Fig. 6A). RT-PCR on cDNA prepared without reverse transcriptase confirmed the absence of detectable DNA in the RNA extracts (data not shown). We concluded that the differential expression of mlg_0216 is affected by aerobic/anaerobic conditions and the oxidation state of arsenic.
FIG. 6.
(A) RT-PCR analysis of 16S rRNA gene (top) and mlg_0216 (bottom) in MLHE-1 after addition of 5 mM arsenate or 5 mM arsenite. Triplicate cultures of MLHE-1 were grown in MLC medium under aerobic or anaerobic conditions with 40 mM acetate or with 40 mM acetate and 20 mM nitrate, respectively. Once the cultures reached mid-log phase, cells were collected, followed by addition of 5 mM arsenate or 5 mM arsenite. After 6 h, the cultures were sampled once again. One representative sample from each triplicate condition is presented in the gel. Lanes 1 and 2 are aerobic samples before and after addition of arsenate. Lanes 3 and 4 are aerobic samples before and after addition of arsenite. Lanes 5 and 6 are anaerobic samples before and after addition of arsenate. Lanes 7 and 8 are anaerobic samples before and after addition of arsenite. Lane 9 is MLHE-1 DNA, lane 10 is Shewanella sp. strain ANA-3 DNA, and lane 11 is the water control. (B) Semiquantitative analysis of arxA gene expression done by subtracting the gel background from the band intensities of the arxA RT-PCR products (bottom) and from the 16S rRNA gene RT-PCR product (top). The black bars indicate relative arxA gene expression levels before addition of arsenate or arsenite, and the white bars indicate arxA gene expression levels 6 h after addition of arsenate or arsenite. Bars and error bars represent averages and standard deviations of triplicate cultures, respectively.
DISCUSSION
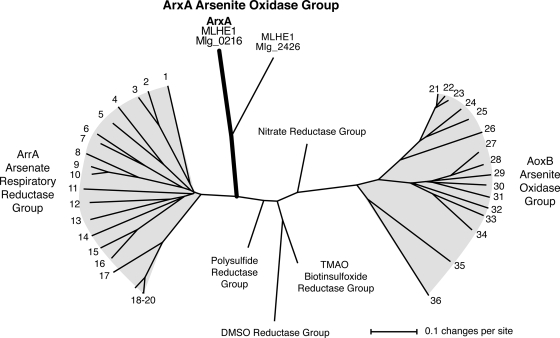
The goal of this study was to determine if the arrA-like genes (mlg_0216 and mlg_2426) of A. ehrlichii strain MLHE-1 are required for chemoautotrophic arsenite oxidation. A genetic system was first established in MLHE-1 and implemented to make insertion mutations in mlg_0216 and mlg_2426. In the growth studies with the two gene disruption mutants, only MLHE1_0216 failed to grow anaerobically by arsenite oxidation coupled to nitrate reduction. Our results indicate that mlg_2426 is not involved in the arsenite oxidation pathway in MLHE-1, thus raising further questions about the physiological role of mlg_2426 in MLHE-1. More importantly, mlg_0216 most likely encodes an arsenite oxidase that is considerably different than AoxB. This conclusion is supported by the distinct position of Mlg_0216 on the phylogenetic tree with other DMSO reductase family molybdenum enzymes (Fig. 7). Richey et al. (20) referred to Mlg_0216 as ArrA for a variety of reasons, such as greater sequence similarity to ArrA than AoxB groups, common biochemical features of ArrA, and the operon structure of mlg_0216 with greater resemblance to arr than aox operons. Despite these observations, we refer to mlg_0216 as arxA because it likely functions as an arsenite oxidase in vivo. The arxA name also reflects the arsenate reductase arsenite oxidase (arxA) bifunctional nature of Mlg_0216 in vitro.
FIG. 7.
Phylogenetic analysis of arsenate respiratory reductases (ArrA), AoxB-type arsenite oxidases, and the ArxA-type arsenite oxidase of MLHE-1. The unrooted tree was constructed using the neighbor-joining method; gaps were ignored in the final phylogeny. The numbering refers to representative amino acid sequences of ArrA and AoxB as follows, where * or ** indicates that the organism is known to respire arsenate or oxidize arsenite, respectively. Arsenate respiratory reductase group (ArrA): 1, Chrysiogenes arsenatis AAU11839*; 2, Geobacter lovleyi ZP_01593421; 3, Geobacter uraniireducens Rf4 ZP_01140714; 4, Bacillus selenitireducens AAQ19491*; 5, Bacillus arsenicoselenatis AAU11841*; 6, Sulfurospirillum barnesii AAU11840*; 7, Wolinella succinogenes NP_906980*; 8, Desulfosporosinus ABB02056*; 9 and 10, Desulfitobacterium YP_520364 and ZP_01372404*, respectively; 11, unidentified deltaproteobacterium MLMS-1 ZP_01288668*; 12, Natranaerobius thermophilus YP_001916826; 13, Halarsenatibacter silvermanii SLAS-1 ACF74513*; 14, Desulfonatronospira thiodismutans ASO3-1 ZP_03737819; 15, “Alkaliphilus metalliredigens” ZP_00800578; 16, Alkaliphilus oremlandii OhILAs ZP_01360543*; 17, Shewanella piezotolerans WP3 YP_002311519; 18 to 20, Shewanella group AAQ01672*, ZP_01704274, and YP_964317. Arsenite oxidase AoxB group: 21, Rhizobium sp. NT-26 AAR05656**; 22, Agrobacterium tumefaciens ABB51928**; 23, Ochrobactrum tritici ACK38267**; 24, Xanthobacter autotrophicus Py2 ZP_01198801; 25, Nitrobacter hamburgensis YP_571843; 26, Roseovarius sp. strain 217 ZP_01034989; 27, Ralstonia sp. strain 22 ACX69823; 28, Alcaligenes faecalis AAQ19838**; 29, Herminiimonas arsenoxydans AAN05581**; 30, Burkholderia multivorans ZP_0157266830; 31, Rhodoferax ferrireducens YP_524325; 32, Thiomonas sp. strain 3As CAM58792**; 33, Pseudomonas sp. strain TS44 ACB05943; 34, Halomonas sp. strain HAL1 ACF77048; 35, Chloroflexus aurantiacus ZP_00356; 36, Thermus thermophilus YP_145366**.
Looking at the open reading frames surrounding arxA, it appears that this gene is part of an operon with five other genes, arxB′ABCDE (Fig. 1). The putative proteins encoded by arxB′ABCDE are more consistent with those commonly associated with arr operons of other prokaryotes. ArxB′ and ArxB are predicted to contain [4Fe-4S] iron-sulfur clusters, which are different from the [2Fe-2S] cluster of the small subunit AoxA. ArxC is a homolog of NrfD (25), a transmembrane protein that is involved in redox interactions with quinones. ArxD is a TorD-like protein, a cytoplasmic chaperone involved in protecting molybdoenzymes from degradation until insertion of molybdenum cofactor (17). Lastly, ArxE resembles a peptidylprolyl isomerase that aids in protein folding (4). A twin-arginine translocation (TAT) sequence (NRRRFLK, amino acids 9 to 36) in the N terminus of ArxA indicates that the protein is exported to the periplasm as a folded protein complex. A periplasmic location would be consistent with ArrA and several other molybdopterin-containing proteins (21). In a number of DMSO reductase family enzymes, the TAT leader sequence is associated with the molybdopterin subunit; however, in the Aox enzyme complex, the TAT sequence is associated with the small Fe-S subunit, AoxA. Further molecular and biochemical work is needed to decipher the interactions of arx gene products and how arsenite oxidation is coupled to nitrate respiration.
Gene expression studies with MLHE-1 arxA showed that its expression was only induced with arsenite and under anaerobic conditions. No induction was observed with arsenate or under aerobic growth conditions (with and without arsenic). The induction pattern with arsenite was similarly observed for the aox genes in the arsenite-oxidizing strain of Agrobacterium tumefaciens; however, these studies were done with aerobically grown cultures (7). The arsenite-dependent regulation of aox in A. tumefaciens is mediated by a two-component signal transduction system, AoxS (sensor) and AoxR (response regulator). The aoxSR genes are located within the aox operon and are required for arsenite oxidation in A. tumefaciens. Though arsenite-dependent regulation in A. tumefaciens is mediated by AoxS and AoxR, it is still unclear if arsenite is the molecule sensed by AoxS. In MLHE-1, putative regulators (encoded by mlg_0218-0220) of the arx operon are located adjacent and in the opposite orientation to arxB′ABCDE. These putative regulators include a periplasmic phosphonate binding protein (Mlg_0218), a two-component signal transduction histidine kinase (Mlg_0219), and a response regulator (Mlg_0220). The latter two predicted regulators resemble AoxS and AoxR of A. tumefaciens and other arsenite oxidizers (12, 23).
Aerobic/anaerobic conditions were also a primary factor that led to the differential expression of arxA. This observation is similar to the respiratory pathway in the arsenate-respiring facultative anaerobe Shewanella sp. strain ANA-3. Murphy et al. (13) showed that genes encoding the anaerobic regulators ArcA (a two-component sensor/response regulator that detects anaerobic conditions by the change in the redox state of the quinone pool [3]) and EtrA (a homolog of the fumarate and nitrate reduction [FNR] regulator [2]) and carbon catabolite repression (cyclic AMP receptor protein [CRP] and adenylate cyclases) affect arsenate respiration in Shewanella sp. strain ANA-3 (13). Upon closer inspection of the MLHE-1 genome, we identified three possible fnr (mlg_0319, mlg_1070, and mlg_2113)- and crp-like genes (mlg_0631, mlg_1115, and mlg_2246). In addition, the MLHE-1 genome also contains two copies of NarQL nitrate response regulatory systems; NarQ is a membrane-bound nitrate sensor protein, and NarL is a DNA binding response regulator (mlg_1005-1006 and mlg_1670 −1669). Moreover, a search of the promoter region of the arx operon revealed possible binding sites for Fnr, CRP, ArcA, and NarL. These observations suggest that, in addition to arsenite, global regulators for metabolism, oxygen sensing, and nitrate sensing may be involved in the regulation of the arx operon.
Questions have been raised regarding the physiological role of arsenite oxidation in energy conservation versus detoxification (24). However, in organisms like MLHE-1, arsenite is used as an electron donor for anaerobic respiration and thus energy conservation. The recent identification of the anoxygenic photosynthetic arsenite oxidizer Ectothiorhodospira strain PHS-1 has expanded the physiological “scope” of arsenite oxidation in energy conservation pathways in prokaryotes. PHS-1 was isolated from a hot spring in Mono Lake (10). The strain appeared to lack an aoxB-type arsenite oxidase. Instead an arrA-like DNA sequence (most similar to the MLHE-1 arxA gene) was identified in PHS-1. Since then, we have sequenced nearly the entire arrA-like sequence of PHS-1 using an inverse PCR approach (unpublished data). Analysis of this sequence shows that it is ∼80% similar to ArxA of MLHE-1. Because MLHE-1 and PHS-1 are in the family Ectothiorhodospiraceae, we hypothesize that the PHS-1 arrA-like sequence functions as an arsenite oxidase for the anaerobic photosynthetic arsenite oxidation pathway.
The identification of mlg_0216 as an arsenite oxidase alternative to aoxB opens up new possibilities to reexamine why certain arsenite-oxidizing strains have eluded PCR detection for the aoxB-type arsenite oxidases. Similarly to MLHE-1, two chemoautotrophic arsenite-oxidizing denitrifiers have been isolated: DAO1 and DAO10 (19). Both isolates are facultative aerobes, are most closely related to Azoarcus (DAO1) and Sinorhizobium (DAO10) soil bacteria, and can completely reduce nitrate to nitrogen gas. PCR analysis using aoxB degenerate primers was used to try to detect aoxB-like genes in DAO1 and DAO10. Strain DAO10 contained an aoxB-like gene ∼73% similar to those of A. tumefaciens and strain NT-26. However, the PCR primer set was not able to detect an aoxB-like gene in strain DAO1 (18). A similar lack of detection was also observed when we tried degenerate aoxB PCR primers sets on MLHE-1 DNA (data not shown). It is tempting to speculate that Azoarcus-like strain DAO1 has an arxA-like arsenite oxidase gene instead of the aoxB type. However, failure to detect aoxB in DAO1 may simply be due to shortcomings in the current aoxB-degenerate primer sets or because the strain uses a completely different arsenite oxidation pathway. The diversity of arsenite oxidases in new isolates and environments may be expanded through future development of arxA-specific PCR primers.
In summary, our results provide strong evidence that mlg_0216 (and not mlg_2426) encodes a “new” type of arsenite oxidase that has closer phylogenetic relatedness to arsenate respiratory reductases than to AoxB arsenite oxidases. This conclusion is also supported by recent work demonstrating the reversible nature of ArxA and ArrA enzymes (i.e., capable of in vitro arsenate reduction or arsenite oxidation) (reviewed in reference 16). As originally noted by Oremland et al. (16), the arxA genes employed by anoxygenic photosynthetic arsenite oxidizers may be ancestral to the AoxB arsenite oxidases and arsenate respiratory reductases (ArrA). Further research on this “new” type of arsenite oxidation pathway in chemoautotrophs and phototrophs will be insightful for understanding the evolution of genes involved in arsenic-based energy conservation reactions.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Ahmann, D., A. L. Roberts, L. R. Krumholz, and F. M. Morel. 1994. Microbe grows by reducing arsenic. Nature 371:750. [DOI] [PubMed] [Google Scholar]
- 2.Grainger, D. C., H. Aiba, D. Hurd, D. F. Browning, and S. J. Busby. 2007. Transcription factor distribution in Escherichia coli: studies with FNR protein. Nucleic Acids Res. 35:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gralnick, J. A., C. T. Brown, and D. K. Newman. 2005. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol. Microbiol. 56:1347-1357. [DOI] [PubMed] [Google Scholar]
- 4.Hodak, H., A. Wohlkonig, C. Smet-Nocca, H. Drobecq, J. M. Wieruszeski, M. Senechal, I. Landrieu, C. Locht, M. Jamin, and F. Jacob-Dubuisson. 2008. The peptidyl-prolyl isomerase and chaperone Par27 of Bordetella pertussis as the prototype for a new group of parvulins. J. Mol. Biol. 376:414-426. [DOI] [PubMed] [Google Scholar]
- 5.Hoeft, S. E., J. S. Blum, J. F. Stolz, F. R. Tabita, B. Witte, G. M. King, J. M. Santini, and R. S. Oremland. 2007. Alkalilimnicola ehrlichii sp nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int. J. Syst. Evol. Microbiol. 57:504-512. [DOI] [PubMed] [Google Scholar]
- 6.Huson, D. H., D. C. Richter, C. Rausch, T. Dezulian, M. Franz, and R. Rupp. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashyap, D. R., L. M. Botero, W. L. Franck, D. J. Hassett, and T. R. McDermott. 2006. Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188:1081-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, B., and L. A. Loeb. 1995. Human immunodeficiency virus reverse transcriptase substitutes for DNA polymerase I in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92:684-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolibachuk, D., A. Miller, and D. Dennis. 1999. Cloning, molecular analysis, and expression of the polyhydroxyalkanoic acid synthase (phaC) gene from Chromobacterium violaceum. Appl. Environ. Microbiol. 65:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulp, T. R., S. E. Hoeft, M. Asao, M. T. Madigan, J. T. Hollibaugh, J. C. Fisher, J. F. Stolz, C. W. Culbertson, L. G. Miller, and R. S. Oremland. 2008. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science 321:967-970. [DOI] [PubMed] [Google Scholar]
- 11.McEwan, A. G., J. P. Ridge, C. A. McDevitt, and P. Hugenholtz. 2002. The DMSO reductase family of microbial molybdenum enzymes; molecular properties and role in the dissimilatory reduction of toxic elements. Geomicrobiol. J. 19:3-21. [Google Scholar]
- 12.Muller, D., C. Medigue, S. Koechler, V. Barbe, M. Barakat, E. Talla, V. Bonnefoy, E. Krin, F. Arsene-Ploetze, C. Carapito, et al. 2007. A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet. 3:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy, J. N., K. J. Durbin, and C. W. Saltikov. 2009. Functional roles of arcA, etrA, cyclic AMP (cAMP)-cAMP receptor protein, and cya in the arsenate respiration pathway in Shewanella sp. strain ANA-3. J. Bacteriol. 191:1035-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy, J. N., and C. W. Saltikov. 2007. The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Shewanella species. J. Bacteriol. 189:2283-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oremland, R. S., S. E. Hoeft, J. M. Santini, N. Bano, R. A. Hollibaugh, and J. T. Hollibaugh. 2002. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Appl. Environ. Microbiol. 68:4795-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oremland, R. S., C. W. Saltikov, F. Wolfe-Simon, and J. F. Stolz. 2009. Arsenic in the evolution of Earth and extraterrestrial ecosystems. Geomicrobiol. J. 26:522-536. [Google Scholar]
- 17.Pommier, J., V. Mejean, G. Giordano, and C. Iobbi-Nivol. 1998. TorD, a cytoplasmic chaperone that interacts with the unfolded trimethylamine N-oxide reductase enzyme (TorA) in Escherichia coli. J. Biol. Chem. 273:16615-16620. [DOI] [PubMed] [Google Scholar]
- 18.Rhine, E. D., S. M. Ni Chadhain, G. J. Zylstra, and L. Y. Young. 2007. The arsenite oxidase genes (aroAB) in novel chemoautotrophic arsenite oxidizers. Biochem. Biophys. Res. Commun. 354:662-667. [DOI] [PubMed] [Google Scholar]
- 19.Rhine, E. D., C. D. Phelps, and L. Y. Young. 2006. Anaerobic arsenite oxidation by novel denitrifying isolates. Environ. Microbiol. 8:899-908. [DOI] [PubMed] [Google Scholar]
- 20.Richey, C., P. Chovanec, S. E. Hoeft, R. S. Oremland, P. Basu, and J. F. Stolz. 2009. Respiratory arsenate reductase as a bidirectional enzyme. Biochem. Biophys. Res. Commun. 382:298-302. [DOI] [PubMed] [Google Scholar]
- 21.Saltikov, C. W., and D. K. Newman. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U. S. A. 100:10983-10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saltikov, C. W., R. A. Wildman, Jr., and D. K. Newman. 2005. Expression dynamics of arsenic respiration and detoxification in Shewanella sp. strain ANA-3. J. Bacteriol. 187:7390-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santini, J. M., and R. N. vanden Hoven. 2004. Molybdenum-containing arsenite oxidase of the chemolithoautotrophic arsenite oxidizer NT-26. J. Bacteriol. 186:1614-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silver, S., and L. T. Phung. 2005. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon, J., and M. Kern. 2008. Quinone-reactive proteins devoid of haem b form widespread membrane-bound electron transport modules in bacterial respiration. Biochem. Soc. Trans. 36:1011-1016. [DOI] [PubMed] [Google Scholar]
- 26.Stolz, J. F., P. Basu, J. M. Santini, and R. S. Oremland. 2006. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60:107-130. [DOI] [PubMed] [Google Scholar]
- 27.Swofford, D. L. 1999. Phylogenetic analysis using parsimony (*and other methods), 4.0b10b ed. Sinauer Associates, Sunderland, MA.
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W; improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widdel, F., and N. Pfennig. 1981. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov., sp. nov. Arch. Microbiol. 129:395-400. [DOI] [PubMed] [Google Scholar]