Abstract
Clostridium perfringens causes several diseases in domestic livestock, including necrotic enteritis in chickens, which is of concern to the poultry industry due to its health implications and associated economic cost. The novel pore-forming toxin NetB is a critical virulence factor in the pathogenesis of this disease. In this study, we have examined the regulation of NetB toxin production. In C. perfringens, the quorum sensing-dependent VirSR two-component signal transduction system regulates genes encoding several toxins and extracellular enzymes. Analysis of the sequence upstream of the netB gene revealed the presence of potential DNA binding sites, or VirR boxes, that are recognized by the VirR response regulator. In vitro binding experiments showed that purified VirR was able to recognize and bind to these netB-associated VirR boxes. Furthermore, using a reporter gene assay, the netB VirR boxes were shown to be functional. Mutation of the virR gene in two avian C. perfringens strains was shown to significantly reduce the production of the NetB toxin; culture supernatants derived from these strains were no longer cytotoxic to Leghorn male hepatoma cells. Complementation with the virRS operon restored the toxin phenotypes to wild type. The results also showed that the VirSR two-component system regulates the expression of netB at the level of transcription. We postulate that in the gastrointestinal tract of infected birds, NetB production is upregulated when the population of C. perfringens cells reaches a threshold level that leads to activation of the VirSR system.
Clostridium perfringens is a Gram-positive, anaerobic, spore-forming rod that secretes many toxins and extracellular enzymes. It is widespread in the environment and in the gastrointestinal tracts of both humans and animals. In addition to human diseases, such as gas gangrene and food poisoning (33, 34), C. perfringens is also the causative agent of several enterotoxemic diseases of animals, including necrotic enteritis (40, 41). In chickens, necrotic enteritis presents as either an acute clinical or subclinical disease and has a significant global economic impact; it is estimated to cost the international poultry industry approximately US$2 billion each year (43, 45). The acute form of this disease is believed to involve several predisposing factors (44) that allow C. perfringens type A to multiply to high numbers in the small intestine, and it results in the concomitant elaboration of extracellular toxins that damage the intestines. Tissue disruption ranges from macroscopic lesions on the small intestinal mucosa to extensive necrosis of the mucosal surface in severe cases (45).
Early studies suggested that alpha-toxin was the major virulence factor involved in the pathogenesis of necrotic enteritis (1-3). Although this toxin has been shown to be critical in human gas gangrene (4), recent virulence tests with chickens using defined chromosomal alpha-toxin mutants clearly demonstrated that it was not essential for necrotic enteritis (20). A novel toxin, NetB, was identified and shown to be a critical virulence factor in this disease. C. perfringens netB mutants were unable to induce disease in chickens, which was reversed by complementation with the wild-type netB gene (19). Both native and recombinant forms of NetB were cytotoxic to chicken Leghorn male hepatoma (LMH) cells in vitro but not to several other cell lines. A gas gangrene strain that does not produce NetB but produces alpha-toxin and perfringolysin O (PFO) was not toxic for LMH cells (19).
At present, no information is available regarding how the production of this key toxin is regulated. In the human gas gangrene isolate, strain 13, the production of alpha-toxin and perfringolysin O, the toxins implicated in gas gangrene, is regulated by the VirSR two-component signal transduction system, which comprises the VirS sensor histidine kinase and its cognate response regulator, VirR (9, 22, 38). Since its initial identification, it has become the best-characterized two-component system in the clostridia. It is a global regulatory network, since it not only regulates toxin production but controls the expression of housekeeping genes (7, 18, 29, 39) and genes encoding several regulatory RNA molecules (7, 31, 39).
The VirSR phosphorelay appears to be mediated by quorum sensing (30). In this system, it is proposed that threshold concentrations of a peptide that is synthesized and secreted by the product of the agrBD genes are detected by the VirS sensor histidine kinase. Autophosphorylation of VirS is followed by phosphotransfer to VirR (9). The VirR response regulator then recognizes and binds independently to two imperfect directly repeated sequences, called the VirR boxes, located upstream of the target genes (12). We have previously shown that VirR directly activates transcription of the perfringolysin O structural gene, pfoA, and that the maintenance of the integrity, spatial organization, and helical phasing of the VirR boxes is crucial for optimal levels of perfringolysin O production (10, 12). VirR boxes are located upstream of several other genes in three different strains of C. perfringens, and we have shown that VirR recognizes and binds to these alternative binding sites in vitro and activates reporter gene expression in vivo (10, 26).
In this article, we report the identification of VirR boxes upstream of the netB gene in the necrotic enteritis strain EHE-NE18, which led to the hypothesis that expression of netB, and hence the subsequent production of the toxin, is regulated by VirSR. Using in vitro binding assays and reporter gene expression assays, we show that these VirR boxes are functional. Mutation of the virR genes of EHE-NE18 and a second necrotic enteritis-causing strain, 56, reveals that the VirSR two-component signal transduction system is involved in the regulation of NetB production. Furthermore, quantitative real-time PCR (qRT-PCR) analysis of RNA isolated from EHE-NE18, its isogenic virR mutant, and a complemented derivative demonstrates that netB expression is regulated at the level of transcription. In summary, these results have revealed another toxin gene that is regulated by the VirSR system, thereby adding to the repertoire of genes in the ever-expanding VirSR regulon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Escherichia coli strains were grown at 37°C in 2× YT agar or broth or SOC broth (36) supplemented with chloramphenicol (30 μg/ml) or erythromycin (150 μg/ml). C. perfringens strains were cultured at 37°C in brain heart infusion broth (Oxoid), Trypticase-peptone-glucose (TPG) broth (35), fluid thioglycolate medium (FTG) (Difco), or nutrient agar supplemented with 30 μg/ml chloramphenicol (NACm30), 7 μg/ml or 50 μg/ml erythromycin (NAEm7 or NAEm50, respectively), or 10 μg/ml rifampin and 10 μg/ml nalidixic acid (NARif10Nal10). For the screening of perfringolysin O (PFO) production, C. perfringens transformants were grown on horse blood agar (HBA) (22). All agar cultures of C. perfringens were incubated in an atmosphere of 10% (vol/vol) H2 and 10% (vol/vol) CO2 in N2.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80Δ lacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdr17(rK− mK+) deoR thi-1 supE44 gyrA96 relA1 | Life Technologies |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 λ− | Invitrogen |
| BL21(DE3)(C43) | F′ ompT hsdSB(rB− mB−) gal dcm (DE3)(C43) | 24 |
| C. perfringens strains | ||
| JIR325 | 13 Nalr Rifr | 22 |
| EHE-NE18 | Australian chicken necrotic enteritis isolate | 20 |
| NE18ΔnetB1 | EHE-NE18 ΔnetB::catP | 19 |
| 56 | Belgian chicken necrotic enteritis isolate | 14 |
| JIR4228 | JIR325 Ω Tn916 (ΔpfoR pfoA colA luxS) | 5 |
| JIR12377 | EHE-NE18 virR Ω targetron | This study |
| JIR12382 | 56 virR Ω targetron | This study |
| Plasmids | ||
| pMTL540F | Clostridial expression vector | 15 |
| pJIR750 | C. perfringens-E. coli shuttle vector, Cmr | 6 |
| pJIR750ai | pJIR750Ωalpha-toxin targetron | 8 |
| pJIR751 | C. perfringens-E. coli shuttle vector, Emr | 6 |
| pJIR1456 | C. perfringens-E. coli shuttle vector with oriT, Cmr | 21 |
| pJIR1877 | pUC18(EcoRI/HindII) harboring the product of PCR with UP/JRP372 (EcoRI/HindIII; 1.0 kb) containing virR and its promoter | 23 |
| pJIR1897 | pJIR750 Ω (HindIII/EcoRI: pJIR1877, 1.0 kb, virR) | 9 |
| pJIR2233 | pJIR750 (HindIII/BamHI) harboring the virR promoter (HindIII/BamHI: pJIR1877, 0.214 kb) | This study |
| pJIR2373 | pfoA reporter gene vector, Emr | 10 |
| pJIR2422 | pJIR2373(EcoRV) harboring annealed complementary primers 17443/17444 (34 bp) (wild-type pfoA VirR boxes) | 10 |
| pJIR2529 | pJIR2233(BamHI/SacI) harboring the virRS operon (BamHI/SacI; 2.1 kb) | This study |
| pJIR3464 | pJIR3754 (FspI/SpeI) harboring the replication region from pMTL540F (NheI/BstZ171; 1.73 kb) | This study |
| pJIR3562 | pJIR3756 (MluI) harboring PfdermB-RAM (MluI; 1.23 kb) | This study |
| pJIR3566 | Clostridial targetron vector derived from pJIR750ai, contains ermB-RAM and lacZα, Cmr | This study |
| pJIR3575 | pJIR2373(EcoRV) harboring annealed complementary primers JRP4051/JRP4052 (34 bp) (netB VirR boxes) | This study |
| pJIR3608 | Group II intron of pJIR3566 retargeted to the 546/547 site of the virR gene | This study |
| pJIR3753 | pJIR750ai (HindIII/SphI) harboring Pgdh promoter region (HindIII/SphI; 207 bp) | This study |
| pJIR3754 | pJIR3753 (XbaI/SphI) harboring a PCR product containing the oriT of pJIR1456 | This study |
| pJIR3756 | Clostridial vector harboring SOE PCR product (SalI/XbaI; 321 bp) containing the ferredoxin promoter (Pfd) fused to the ermB-RAM (PfdermB-RAM) | This study |
Molecular techniques.
Plasmid DNA from E. coli cells was routinely isolated using an alkaline lysis method (25). When DNA was used for sequencing, an additional polyethylene glycol precipitation step was included, as outlined in the instructions of the Prism Ready Reaction DyeDeoxy terminator cycle sequencing kit (Applied Biosystems). Restriction endonucleases and other enzymes were used as specified by the manufacturer (Roche Diagnostics or New England Biolabs). All oligonucleotide primers are listed in Table 2.
TABLE 2.
Oligonucleotides used in this work
| Primer or use | Sequence (5′-3′) | Location or usea |
|---|---|---|
| Cloning of VirR boxes | ||
| JRP4051 | ACCAGTTATGTATAAATTTTGACCAGTTTTACCA | VirR boxes upstream of netB (+) |
| JRP4052 | TGGTAAAACTGGTCAAAATTTATACATAACTGGT | VirR boxes upstream of netB (−) |
| Gel mobility shift analysis | ||
| JRP589 | GGAACTCATATTATAATTGG | Upstream of pfoA VirR boxes (+) |
| JRP618 | CTCTAATTTTTTCTTTTCCC | Downstream of pfoA VirR boxes (+) |
| Construction of pJIR3566 | ||
| JRP2712 | ATGATCAAGCTTGCATGCAAAAAATAAACTGAGAAAATGATATAC | gdh promoter (+) |
| JIR2713 | ATGATCAAGCTTCATACCTAATTTATCACATGC | gdh promoter (−) |
| JRP4613 | ACGCGTCGGAAACGTAAAAGAAGTTATGGAAATAAG | Construction of ermB-RAM cassette |
| JRP4614 | GAACATAATGCTCATGTAATCACTCCTTCTTAATTAC | Construction of ermB-RAM cassette |
| JRP4615 | GAGTGATTACATGAGCATTATGTTCAGTAAGGTCGTTAATC | Construction of ermB-RAM cassette |
| JRP4616 | CTTGGGTTAATTGAGGCCTGAGTATAAGGTGACTTATAC | Construction of ermB-RAM cassette |
| JRP4617 | CTCAGGCCTCAATTAACCCAAGAACAAAAATATAAAATATTCTC | Construction of ermB-RAM cassette |
| JRP4618 | ACGCGTGGGGAATTATTTCCTCCCGTTAAATAATAG | Construction of ermB-RAM cassette |
| JRP4619 | GAGTATTTATGAAAAGCGG | Replace native promoter of ermB-RAM with pasfdx promoter region |
| JRP4620 | ATACAATAAGTTATGAGAAGGAGTGATTACATGAGCATTATG | Replace native promoter of ermB-RAM with pasfdx promoter region |
| JRP4621 | GTCGACACGCGTGAAGATTTAGGATTTACTGTAAT | Replace native promoter of ermB-RAM with pasfdx promoter region |
| JRP4622 | GTAATCACTCCTTCTCATAACTTATTGTATCATGTTTTTAAAC | Replace native promoter of ermB-RAM with pasfdx promoter region |
| JRP4042 | AAAAAAGCTTCAGGAAACAGCTATGACCATGATTACGCCTAGCTTCCATGCCTGC | Amplification of lacZα (+) |
| JRP4623 | AAAAAATGTACAGGTGTTGGCGGGTGTCGGG | Amplification of lacZα (−) |
| Targetron | ||
| JRP4106 | TGAACGCAAGTTTCTAATTTCGGTTTTAAGTCGATAGAGGAAAGTGTCT | EBS2 primer to retarget intron to virR |
| JRP4107 | AAAAAAGCTTATAATTATCCTTACTTAACAATAAGGTGCGCCCAGATAGGGTG | IBS primer to retarget intron to virR |
| JRP4108 | CAGATTGTACAAATGTGGTGATAACAGATAAGTCAATAAGGGTAACTTACCTTTCTTTGT | EBS1 primer to retarget intron to virR |
| JRP3867 | CGAAATTAGAAACTTGCGTTCAGTAAAC | Targetron universal primer |
| JRP4240 | CGCGTCGGAAACGTAAAAGAAGTTATGGAAATAAG | Upstream of ermB RAM (+) |
| JRP4241 | CTATTATTTAACGGGAGGAAATAATTCCCCACGCGT | Downstream of ermB RAM (−) |
| JRP182 | GTTATGAAGTTCGTGCTTTTAG | Upstream of targetron insertion within virR (+); also used to generate virR probe |
| JRP2813 | ACGCGTTCATTAACATATTAAATCCCC | Downstream of targetron insertion within virR (−) |
| JRP111 | GTATTATTACCTTTCTCTCA | virR probe (−) |
| JRP2633 | CCGGGATCCTTAGGGTAACAAAAAACACC | catP probe (−) |
| JRP2142 | CTCAGTACTGAGAGGGAACTTAGATGGTAT | catP probe (+) |
| JRP3947 | GGGAACGAAACGAAAGCG | Targetron probe (+) |
| JRP3948 | CGTAATAAATATCTGGAC | Targetron probe (−) |
| JRP2369 | AATAAGTAAACAGGTAACGTCT | ermB probe (+) |
| JRP2370 | GCTCCTTGGAAGCTGTCAGTAG | ermB probe (−) |
| qRT-PCR | ||
| JRP2479 | CCATCTGTTTTTATATCTGCTCCAGTA | Within rpoA (+) |
| JRP2480 | GGAAGGTGAAGGACCAAAAACTATT | Within rpoA (−) |
| JRP4707 | AAATATACTTCTAGTGATACCGCTTCACA | Within netB (+) |
| JRP4708 | GAGGATCTTCAATAAATGTTCCACTTAA | Within netB (−) |
+, sense primer; −, antisense primer.
Competent E. coli (17) and C. perfringens (37) cells were prepared and transformed as described previously unless otherwise indicated. C. perfringens genomic DNA was isolated from 5 ml FTG broth cultures as was done previously (28). PCR amplification was carried out as described previously (10). PCR products were either purified directly using the QIAquick PCR purification kit (Qiagen) or extracted from agarose gels using the QIAquick gel extraction kit (Qiagen), according to the manufacturer's instructions.
Cloning of the netB VirR boxes into pJIR2373 was carried out as described previously (10). The resultant construct, pJIR3573, was used to transform the C. perfringens pfoA mutant, JIR4228 (Table 1), to yield JIR12345.
Construction of the vector used in targetron mutagenesis, pJIR3566.
As part of another ongoing study, a new targetron vector was constructed for the mutagenesis experiments reported here. The vector, pJIR3566, utilizes a clostridial promoter and contains an ermB retrotransposition-activated marker (ermB-RAM). To construct pJIR3566, the Pgdh promoter region from Clostridium difficile strain 630 was PCR amplified using primers JRP2712 and JRP2713. The resultant 207-bp fragment was then digested with HindIII and SphI and cloned into the targetron vector pJIR750ai (8), generating pJIR3753. The RP4 oriT region was PCR amplified from plasmid pJIR1456 (21), cloned into pCR2.1-TOPO (Invitrogen), digested with SphI and SpeI, and subcloned into the XbaI and SphI sites of pJIR3753, resulting in pJIR3754. The replication region of pMTL540F (15) was then excised using NheI and BstZ17I and subcloned into the FspI and SpeI sites of pJIR3754, leading to the construction of pJIR3464.
To facilitate one-step selection of chromosomal integrants, an ermB retrotransposition-activated marker (ermB-RAM was constructed by a series of stepwise splice overlap extension (SOE) PCRs (16) and cloning reactions that led to the insertion of the td group I intron and its minimal exon sequences (13) in reverse orientation within the ermB coding region, immediately downstream of the ermB start codon (Table 2). The native promoter of the ermB-RAM cassette was subsequently replaced with the strong Clostridium pasteurianum ferredoxin promoter (Pfd) (pJIR3756) to maximize expression of ermB, and the modified cassette was cloned into the MluI site of pJIR3464 to give pJIR3562. Finally, to facilitate blue/white screening of retargeted targetron elements, the 5′ (retargeted) end of the group II intron within the resultant plasmid, pJIR3562, was replaced with the lacZα gene, leading to the construction of the final clostridial targetron plasmid, pJIR3566.
Construction of the virR targetron plasmid.
To identify potential targetron insertion sites, the nucleotide sequence of virR was submitted to the Sigma TargeTron design site (http://www.sigma-genosys.com/targetron/). Of the predicted sites, insertion into the sense strand at position 546/547 from the ATG start codon in virR was selected for targetron modification. To retarget the group II intron, primer-mediated mutation by PCR was carried out with the IBS, EBS2, EBS1/δ, and EBS universal primers (Table 2) in accordance with the instructions from the TargeTron gene knockout system (Sigma-Aldrich), with the exception that Phusion High-Fidelity DNA polymerase (NEB) and FailSafe buffer E (Epicentre Biotechnologies) were used in place of the JumpStart REDTaq Ready mix. The 350-bp gel-extracted retargeting PCR product was digested with HindIII and BsrGI and ligated into pJIR3566 DNA that had been digested with the same enzymes and alkaline phosphatase treated. The ligation mixture was used to transform E. coli TOP10 cells (Invitrogen), and plasmids from a selection of the resultant transformants were isolated and sequenced. A vector, pJIR3608, that contained the desired altered nucleotides, was chosen for subsequent mutagenesis studies.
Construction of virR mutants using the Targetron mutagenesis system.
Approximately 5 μg of pJIR3608 was used to transform C. perfringens strains EHE-NE18 and 56 by electroporation. Transformed cells were inoculated into 20-ml brain heart infusion (BHI) broths and incubated overnight at 37°C. The resultant cultures were subsequently passaged twice, over a 24-h period, in 20 ml of BHI broth. Transformants containing potential intron insertions were selected on NAEm7. Following overnight incubation at 37°C, selected colonies were patched onto NACm30 and NAEm7. Transformants that were resistant to erythromycin but sensitive to chloramphenicol were chosen for further analysis, since they represented derivatives that contained an inserted intron but were cured of pJIR3608. These transformants were also screened on HBA to assess the effect of the mutation on perfringolysin O production.
To show that the intron had inserted into the virR gene, genomic DNA was isolated as described previously (28) and analyzed by PCR with the primer pairs JRP2813/JRP182 or JRP4240/JRP4241. The former primer pair flanks the site of insertion within the virR gene, while the latter primer pair amplifies the ermB-RAM within the intron. To confirm the PCR results, Southern hybridization analysis was carried out. Briefly, genomic DNA of each strain was digested with BglII, separated by electrophoresis, transferred to a nylon membrane (GE Healthcare), and probed with PCR-amplified, digoxigenin (DIG)-labeled virR, ermB, catP, or intron-specific probes (Table 2). Blots were developed as per the manufacturer's instructions.
Perfringolysin O.
C. perfringens strains were grown to late log phase before culture supernatants were collected by centrifugation as described previously (4). Perfringolysin O activity was determined by measuring the hemolysis of horse red blood cells in a doubling dilution assay, as described previously (42). The perfringolysin O titer was defined as the reciprocal of the last well that showed complete hemolysis, which was indicated by a significant decrease in absorbance at 570 nm. The PFO titers represent the means obtained from duplicate assays using supernatants derived from three independent cultures of each strain.
Gel mobility shift analysis.
The 183-bp DNA target fragments (12) were PCR amplified with oligonucleotides JRP589 and JRP618 using the pfoA reporter constructs as templates. These DNA fragments subsequently were labeled with digoxigenin-11-ddUTP (Roche) as per the manufacturer's instructions. Gel mobility shift assays were performed with purified VirR protein as described previously (10, 12).
Isolation of RNA and qRT-PCR.
EHE-NE18 cells were harvested at late logarithmic growth phase. Total RNA was isolated using the TriZol reagent (Invitrogen) as described previously (11). Conversion of 2 μg of RNA to cDNA and quantitative real time PCR (qRT-PCR) analysis were carried out as before (9) using rpoA or netB primers (Table 2). Total RNA or genomic DNA was isolated from at least three biological replicates from each strain and assayed in triplicate. The values obtained were normalized to that of the rpoA gene for each strain, and the results were expressed as a proportion of that for the wild type.
Western blot analysis.
Two methods were used to analyze the production of NetB toxin. To determine when NetB toxin was produced, culture supernatants were harvested throughout the growth of EHE-NE18 in TPG broth. Each sample was separated by SDS-PAGE on a NuPAGE Novex 4 to 12% Bis-Tris gel (Invitrogen) in NuPAGE morpholineethanesulfonic acid (MES) SDS running buffer (Invitrogen). Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (PALL) and probed with rabbit polyclonal rNetB antiserum (19). Blots were developed with the ECL Western blotting kit (Amersham Biosciences) as per the manufacturer's instructions.
To examine the production of NetB by EHE-NE18 and 56 virR mutants, culture supernatants were harvested at late logarithmic growth phase. The total protein concentration of the supernatants was determined using the bicinchoninic acid (BCA) protein assay kit. Approximately 200 μg of protein from each sample was separated by 12% SDS-PAGE. Proteins were then transferred and immobilized onto a nitrocellulose membrane (Millipore). The membrane was probed with polyclonal rabbit NetB antiserum (1:200) (19), followed by diluted (1:2,000) goat anti-rabbit antiserum secondary antibody (Millipore). Bound antibodies were detected using the Western Lightning Plus chemiluminescence reagent (Perkin Elmer) as per the manufacturer's instructions. Chemiluminescence was detected using X-ray film (Fujifilm).
Cell line and cytotoxicity assay.
The chicken hepatoma cell line LMH (ATCC CRL-2117) was maintained in Earl's minimum essential medium (EMEM) supplemented with l-glutamine, 10% fetal calf serum (FCS), 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), penicillin (100 U/ml), streptomycin (100 μg/ml), and Fungizone (100 μg/ml) (LMH cells also require 0.2% gelatin for adherence to surfaces) and incubated in a humidified environment of 5% CO2 at 37°C.
To test for cytotoxicity, LMH cells were cultured to 70% confluence in 96-well plates (Nunc) grown in EMEM growth medium, with 5% FCS and without phenol red, at 37°C. C. perfringens strains were grown in TPG broth to a turbidity at 600 nm of 0.6. Culture supernatants were obtained by centrifugation at 18,000 × g for 10 min and then dialyzed in phosphate-buffered saline. Volume-normalized, dialyzed culture supernatants were added to the LMH cell medium and incubated for 4 h at 37°C. Lactate dehydrogenase (LDH) release into the supernatant was measured as an indicator of cytolysis using the Cyto-Tox (Promega) kit and expressed as a percentage of cytotoxicity.
RESULTS
VirR activates pfoA reporter gene expression by binding to netB VirR boxes.
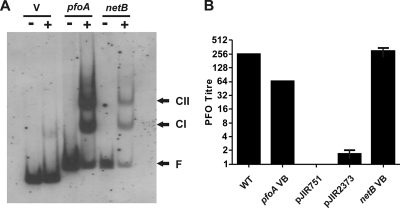
Sequence analysis of the netB promoter region of EHE-NE18 revealed the presence of potential VirR boxes upstream of the putative −35 box (Fig. 1 A). The sequence and spatial organization of these potential VirR binding sites were similar to those of the VirR boxes located upstream of the pfoA (10), vrr, virU, virT, and ccp genes (31), all of which have been shown to be recognized by VirR (10) (Fig. 1B). Gel mobility shift experiments using the purified VirR protein and DIG-labeled target DNA demonstrated that VirR was able to recognize and bind to the netB VirR boxes in vitro. The two shifted bands, complex I (CI) and complex II (CII), were similar to those observed with the control pfoA VirR box region (Fig. 2 A).
FIG. 1.
Identification of VirR boxes. (A) Nucleotide sequence of the region upstream of the netB gene in strain EHE-NE18. The start of the netB coding sequence is indicated by the bent arrow, with the sequence in bold. The putative ribosome binding site is indicated by the asterisks, while the potential −10 and −35 boxes of the VirR-dependent promoter are labeled as −10 and −35, respectively and are underlined and shown in bold. The VirR boxes are in bold and boxed and are labeled VB1 and VB2. (B) Comparison of the sequence and spatial organization of the VirR boxes of the pfoA, vrr, virU, virT, ccp, and netB genes. The VirR boxes are in bold, boxed, and labeled VB1 and VB2. The virRS-dependent promoters are in bold, underlined, and denoted by −10 and −35. The promoters of vrr, virU, virT, and ccp were determined previously (31).
FIG. 2.
Functional analysis of VirR boxes. (A) Gel mobility shift analysis of the VirR boxes upstream of netB. The DIG-labeled DNA targets containing the VirR boxes upstream of pfoA or netB were incubated in the absence (−) or presence (+) of 1 μg of purified VirR. The pJIR2373 vector control is denoted by V. The free DNA (F), CI, and CII bands are labeled and indicated by the arrows. (B) Perfringolysin O activity of reporter constructs. Perfringolysin O (PFO) titers of the JIR325 wild-type strain (WT), JIR4228 derivatives carrying reporter constructs containing the VirR boxes located upstream of the pfoA gene (pfoA VB) and the netB gene (netB VB), and JIR4228 derivatives with the shuttle vector (pJIR751) and the reporter gene vector (pJIR2373) are shown. The error bars represent the standard error of the mean; n = 3.
To determine if the netB VirR boxes were functional, pfoA reporter plasmids were constructed as described previously (10) and used to transform the JIR325-derived pfoA mutant, JIR4228 (5). Perfringolysin O activity was used to measure the expression of the pfoA reporter gene. As expected, the results showed that the positive-control strain, which carried the pfoA VirR boxes cloned upstream of the reporter gene, produced levels of perfringolysin O activity that were similar to those of the wild type, while no activity was detected from the negative-control strain harboring the shuttle vector, pJIR751 (Fig. 2B). Background levels of perfringolysin O activity were observed from the strain carrying the pJIR2373 pfoA reporter gene vector, as previously observed (10). In contrast, a wild-type level of perfringolysin O activity was observed when the netB VirR boxes were cloned upstream of the reporter gene (Fig. 2B).
NetB is produced at high cell density.
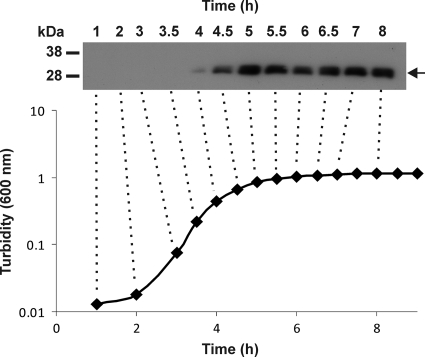
To determine when NetB is produced, Western blot analysis using polyclonal NetB antisera was carried out on culture supernatants that were isolated at various time points during the growth of strain EHE-NE18. The results showed that under these experimental conditions, NetB production begins 4 h after inoculation (logarithmic growth phase), with increasing production at later time points (Fig. 3).
FIG. 3.
Analysis of NetB toxin production in strain EHE-NE18. The growth of EHE-NE18 in TPG broth was monitored at 30-min intervals by measuring the turbidity at 600 nm. Culture supernatants were harvested at regular time points and analyzed by Western blotting using a polyclonal NetB antibody. The NetB protein is designated by the arrow, and the marker sizes are shown in kDa. The dotted lines link the selected culture supernatants to the corresponding time points.
Construction of virR mutants of EHE-NE18 and 56.
Previous work showed that VirSR-regulated genes that have correctly spaced and positioned VirR boxes in the upstream promoter region are directly regulated by VirR (10, 12, 26, 31). To determine whether VirR regulated netB expression, the virR genes of two chicken necrotic enteritis-causing isolates, EHE-NE18 and 56, were insertionally inactivated by use of targetron technology (47). To aid in the construction of these mutants, we used a clostridial targetron vector, pJIR3566, which replicates in the clostridia, utilizes a strong clostridial promoter to drive targetron expression, and contains a retrotransposition-activated marker (47) that encodes erythromycin resistance to facilitate one-step selection of chromosomal integrants. Plasmid pJIR3566 also facilitates blue/white selection of retargeted introns due to the inclusion of the lacZα gene fragment between the HindIII and BsrGI restriction sites, which are utilized during the intron “retargeting” procedure.
Of the predicted targetron sites, the insertion site within the region encoding the VirR DNA binding domain (FxRxHrS motif) was selected, since we previously demonstrated that mutation of this motif eliminated VirR function (23). The vector containing the virR targetron, pJIR3608, was introduced into EHE-NE18 and 56 by electroporation. As a result of the presence of the ermB-RAM, transformants containing potential insertions were selected on NAEm7. Verification of targetron insertion into the desired site in the resultant mutants was obtained by PCR and Southern blotting (data not shown).
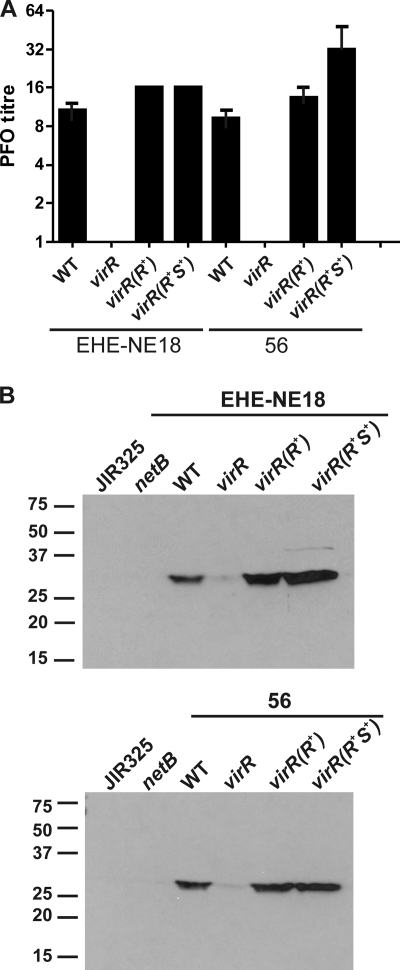
Mutation of the virR gene in the gas gangrene isolate strain 13 led to elimination of perfringolysin O production (38). Similar results were obtained with the EHE-NE18- and 56-derived virR mutants. When grown on HBA, the mutants showed no hemolytic activity (data not shown). This phenotype was confirmed by quantitative perfringolysin O assays. Compared to the wild-type strains, the virR mutants of EHE-NE18 and 56, JIR12405 and JIR12409, respectively, did not produce any detectable levels of perfringolysin O. Production of this toxin was restored to wild-type levels when the virR mutations were successfully complemented in trans by virR alone or by the wild-type virRS operon (Fig. 4 A). The assay results confirmed that the targetron insertion had successfully inactivated VirR function.
FIG. 4.
Analysis of toxin production in isogenic derivatives. (A) Analysis of perfringolysin O production. The perfringolysin O (PFO) titers of the wild-type strains EHE-NE18 and 56 (WT), virR mutant derivatives carrying the pJIR750 vector (virR), and virR mutants complemented by virR [virR(R+)] or the virR/S operon [virR(R+S+)] in trans on pJIR1897 and pJIR2529, respectively, are shown. The error bars represent the standard errors of the means (n = 3). (B) Western blot analysis of NetB production by virR mutants. Culture supernatants were isolated from JIR325, the EHE-NE18 netB mutant (netB), the wild-type strains (WT), virR mutants (virR), and virR mutants complemented with virR [virR(R+)] or the virRS operon [virR(R+S+)]. Proteins from cell culture supernatants (200 μg) were separated by SDS-PAGE and subsequently transferred to nitrocellulose. The NetB protein was visualized by use of polyclonal NetB antisera followed by chemiluminescence detection. The wild-type strains from which the virR mutants were derived are indicated above each blot. The protein size markers (kDa) are indicated on the side of each blot.
VirR regulates NetB production in necrotic enteritis strains EHE-NE18 and 56.
Culture supernatants were assayed to determine the effect of the virR mutations on NetB production. Western blot analysis using polyclonal NetB antisera demonstrated that NetB was being produced by both EHE-NE18 and 56. In contrast, no NetB was produced by the previously isolated EHE-NE18 netB mutant (Table 1) and the strain 13 derivative, JIR325. Very low, almost undetectable levels of NetB were observed with the virR mutants of both strains. The production of the NetB protein was restored when the mutation was complemented in trans with the wild-type virR genes or with the virRS operon (Fig. 4B). These results demonstrated that the production of NetB in these strains was positively regulated by the VirSR system.
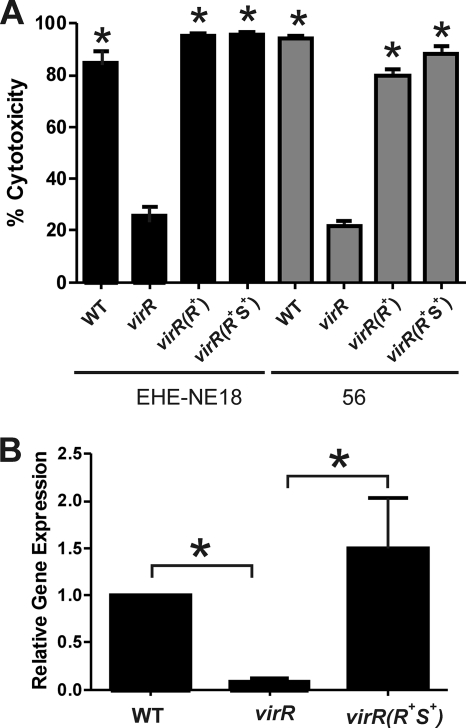
To quantitate the level of NetB toxin activity, supernatants were analyzed in an LMH cell cytotoxicity assay, using lactate dehydrogenase (LDH) release from the cells as an indicator of cytolysis (19). The level of cytotoxicity observed with culture supernatants derived from the virR mutants was significantly reduced compared to the wild-type controls. In contrast, effectively wild-type levels of cytotoxicity were observed when supernatants from the complemented strains were tested (Fig. 5 A). These results are consistent with those obtained in the Western blots and confirmed that NetB production was VirSR dependent.
FIG. 5.
Effect of virR mutation on NetB cytotoxicity and netB expression. (A) LMH cell cytotoxicity assay. Culture supernatants were isolated from JIR325, the wild-type strains (WT), virR mutants (virR), and virR mutants complemented with virR [virR(R+)] or the virRS operon [virR(R+S+)]. The amount of cytotoxicity induced by NetB in each cell supernatant is expressed as a percentage and is indicated by the black bars (strain EHE-NE18) or gray bars (strain 56). The error bars represent the standard deviations, and the asterisks (*) denote a P value of <0.05 compared to results for the respective virR mutants, as calculated by nonparametric, two-tailed, Mann-Whitney t tests. (B) Quantitative real-time PCR analysis of netB transcription. Total RNA from EHE-NE18 (WT), the EHE-NE18 virR mutant (virR), and its complemented derivative [virR(R+S+)] was isolated from four biological replicates and assayed in triplicate. The expression levels of netB (black bars) in each strain relative to WT levels normalized to the rpoA gene (relative gene expression) are shown. The error bars represent the standard errors of the means, and the asterisks (*) represent a P value of <0.05 calculated by two-tailed Student's t test.
To determine if regulation was mediated at the transcriptional level, the amount of netB transcript in EHE-NE18, its virR mutant, and the virR mutant complemented with the virRS operon was quantitated by use of qRT-PCR. The results showed that netB transcription in the virR mutant was significantly reduced compared to that in the wild type but that expression was restored to wild-type levels upon complementation (Fig. 5B).
DISCUSSION
In this study, we identified potential VirR boxes upstream of the netB gene in the chicken necrotic enteritis-causing strain, EHE-NE18. Comparison of the VirR boxes found upstream of the pfoA gene with those associated with the netB gene revealed some sequence variation, but despite these differences, VirR was still able to recognize and bind to the netB VirR boxes in vitro and stimulate the expression of the pfoA reporter gene in vivo. Taken together, these results indicated that the VirR boxes upstream of netB were functional and suggested that netB transcription was regulated by the VirSR two-component signal transduction system. This hypothesis was confirmed by the construction of virR mutant derivatives of two necrotic enteritis strains, EHE-NE18 and 56, and their complementation by either virR or virRS in trans. Reduced NetB production was observed in the virR mutants and restored in the complemented derivatives. Further analysis showed that the low level of NetB produced in the virR mutant of EHE-NE18 was consistent with the significantly reduced level of netB transcript in comparison to those of the wild-type and complemented strains. These experiments provide clear evidence that NetB toxin production is regulated at the transcriptional level by the VirSR system.
These findings have significant implications for our understanding of the pathogenesis of avian necrotic enteritis infections. The VirSR system may function in a manner similar to that of the staphylococcal Agr quorum sensing system (27). Recent studies have indicated that in C. perfringens strain 13, the VirSR system is important in sensing an extracellular signal that leads to the transcriptional activation of toxin genes (30). In the staphylococcal Agr system, the extracellular signal is an autoinducing peptide that is a derivative of an AgrD propeptide that is modified by AgrB upon secretion (32). The Agr system represents a quorum sensing system that is responsive to cell density. We have identified similar agrB and agrD genes in the strain EHE-NE18 genome (data not shown), and other workers have proposed that in strain 13, the product of these genes is the autoinducing peptide that activates the VirSR system (30). Furthermore, in this study we have shown that NetB is not produced constitutively; it is produced only when the cells reach late logarithmic phase. Therefore, we postulate that VirSR-dependent NetB toxin production in C. perfringens is responsive to cell density.
Based on the data obtained in studies on strain 13 (30, 46), the results presented here, and our understanding of the pathogenesis of avian necrotic enteritis (45), we suggest that in the normal avian gastrointestinal tract, the low levels of C. perfringens cells are not sufficient to lead to induction of the VirSR system and significant NetB production. However, when the birds are suddenly changed to a high-energy, protein-rich diet, the growth of C. perfringens is stimulated. Therefore, its population density increases and the amount of autoinducing peptide secreted into the lumen of the gastrointestinal tract increases to the point where its concentration is high enough to lead to the induction of NetB toxin production by the induction of VirS, which subsequently activates the VirR response regulator. This mechanism would represent a highly efficient environmental adaption, since cytolytic toxins like NetB would be produced only when the C. perfringens population density was high and there was an increased probability of subsequent nutrient limitation. We propose that as with other C. perfringens toxins, the primary function of NetB is to increase nutrient availability by releasing complex macromolecules from cells of the host, not to cause any disease pathology.
To date, most studies on the VirSR system have been carried out on human gas gangrene or food poisoning strains of C. perfringens. Recently other workers showed that exposure of a C. perfringens type C strain to CaCo2 cells leads to VirSR-dependent upregulation of β-toxin and perfringolysin O production (46), and we have now shown that NetB toxin production in avian isolates of C. perfringens is VirSR regulated. Taken together, these studies demonstrate that the regulation of toxin production by the VirSR system is not limited to human isolates but also extends to strains that cause disease in animals. Despite very significant differences in the diseases that these strains cause and the hosts that are affected, the method by which toxin production is regulated is essentially the same. These findings once more illustrate the global nature of this quorum-sensing VirSR two-component regulatory system and can again be interpreted on the basis that the major reason that C. perfringens strains produce specific hydrolytic and cytolytic toxins is to provide host-derived nutrients under conditions of high cell density.
Acknowledgments
This research was supported by the Australian Research Council Centre of Excellence in Structural and Functional Microbial Genomics with funding from the Australian Research Council and by grants from the Australian National Health and Medical Research Council and the Australian Poultry Cooperative Research Centre.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 10 May 2010.
REFERENCES
- 1.Al-Sheikhly, F., and R. B. Truscott. 1977. The interaction of Clostridium perfringens and its toxins in the production of necrotic enteritis of chickens. Avian Dis. 21:256-263. [PubMed] [Google Scholar]
- 2.Al-Sheikhly, F., and R. B. Truscott. 1977. The pathology of necrotic enteritis of chickens following infusion of broth cultures of Clostridium perfringens into the duodenum. Avian Dis. 21:230-240. [PubMed] [Google Scholar]
- 3.Al-Sheikhly, F., and R. B. Truscott. 1977. The pathology of necrotic enteritis of chickens following infusion of crude toxins of Clostridium perfringens into the duodenum. Avian Dis. 21:241-245. [PubMed] [Google Scholar]
- 4.Awad, M. M., A. E. Bryant, D. L. Stevens, and J. I. Rood. 1995. Virulence studies on chromosomal α-toxin and θ-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of α-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15:191-202. [DOI] [PubMed] [Google Scholar]
- 5.Awad, M. M., and J. I. Rood. 1997. Isolation of α-toxin, θ-toxin and κ-toxin mutants of Clostridium perfringens by Tn916 mutagenesis. Microb. Pathog. 22:275-284. [DOI] [PubMed] [Google Scholar]
- 6.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:223-235. [DOI] [PubMed] [Google Scholar]
- 7.Banu, S., K. Ohtani, H. Yaguchi, T. Swe, S. T. Cole, H. Hayashi, and T. Shimizu. 2000. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol. Microbiol. 35:854-864. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y., B. A. McClane, D. J. Fisher, J. I. Rood, and P. Gupta. 2005. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl. Environ. Microbiol. 71:7542-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, J. K., M. M. Awad, S. McGowan, and J. I. Rood. 2009. Functional analysis of the VirSR phosphorelay from Clostridium perfringens. PLoS One 4:e5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, J. K., B. Dupuy, D. S. Deveson, and J. I. Rood. 2004. The spatial organization of the VirR boxes is critical for VirR-mediated expression of the perfringolysin O gene, pfoA, from Clostridium perfringens. J. Bacteriol. 186:3321-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, J. K., and J. I. Rood. 2000. Glutamate residues in the putative transmembrane region are required for the function of the VirS sensor histidine kinase from Clostridium perfringens. Microbiology 146:517-525. [DOI] [PubMed] [Google Scholar]
- 12.Cheung, J. K., and J. I. Rood. 2000. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 182:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu, F. K., G. F. Maley, D. K. West, M. Belfort, and F. Maley. 1986. Characterization of the intron in the phage T4 thymidylate synthase gene and evidence for its self-excision from the primary transcript. Cell 45:157-166. [DOI] [PubMed] [Google Scholar]
- 14.Gholamiandehkordi, A. R., L. Timbermont, A. Lanckriet, W. Van Den Broeck, K. Pedersen, J. Dewulf, F. Pasmans, F. Haesebrouck, R. Ducatelle, and F. Van Immerseel. 2007. Quantification of gut lesions in a subclinical necrotic enteritis model. Avian Pathol. 36:375-382. [DOI] [PubMed] [Google Scholar]
- 15.Heap, J. T., O. J. Pennington, S. T. Cartman, G. P. Carter, and N. P. Minton. 2007. A universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452-464. [DOI] [PubMed] [Google Scholar]
- 16.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 18.Kawsar, H. I., K. Ohtani, K. Okumura, H. Hayashi, and T. Shimizu. 2004. Organization and transcriptional regulation of myo-inositol operon in Clostridium perfringens. FEMS Microbiol. Lett. 235:289-295. [DOI] [PubMed] [Google Scholar]
- 19.Keyburn, A. L., J. D. Boyce, P. Vaz, T. L. Bannam, M. E. Ford, D. Parker, A. Di Rubbo, J. I. Rood, and R. J. Moore. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keyburn, A. L., S. A. Sheedy, M. E. Ford, M. M. Williamson, M. M. Awad, J. I. Rood, and R. J. Moore. 2006. Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect. Immun. 74:6496-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyras, D., and J. I. Rood. 1998. Conjugative transfer of RP4-oriT shuttle vectors from Escherichia coli to Clostridium perfringens. Plasmid 39:160-164. [DOI] [PubMed] [Google Scholar]
- 22.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 23.McGowan, S., I. S. Lucet, J. K. Cheung, M. M. Awad, J. C. Whisstock, and J. I. Rood. 2002. The FxRxHrS motif: a conserved region essential for DNA binding of the VirR response regulator from Clostridium perfringens. J. Mol. Biol. 322:997-1011. [DOI] [PubMed] [Google Scholar]
- 24.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 25.Morelle, G. 1989. A plasmid extraction procedure on a miniprep scale. Focus 11:7-8. [Google Scholar]
- 26.Myers, G. S. A., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick, R. P., and E. Geisinger. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541-564. [DOI] [PubMed] [Google Scholar]
- 28.O'Connor, J. R., D. Lyras, K. A. Farrow, V. Adams, D. R. Powell, J. Hinds, J. K. Cheung, and J. I. Rood. 2006. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 61:1335-1351. [DOI] [PubMed] [Google Scholar]
- 29.Ohtani, K., H. Takamura, H. Yaguchi, H. Hayashi, and T. Shimizu. 2000. Genetic analysis of the ycgJ-metB-cysK-ygaG operon negatively regulated by the VirR/VirS system in Clostridium perfringens. Microbiol. Immunol. 44:525-528. [DOI] [PubMed] [Google Scholar]
- 30.Ohtani, K., Y. Yuan, S. Hassan, R. Wang, Y. Wang, and T. Shimizu. 2009. Virulence gene regulation by the agr system in Clostridium perfringens. J. Bacteriol. 191:3919-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okumura, K., K. Ohtani, H. Hayashi, and T. Shimizu. 2008. Characterization of genes regulated directly by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 190:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu, R., W. Pei, L. Zhang, J. Lin, and G. Ji. 2005. Identification of the putative staphylococcal AgrB catalytic residues involving the proteolytic cleavage of AgrD to generate autoinducing peptide. J. Biol. Chem. 280:16695-16704. [DOI] [PubMed] [Google Scholar]
- 33.Rood, J. I. 2007. Clostridium perfringens and histotoxic disease, p. 753-770. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: a handbook on the biology of bacteria, 3rd ed., vol. 4. Bacteria: firmicutes, cyanobacteria. Springer, New York, NY. [Google Scholar]
- 34.Rood, J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333-360. [DOI] [PubMed] [Google Scholar]
- 35.Rood, J. I., E. A. Maher, E. B. Somer, E. Campos, and C. L. Duncan. 1978. Isolation and characterization of multiple antibiotic-resistant Clostridium perfringens strains from porcine feces. Antimicrob. Agents Chemother. 13:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd edition ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Scott, P. T., and J. I. Rood. 1989. Electroporation-mediated transformation of lysostaphin-treated Clostridium perfringens Gene 82:327-333. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu, T., W. Ba-Thein, M. Tamaki, and H. Hayashi. 1994. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J. Bacteriol. 176:1616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu, T., H. Yaguchi, K. Ohtani, S. Banu, and H. Hayashi. 2002. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 43:257-265. [DOI] [PubMed] [Google Scholar]
- 40.Songer, J. G. 2005. Clostridial diseases in domestic animals, p. 527-542. In P. Dürre (ed.), Handbook on clostridia. CRC Press, Boca Raton, FL.
- 41.Songer, J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens, D. L., J. Mitten, and C. Henry. 1987. Effects of α and θ toxins from Clostridium perfringens on human polymorphonuclear leukocytes. J. Infect. Dis. 156:324-333. [DOI] [PubMed] [Google Scholar]
- 43.Van der Sluis, W. 2000. Clostridial enteritis is an often underestimated problem. World Poultry J. 16:42-43. [Google Scholar]
- 44.Van Immerseel, F., J. De Buck, F. Pasmans, G. Huyghebaert, F. Haesebrouck, and R. Ducatelle. 2004. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 33:537-549. [DOI] [PubMed] [Google Scholar]
- 45.Van Immerseel, F., J. I. Rood, R. J. Moore, and R. W. Titball. 2009. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. 17:32-36. [DOI] [PubMed] [Google Scholar]
- 46.Vidal, J. E., K. Ohtani, T. Shimizu, and B. A. McClane. 2009. Contact with enterocyte-like Caco-2 cells induces rapid upregulation of toxin production by Clostridium perfringens type C isolates. Cell Microbiol. 11:1306-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong, J., M. Karberg, and A. M. Lambowitz. 2003. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res. 31:1656-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]