Abstract
We recently discovered a critical role for type I interferon (IFN) in the development of murine Lyme arthritis. Borrelia burgdorferi-mediated induction of IFN-responsive genes by bone marrow-derived macrophages (BMDMs) was dependent upon a functional type I IFN receptor but independent of Toll-like receptor 2 (TLR2), TLR4, TLR9, and the adapter molecule MyD88. We now demonstrate that induction of the IFN transcriptional profile in B. burgdorferi-stimulated BMDMs occurs independently of the adapter TRIF and of the cytoplasmic sensor NOD2. In contrast, B. burgdorferi-induced transcription of these genes was dependent upon a rapid STAT1 feedback amplification pathway. IFN profile gene transcription was IRF3 dependent but did not utilize B. burgdorferi-derived DNA or DNase-sensitive ligands. Instead, IFN-responsive gene expression could be induced by B. burgdorferi-derived RNA. Interferon regulatory factor 3 (IRF3)-dependent IFN profile gene transcription was also induced by sonicated bacteria, by the lipoprotein OspA, and by factors released into the BSKII medium during culture of B. burgdorferi. The IFN-stimulatory activity of B. burgdorferi culture supernatants was not destroyed by nuclease treatment. Nuclease digestion also had no effect on IFN profile induction mediated by sonicated B. burgdorferi. Thus, B. burgdorferi-derived RNA, OspA, and non-nucleic acid ligands present in both sonicated bacteria and B. burgdorferi culture medium contribute to type I IFN-responsive gene induction. These findings suggest that B. burgdorferi invasion of joint tissue and the resultant type I IFN induction associated with Lyme arthritis development may involve multiple triggering ligands.
Type I interferons (IFNs) have long been recognized as potent antiviral cytokines and have also been associated with the response to several intracellular bacterial and protozoan pathogens, including Listeria monocytogenes, Francisella tularensis, and Trypanosoma cruzi (15, 20, 23, 47). Recent reports have also linked type I IFN production with host defense to extracellular bacterial pathogens, such as group A and group B streptococci (11, 14). Type I IFN production has been implicated in the development of autoimmune diseases, including systemic lupus erythematosus (SLE) and juvenile idiopathic arthritis (JIA), and in arthritis associated with type I IFN treatment of hepatitis C and multiple sclerosis patients (13, 28, 40, 44, 52). In addition, we recently uncovered a novel role for type I IFN in the development of subacute murine arthritis caused by the Lyme disease spirochete Borrelia burgdorferi (30).
Production of type I IFN can occur as a direct downstream consequence of pattern recognition receptor (PRR) ligation of pathogen-associated molecular patterns (PAMPs). The best-characterized pathways that culminate in the transcription of type I IFNs involve Toll-like receptor (TLR) ligation of bacterium-derived lipopolysaccharide (LPS), CpG DNA motifs, or virus-derived nucleic acids. Signal transduction through most TLRs involved in type I IFN production proceeds through the adapter molecule MyD88. Exceptions include the double-stranded RNA (dsRNA)-sensing TLR3- and TLR4-mediated type I IFN transcription, both of which signal through TRIF. Other PRRs that sense RNA viruses and result in transcription of type I IFNs include the RIG-I-like receptor (RLR) family members RIG-I, MDA5, and LGP2, all of which utilize the adapter molecule MAVS (also known as VISA, CARDIF, or IPS-1) for further signal transduction events (21, 45). Muramyl dipeptide (MDP) derived from Listeria monocytogenes and N-glycolyl-MDP derived from Mycobacterium tuberculosis were recently demonstrated to induce type I IFN transcription as a consequence of signaling through the NOD-like receptor (NLR) NOD2 (16, 23, 34). Although the signal transduction pathways driving NOD2-dependent transcription of type I IFN have not yet been elucidated for L. monocytogenes-activated macrophages (16, 23), M. tuberculosis-derived N-glycolyl-MDP utilizes the adapter molecule RIP2 to trigger type I IFN production in a TBK1-, interferon regulatory factor 3 (IRF3)-, and IRF5-dependent manner (34).
Although TLRs and RLRs utilize different adapter molecules, both pathways culminate in the downstream production of type I IFNs through the activation of the IRF kinases TBK1 and/or IKKɛ1 and the subsequent phosphorylation of the transcription factors IRF3 and/or IRF7, depending on the cell type and pathogen involved. Type I IFNs can then amplify the production of more type I IFN and IFN-responsive genes through ligation of the type I IFN receptor and the resultant downstream activation of JAK-STAT-mediated signal transduction events (21, 45).
Previous reports from both our laboratory and those of colleagues have demonstrated that B. burgdorferi stimulation results in IFN-responsive gene induction in joint tissue of infected mice, in skin and blood from Lyme disease patients, and in cultured macrophages/monocytes derived from humans and mice (30, 38, 39, 49, 50). We recently reported that induction of IFN-responsive genes in Borrelia burgdorferi-stimulated murine bone marrow-derived macrophages (BMDMs) was mediated by type I IFN, as transcription of IFN-responsive genes was abolished in type I IFN receptor-deficient (IFNAR1−/−) BMDMs. Intriguingly, although B. burgdorferi is an extracellular bacterial pathogen whose primary mode of PRR signaling involves lipoprotein interaction with TLR1/TLR2 heterodimers, transcription of IFN-responsive genes was TLR2, TLR4, TLR9, and MyD88 independent (17, 30, 50).
Although MyD88-independent induction of type I IFN-responsive genes has been reported for numerous intracellular viruses and bacteria (21, 45), two recent publications have described a MyD88-independent transcription of type I IFN by extracellular bacterial pathogens. Stimulation of macrophages by both group A streptococci (GAS) and group B streptococci (GBS) triggers a MyD88-independent, IRF3-dependent type I IFN response (11, 14). It was also reported that type I IFN induction by BMDMs could be mediated by the cytosolic delivery of GBS DNA (11). Intriguingly, a recent publication described the utilization of RIG-I and MDA5 macrophage receptors by the intracellular bacterium Legionella pneumophila for the MyD88-independent induction of a type I IFN response (31). In contrast, neither the B. burgdorferi ligand(s) that triggers IFN profile induction by macrophages nor the signal transduction pathways utilized by activated macrophages to transcribe type I IFN-responsive genes have been identified. This report provides insight into the nature of the B. burgdorferi ligand(s) and the signal transduction pathways utilized by macrophages in the induction of type I IFN-responsive gene transcripts.
MATERIALS AND METHODS
Bacteria and mice.
C3H/HeN and C57BL/6 (B6) mice were purchased from Charles River Laboratories, and C3H/HeJ, B6 Nod2−/− (B6.129S1-Nod2tm1Flv/J), and B6 TRIF−/− (C57BL/6J-Ticam1Lps2/J) mice were obtained from the Jackson Laboratory. B6 MyD88−/− mice (1) were provided by S. Akira (Hyogo College of Medicine, Japan). C3H STAT1−/− (9) mice were a gift from C. Brown (University of Missouri, Columbia, MO), and IRF3−/− femurs (41) were provided by D. T. Golenbock and K. A. Fitzgerald (University of Massachusetts Medical School, Worchester, MA).
The clonal B. burgdorferi strain N40 (cN40), provided by S. Barthold (University of California, Davis, CA), was utilized in all studies (6). Spirochetes were cultured in BSKII medium (4) containing 6% rabbit serum (Sigma, St. Louis, MO) prepared under endotoxin-free conditions at 32°C.
Reagents.
LPS from Escherichia coli J5 (Rc) was purchased from List Biological Laboratories (Campbell, CA) and repurified to eliminate endotoxin protein contaminants, as described previously (18). The phosphorothioate-modified class B CpG oligonucleotide 1668 (TCCATGACGTTCCTGATGCT) was synthesized by the University of Utah Oligonucleotide/Peptide Synthesis Core facility (Salt Lake City, UT). Poly(dI:dC) was obtained from GE Healthcare Life Sciences (Piscataway, NJ), and MDP was purchased from Sigma. The construction, expression, and purification of recombinant lipidated B. burgdorferi OspA were described elsewhere (51). Recombinant interleukin-6 (IL-6) and IL-12 proteins utilized as standards in sandwich enzyme-linked immunosorbent assays (ELISAs), as well as corresponding capture and biotinylated detection antibodies, were obtained from BD Pharmingen (San Diego, CA). Horseradish peroxidase (HRP)-avidin was purchased from Vector Laboratories (Burlingame, CA). DNase I was obtained from New England BioLabs (Ipswich, MA), and RNase A was purchased from Roche Applied Science (Indianapolis, IN).
BMDM cell culture.
BMDMs were isolated from the femurs and tibias of mice (29) and cultured in RPMI 1640 (Invitrogen Life Science, Carlsbad, CA) containing 30% L929 culture supernatant and 20% horse serum (HyClone, Logan, UT) at 37°C with 5% CO2. After reaching confluence, BMDMs were harvested and replated at a density of 7.5 × 105/ml in 12-well dishes in 1 ml of serum-free RPMI 1640 containing 1% Nutridoma (Roche Applied Science). Nutridoma was utilized in lieu of medium optimized to ensure the greatest viability of B. burgdorferi upon addition to BMDMs (22), because the IFN response was optimal in serum-free media. The MycoProbe Mycoplasma detection kit (R&D Systems, Minneapolis, MN) was utilized to confirm that the L929 culture supernatants and all other cell culture media were free of contaminating Mycoplasma species.
Following an overnight incubation, the medium was removed and cells were stimulated in 12-well plates for 6 or 24 h in 1 ml of medium containing 7.4 × 106/ml live (multiplicity of infection [MOI], 10) or 5 μg/ml sonicated B. burgdorferi organisms. Prior to use in BMDM experiments, live bacteria were washed twice with phosphate-buffered saline (PBS) to remove BSKII components (30). B. burgdorferi viability was at least 85 to 90%, as assessed by Petroff-Hauser counting of motile spirochetes via dark-field microscopy. Sonicated cN40 was prepared as described elsewhere (42). In some experiments, other stimuli, such as LPS or poly(dI:dC), were added to separate wells, as indicated in the appropriate figure legend(s). Upon assay completion, cellular supernatants were removed for IL-6 and IL-12 cytokine assessment by sandwich ELISA, as described elsewhere (10).
Nuclease digestion.
Nucleic acids were prepared from log-phase cultures of B. burgdorferi using standard procedures for DNA (3) and Trizol for RNA (2). Two micrograms of B. burgdorferi genomic DNA, RNA, or live or sonicated bacteria was digested with DNase I or RNase A according to the manufacturer's specifications.
B. burgdorferi culture supernatant.
Supernatant from late-logarithmic-phase cultures of B. burgdorferi was collected via centrifugation and was then applied to 0.2-μm filters (Corning Life Sciences, Lowell, MA) to remove any remaining bacterial cellular debris. Volumes of recovered B. burgdorferi culture supernatant ranging from 50 μl to 150 μl were mixed with serum-free medium to total 1 ml and applied to BMDMs. Control wells received corresponding volumes of sterile complete BSKII medium that had been incubated at 32°C without the inclusion of B. burgdorferi.
RNA isolation and quantitative reverse transcription-PCR (RT-PCR).
RNA was isolated from cultured BMDMs at 6 or 24 h poststimulation by homogenization in Trizol reagent (Invitrogen Life Technologies), as specified by the manufacturer. Five micrograms of RNA was reverse transcribed into cDNA, and transcripts for genes of interest were amplified with the LightCycler 480 (LC480) SYBR green I master mix and an LC480 LightCycler (Roche Applied Science), as previously described (32). The copy number of the gene of interest was calculated from the starting template sample and normalized to 1,000 copies of the mouse β-actin housekeeping gene, as detailed elsewhere (32). The following primer pairs span introns (except for Ifit2) and were utilized for PCR amplification in this study: β-actin gene, 5′-GTAACAATGCCATGTTCAAT-3′ (forward) and 5′-CTCCATCGTGGGCCGCTCTAG-3′ (reverse); Cxcl9, 5′-TTGGGCATCATCTTCCTGGAGCAG-3′ (forward) and 5′-GAGGTCTTTGAGGGATTTGTAGTGG-3′ (reverse); Cxcl10, 5′-GAAATCATCCCTGCGAGCCTATCC-3′ (forward) and 5′-GCAATTAGGACTAGCCATCCACTGGG-3′ (reverse); Gbp2, 5′-CTACCGCACAGGCAAATCCTAC-3′ (forward) and 5′-GTCATTCTGGTTGTCACCCTTCTC-3′ (reverse); Ifit1, 5′-ATGGGAGAGAATGCTGATGGTG-3′ (forward) and 5′-TGTCAAGGAACTGGACCTGCTC-3′ (reverse); Ifit2, 5′-CAACGAGTAAGGAGTCACTGGAGAG-3′ (forward) and 5′-TTGCTGGATGAAGCCCTCAG-3′ (reverse); Tyki, 5′-AGGAGGTCCAGAAAGGGAAGTTC-3′ (forward) and 5′-TATGGCGTAGGTGGCTGTGCTATG-3′ (reverse); and Igtp, 5′-TAGAGCAGACCCACAGAGTTCAGG-3′ (forward) and 5′-CAGCAGTCATAGATTTAGACCACGG-3′.
B. burgdorferi RNA content was assessed using the previously published 16S rRNA gene oligonucleotide primers (33).
RNA interference (RNAi) silencing of Irf3.
The Amaxa nucleofector Y-001 program and mouse macrophage nucleofector kit (Lonza Walkersville Inc.) were utilized for the transfection of BMDMs with small interfering RNAs (siRNAs), as previously described (53). Briefly, 80 pmol of SMARTpool Irf3 siRNAs or of control, SMARTpool scrambled siRNAs (Dharmacon, Chicago, IL) was added to 2 × 106 BMDMs resuspended in 100 μl of mouse macrophage nucleofector solution. Immediately following transfection, BMDMs were placed in complete RPMI medium (2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol [2-ME]) containing 20% fetal calf serum (FCS), transferred to bacteriological petri dishes, and incubated at 37°C in a 5% CO2 incubator for 48 h. Transfected cells were then replated in 1% Nutridoma at a density of 3 × 105 cells/well in 24-well plates and incubated overnight. The appropriate stimulus was added to each well, and cellular RNA was extracted at 6 h poststimulation. In some experiments, these protocols were used for the forced cytosolic delivery of B. burgdorferi sonicate or genomic DNA to BMDMs.
Statistics.
A paired, two-tailed Student t test was utilized for two-group comparisons of parametric data exhibiting no differences in standard deviation. An unpaired, two-tailed Student t test was employed for data where pairing was ineffective, and for categorical data exhibiting differences in standard deviation, the Welch correction was applied to the t test. The Mann-Whitney U test was utilized for assessment of nonparametric data. A one-way analysis of variance (ANOVA) with the Tukey-Kramer multiple-comparison test utilized as the posttest was employed for all multigroup comparisons. The threshold utilized to assess statistical significance for all tests was a P value of <0.05. GraphPad InStat3 version 3.0b for MacIntosh (GraphPad Software, San Diego, CA) was used for all statistical analyses.
RESULTS
Induction of IFN-responsive transcripts by BMDMs in response to B. burgdorferi does not require the adapter TRIF or the peptidoglycan sensor NOD2.
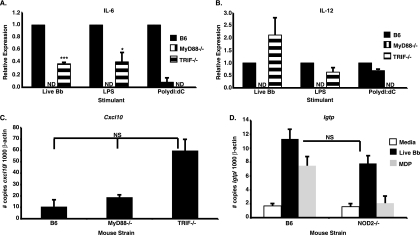
We previously found that many components of the IFN-inducible gene profile found in joints of infected C3H mice were also induced following B. burgdorferi stimulation of BMDMs from wild-type and mutant mice. Importantly, BMDMs from both C3H and B6 mice display this response in vitro, although the magnitude of the response was greater in cells from C3H mice than in those from B6 mice (26, 30). The finding that IFN-responsive gene induction by BMDMs was MyD88 independent (30) suggested the possibility that the alternative TLR adapter, TRIF, could be involved in IFN-responsive gene induction by B. burgdorferi. Therefore, BMDMs obtained from wild-type B6 mice or from B6 mice with gene disruptions in MyD88 or TRIF were stimulated with live B. burgdorferi for 6 h. In agreement with previously published reports (7, 8, 12, 25), the cytokines IL-6 and IL-12 were detected in the supernatants of B6 BMDMs stimulated with B. burgdorferi but not in those obtained from MyD88−/− BMDMs. As anticipated, B6 BMDMs produced IL-6 and IL-12 in response to LPS and poly(dI:dC), whereas MyD88−/− BMDMs failed to secrete these cytokines following LPS administration. TRIF−/− BMDMs were unable to produce cytokines in response to poly(dI:dC) stimulation but responded to LPS, albeit at lower levels than the wild type (Fig. 1 A and B). Intriguingly, TRIF−/− BMDMs produced significantly less IL-6 than wild-type B6 BMDMs in response to B. burgdorferi stimulation (Fig. 1A). Although we are not aware of any previously published reports on IL-6 secretion by B. burgdorferi-infected TRIF−/− BMDMs, reduced BMDM IL-6 production in response to both E. coli and Pseudomonas aeruginosa has been previously documented (19, 37). IL-12 levels were also reduced following B. burgdorferi stimulation of TRIF−/− BMDMs, but this decrease did not achieve statistical significance (Fig. 1B). Both MyD88−/− and TRIF−/− BMDMs efficiently transcribed IFN-responsive genes following B. burgdorferi stimulation, as depicted in Fig. 1C for the representative IFN-inducible transcript Cxcl10. The “hyperinduction” of transcripts in TRIF−/− mice, although not statistically significant, was a consistent finding and possibly reflects streamlined activation of existing signaling pathways in the absence of competing pathways (unpublished observations with other mutant BMDMs). These data indicate that transcription of IFN-responsive genes by B. burgdorferi-stimulated BMDMs is not TRIF dependent and suggests that the extracellular spirochete may be utilizing a non-TLR receptor and signaling pathway previously associated with intracellular bacteria.
FIG. 1.
B. burgdorferi induction of IFN-responsive genes is independent of TRIF and NOD2. (A and B) Wild-type B6, MyD88−/−, or TRIF−/− BMDMs were stimulated with 7.4 × 106/ml live B. burgdorferi organisms (Bb), 100 ng/ml LPS, or 20 μg/ml poly(dI:dC) for 6 h. ND, not detected. The production of IL-6 (A) or IL-12 (B) was assessed by ELISA. Data indicate the means ± standard errors of the means (SEM) and are pooled from two independent experiments (n = 4). Statistical significance was assessed via a paired, two-tailed Student t test. ***, P < 0.001 for IL-6 levels obtained for TRIF−/− BMDMs stimulated with live B. burgdorferi versus comparably treated B6 BMDMs; *, P < 0.05 for LPS-treated TRIF−/− BMDMs versus similarly stimulated B6 BMDMs. In panel B, IL-12 levels for live B. burgdorferi (P = 0.1996)- or LPS (P = 0.1010)-stimulated TRIF−/− BMDMs were not significantly different from those for comparably treated B6 BMDMs. (C) Cellular RNA from B6, MyD88−/−, or TRIF−/− BMDMs was analyzed by quantitative RT-PCR for IFN-responsive gene transcription at 6 h after B. burgdorferi stimulation. Data are depicted as the means ± SEM and are pooled from two independent experiments (n = 4). Statistical significance was assessed via an unpaired, two-tailed Student t test with the Welch correction. Transcript levels for Cxcl10 are depicted as the fold change induction obtained for each normalized transcript following B. burgdorferi stimulation (7.4 × 106/ml) relative to values obtained for unstimulated cells. Although B. burgdorferi-induced IFN-responsive gene expression was elevated in TRIF−/− BMDMs, this increase was not statistically significant (NS) compared with values obtained for B6 (P = 0.0519) and MyD88−/− (P = 0.0777) BMDMs. (D) Cellular RNA from B6 or NOD2−/− BMDMs was analyzed by quantitative RT-PCR for IFN-responsive gene transcription at 24 h after B. burgdorferi stimulation. Data are depicted as the means ± SEM and are pooled from three independent experiments (n = 6). Statistical significance was assessed via the Mann-Whitney U test. Igtp transcript levels obtained from B. burgdorferi (7.4 × 106/ml)- or MDP (10 ng/ml)-stimulated B6 or NOD2−/− BMDMs are displayed as the number of copies normalized to β-actin. The Igtp expression levels obtained for B6 and NOD2−/− BMDMs following B. burgdorferi stimulation were not significantly different (NS) (P = 0.1320).
It was recently reported that Listeria monocytogenes- and Mycobacterium tuberculosis-mediated signaling through the NLR NOD2, which is known to be involved in inflammasome formation and signaling, results in increased TLR-independent induction of IFN-β (16, 23, 34). Since B. burgdorferi seemingly utilizes a TLR-independent receptor on BMDMs for the induction of IFN-responsive transcripts, we next asked whether NOD2 mediates transcription of these genes. Wild-type B6 and NOD2−/− BMDMs were stimulated for 24 h with live B. burgdorferi or with the NOD2 ligand MDP, cellular RNA was extracted, and IFN-responsive gene transcripts were quantified. MDP and B. burgdorferi stimulation of B6 BMDMs resulted in transcription of IFN-responsive genes. MDP stimulation of NOD2−/− BMDMs failed to induce transcription of IFN profile genes, as shown for the representative transcript Igtp (Fig. 1D). Induction of IFN profile genes was also observed in B. burgdorferi-treated NOD2−/− BMDMs (Fig. 1D), indicating that NOD2 is not required for B. burgdorferi-induced transcription of IFN-responsive genes.
STAT1 is required for B. burgdorferi-mediated induction of IFN-responsive gene transcription by murine BMDMs.
Feedback amplification of type I IFNs and transcription of IFN-responsive genes are initiated via signaling through the type I IFN receptor and are dependent upon subsequent activation of JAK-STAT pathway members. STAT1 is the major transcriptional activator recruited for this pathway in macrophages (45). We previously demonstrated that IFN-responsive gene induction in B. burgdorferi-stimulated BMDMs was entirely dependent on a functional type I IFN receptor (30), and therefore we sought to assess whether STAT1 was critical for feedback amplification. IFN-responsive gene transcripts, as illustrated in Table 1 for the representative genes Cxcl9, Cxcl10, Gbp2, and Ifit1, were readily induced in wild-type C3H BMDMs stimulated with B. burgdorferi. In stark contrast, IFN-responsive gene expression was greatly reduced in BMDMs from C3H mice harboring the STAT1 gene ablation, as assayed at 24 h poststimulation (Table 1). Transcription of IFN profile genes was also observed in B. burgdorferi-activated TLR4-defective C3H/HeJ BMDMs (36, 48), indicating that IFN-responsive transcript induction was not due to potential endotoxin contamination of cultured bacteria (data not shown). Although low-level IFN-responsive gene transcripts were detectable by 1 h in wild-type BMDMs, transcription of all genes examined was greatly reduced in STAT1−/− BMDMs, even at 1 h poststimulation (data not shown). Collectively, these data indicate that B. burgdorferi-mediated induction of type I IFN-responsive genes, including those reported to be involved in the earliest transcriptional response to Listeria monocytogenes, such as Ifit1 (23), is entirely dependent upon an early feedback amplification loop that proceeds through STAT1.
TABLE 1.
IFN-responsive gene induction by B. burgdorferi-stimulated BMDMs is STAT1 dependent
| Transcript | Levela (mean ± SEM) with BMDM genotype and stimulant: |
|||
|---|---|---|---|---|
| C3H/HeNb |
C3H STAT1−/−c |
|||
| Medium | Live B. burgdorferi | Medium | Live B. burgdorferi | |
| Cxcl9 | 0.2 ± 0.1 | 0.6 ± 0.1** | 0.0 ± 0.0 | 0.0 ± 0.0** |
| Cxcl10 | 2.6 ± 1.5 | 80.9 ± 10.8** | 0.3 ± 0.2 | 0.3 ± 0.1** |
| Gpb2 | 15.0 ± 4.6 | 110.6 ± 9.8** | 0.8 ± 0.2 | 1.6 ± 0.4** |
| Ifit1 | 13.9 ± 6.1 | 175.3 ± 35.6** | 0.1 ± 0.0 | 0.4 ± 0.1** |
Number of copies normalized to 1,000 copies of the mouse β-actin housekeeping gene. Data are pooled from three independent experiments (n = 6). Statistical significance was assessed by the Mann-Whitney U test.
**, P < 0.01 for live B. burgdorferi versus medium.
**, P < 0.01 for STAT1−/−/live B. burgdorferi versus C3H/HeN/live B. burgdorferi.
IFN-responsive gene induction by B. burgdorferi-stimulated BMDMs is IRF3 dependent.
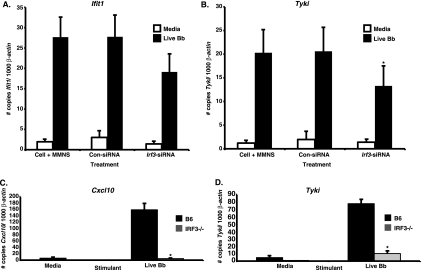
IRF3-dependent signal transduction pathways are utilized by the majority of both extracellular and intracellular bacteria to trigger IFN-responsive gene transcription by macrophages (11, 14, 15, 23). For this reason, we assessed whether B. burgdorferi-mediated induction of IFN profile gene transcription by BMDMs is IRF3 dependent. Silencing of Irf3 expression in C3H BMDMs by small interfering RNAs reduced, but did not abrogate, Ifit1 and Tyki transcript levels in B. burgdorferi-stimulated BMDMs compared with values obtained from cells treated with a pool of control, scrambled siRNAs (Fig. 2 A and B). To further address the involvement of IRF3, we next examined IFN-responsive gene expression in B. burgdorferi-stimulated BMDMs from IRF3−/− B6 mice. Transcription of the representative IFN profile genes Cxcl10 and Tyki was nearly abrogated in IRF3−/− BMDMs, as these genes exhibited 97% and 86% decreases in expression levels, respectively, compared with values obtained for wild-type B6 BMDMs (Fig. 2C and D). Taken together, these data indicate that induction of IFN-responsive gene transcription by B. burgdorferi-stimulated BMDMs is IRF3 dependent.
FIG. 2.
B. burgdorferi-mediated induction of IFN-responsive genes is IRF3 dependent. (A and B) siRNA-mediated silencing of Irf3. Cultured C3H/HeN BMDMs were transfected with control siRNA (Con-siRNA), with Irf3-specific siRNA (Irf3-siRNA), or with transfection medium alone (MMNS) using the Amaxa nucleofector. At 72 h after transfection, cells were stimulated with 7.4 × 106/ml live B. burgdorferi organisms (Bb) for 6 h. Cellular RNA was assessed for IFN-responsive gene expression by RT-PCR, as shown for Ifit1 (A) and Tyki (B). Data are shown as the means ± SEM and are pooled from four independent experiments (n = 5). For Ifit1, P = 0.2491 via a two-tailed, unpaired Student t test for live B. burgdorferi-stimulated Irf3-siRNA versus control siRNA. For Tyki, *, P < 0.05 via a two-tailed, paired Student t test for live B. burgdorferi-stimulated Irf3-siRNA versus control siRNA. (C and D) B. burgdorferi stimulation of IRF3−/− BMDMs. BMDMs obtained from B6 or IRF3−/− mice were stimulated with 7.4 × 106/ml live B. burgdorferi organisms for 6 h. RT-PCR results obtained for the representative IFN-responsive transcripts Cxcl10 (C) and Tyki (D) are displayed as the number of copies of the gene of interest normalized to 1,000 copies of the mouse β-actin housekeeping gene. Data are displayed as the means ± SEM and are pooled from two independent experiments (n = 4). Differences between IRF3−/− and B6 live B. burgdorferi-stimulated BMDMs were determined via the Mann-Whitney U test *, P < 0.05.
IFN-responsive gene transcription can be induced by B. burgdorferi RNA.
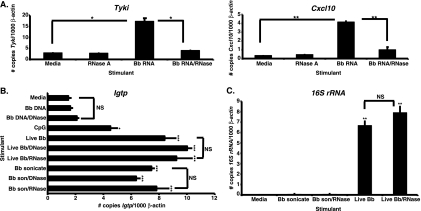
Previous studies utilizing group B streptococci indicated that IRF3-dependent induction of IFN-β by BMDMs was mediated by the cytosolic delivery of GBS DNA (11), whereas L. pneumophila utilizes the RNA-sensing receptors RIG-I and MDA5 for type I IFN gene transcription (31). These findings prompted us to examine whether B. burgdorferi-derived nucleic acid acts as the ligand triggering IRF3-dependent IFN-inducible gene expression by BMDMs. C3H BMDMs were stimulated with B. burgdorferi genomic DNA, RNA, live bacteria, or sonicated bacteria in both the presence and absence of nucleases, and the effects of these stimuli on IFN profile induction were assessed at 6 h poststimulation. As illustrated in Fig. 3 A for the IFN-responsive transcripts Tyki and Cxcl10, the addition of B. burgdorferi RNA resulted in significant induction of IFN-responsive gene transcription. The stimulatory activity mediated by B. burgdorferi RNA was reduced upon incubation with RNase A (Fig. 3A). BMDM stimulation with B. burgdorferi genomic DNA did not result in IFN-inducible gene transcription, as shown in Fig. 3B for Igtp. In contrast, incubation with live or sonicated B. burgdorferi or with CpG DNA resulted in robust induction of IFN-responsive transcripts. As expected, DNase digestion of B. burgdorferi genomic DNA did not affect IFN-inducible gene expression, as the DNA does not act as a type I IFN-stimulatory ligand. Although DNase I and RNase A effectively digested B. burgdorferi genomic DNA and RNA, respectively (data not shown), neither DNase I nor RNase A digestion of live or sonicated B. burgdorferi caused a decrease in IFN-inducible transcript levels (Fig. 3B). In addition, forced cytosolic delivery of B. burgdorferi genomic DNA (0.7 ± 0.1 GBP/1,000 β-actin copies) via Amaxa nucleofector-mediated transfection did not result in the induction of IFN-responsive gene transcription by BMDMs, even though both sonicate (1.3 ± 0.8) and DNase I (2.1 ± 0.1)- or RNase A (2.2 ± 0.4)-digested sonicate preparations delivered in this fashion triggered IFN profile transcription. These results indicate that B. burgdorferi RNA, but not genomic DNA, is utilized as a ligand for the induction of IFN-responsive gene transcription by BMDMs.
FIG. 3.
B. burgdorferi-derived RNA induces IFN profile gene transcription by BMDMs. C3H/HeJ BMDMs were treated for 6 h with 2 μg B. burgdorferi (Bb) RNA, genomic DNA, RNase A-digested B. burgdorferi RNA, DNase I-digested B. burgdorferi DNA, 0.7 μM CpG oligonucleotide, 7.4 × 106/ml live B. burgdorferi organisms, live B. burgdorferi digested with DNase I or RNase A, 5 μg/ml sonicated B. burgdorferi, or sonicate digested with DNase I or RNase A. RT-PCR transcripts are displayed as the number of copies of the gene of interest normalized to 1,000 copies of the mouse β-actin housekeeping gene. (A) B. burgdorferi RNA stimulates IFN-responsive gene transcription by BMDMs. Transcript levels for Tyki and Cxcl10 are shown. Data are depicted as the means ± SEM and are representative of two independent experiments (n = 3). Statistical significance was assessed via the two-tailed, unpaired Student t test with Welch correction. *, P < 0.05 for B. burgdorferi RNA versus medium and for B. burgdorferi RNA/RNase versus B. burgdorferi RNA. **, P < 0.01 for B. burgdorferi RNA versus medium and for B. burgdorferi RNA/RNase versus B. burgdorferi RNA. (B) B. burgdorferi-derived DNA is not a ligand for BMDM IFN profile gene induction. Transcript levels for Igtp are shown. Data are depicted as the means ± SEM and are representative of four independent experiments (n = 3). Statistical analysis was performed using one-way ANOVA with the Tukey-Kramer multiple comparisons posttest. Significant differences for stimulated samples compared to medium alone: *, P < 0.05; ***, P < 0.001. Brackets indicate that there are no significant (NS) differences between live B. burgdorferi- or sonicate-stimulated samples versus the corresponding nuclease-digested treatment groups or for B. burgdorferi genomic DNA-stimulated versus B. burgdorferi DNA/DNase-digested samples. (C) Treatment with RNase A fails to destroy the 16S rRNA content in live B. burgdorferi. Transcript levels for B. burgdorferi 16S rRNA are shown. Data are shown as the means ± SEM and are representative of two independent experiments (n = 3). Statistical significance was assessed via the two-tailed, unpaired Student t test with the Welch correction. Differences between live B. burgdorferi- or live B. burgdorferi/RNase-stimulated BMDMs and medium: **, P < 0.01. Brackets indicate that there are no significant (NS) differences (P = 0.1529) between live B. burgdorferi- and live B. burgdorferi/RNase-digested samples.
To determine why the IFN-stimulatory activity of live and sonicated B. burgdorferi was not destroyed by RNase A digestion, the 16S rRNA transcript levels of both live and sonicated B. burgdorferi and of RNase-digested samples were determined by RT-PCR. As expected, abundant 16S rRNA transcript was detected in live B. burgdorferi. Surprisingly, RNase A treatment failed to alter the 16S rRNA transcript levels obtained for live B. burgdorferi (Fig. 3C). In contrast to live B. burgdorferi, sonicated bacteria did not contain 16S rRNA, so RNase digestion had no effect (Fig. 3C). Collectively, our results indicate that B. burgdorferi RNA is one of at least two ligands capable of stimulating IFN profile transcription, since sonicate lacks 16S rRNA but still readily induces IFN gene expression.
B. burgdorferi culture supernatant contains components that trigger IRF3-dependent IFN-responsive gene transcription by BMDMs.
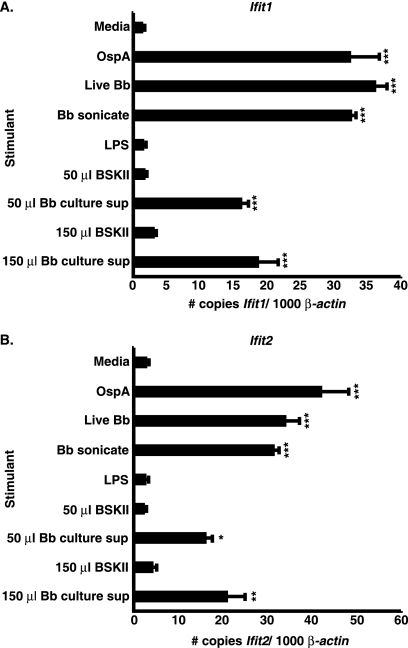
Since B. burgdorferi sonicate, which is likely to include lipoproteins, contains a non-nucleic acid cell-associated component that mediates BMDM IFN profile induction, we next investigated whether OspA, a major lipoprotein induced during in vitro cultivation of B. burgdorferi (5), could elicit IFN-responsive gene transcription by BMDMs. As expected, addition of live or sonicated B. burgdorferi to TLR4-defective C3H/HeJ BMDMs resulted in increased IFN-responsive gene transcription, whereas LPS failed to activate the expression of these genes. Intriguingly, the addition of lipidated recombinant OspA (51) resulted in significant IFN profile gene transcription, as depicted in Fig. 4 for Ifit1 (Fig. 4A) and Ifit2 (Fig. 4B). We next assessed whether a B. burgdorferi-derived soluble component(s) could act as a ligand capable of eliciting IFN-responsive transcript induction from BMDMs. Volumes ranging from 50 to 150 μl of B. burgdorferi supernatant collected from late-logarithmic-phase cultures were mixed with serum-free cell culture medium and applied to cultured BMDMs obtained from C3H/HeJ mice. Equivalent volumes of bacterium-free BSKII medium served as controls for potential induction of IFN-responsive transcripts by BSKII components. Fifty microliters of culture supernatant resulted in increased IFN-responsive gene transcript levels, which was not seen in BMDMs treated with control BSKII medium. Addition of larger volumes of culture supernatant (shown in Fig. 4A and B for 150 μl) did not result in a further increase in IFN profile gene transcript levels, indicating that the stimulatory activity was saturated with 50 μl of sample (Fig. 4A and B). Finally, we asked whether nucleic acids potentially released into the culture medium during B. burgdorferi growth were responsible for the IFN-inductive capacity of the culture supernatant. As demonstrated in Table 2 for Cxcl10 and Tyki, neither DNase nor RNase digestion destroyed the IFN-stimulatory activity mediated by B. burgdorferi culture supernatant. These results suggest that there is a third ligand that is capable of inducing IFN-responsive gene transcription by BMDMs, as cultured B. burgdorferi releases a non-nucleic acid factor that stimulates IFN profile gene expression.
FIG. 4.
Soluble components of B. burgdorferi spent medium govern IFN-responsive gene transcription by BMDMs. Cultured C3H/HeJ BMDMs were stimulated for 6 h with 5 μg/ml recombinant lipidated OspA, 7.4 × 106/ml live B. burgdorferi organisms (Bb), 5 μg/ml sonicated B. burgdorferi, 10 ng/ml LPS, 50 to 150 μl of B. burgdorferi culture supernatant, or 50 to 150 μl of clean BSKII medium. Data are depicted as the means ± SEM and are representative of two independent experiments (n = 3). Statistical analysis was performed using one-way ANOVA with the Tukey-Kramer multiple comparisons posttest. Differences in Ifit1(A) and Ifit2 (B) transcript levels between stimulated and unstimulated samples: *, P < 0.05; **, P < 0.01: ***, P < 0.001.
TABLE 2.
Nuclease treatment does not alter the IFN-stimulatory activity of B. burgdorferi culture supernatants
| Stimulant | Transcript level (mean ± SEM)a |
|
|---|---|---|
| Cxcl10 | Tyki | |
| Medium | 0.4 ± 0.0 | 3.0 ± 0.1 |
| B. burgdorferi RNAb | 4.2 ± 0.1*** | 17.3 ± 1.5* |
| B. burgdorferi RNA + RNasec | 1.0 ± 0.3** | 4.1 ± 0.2* |
| BSKII | 0.5 ± 0.1 | 2.6 ± 0.1 |
| B. burgdorferi culture supernatantd | 3.4 ± 0.1** | 9.0 ± 0.5** |
| B. burgdorferi supernatant + DNase | 5.2 ± 0.7 | 10.6 ± 2.1 |
| B. burgdorferi supernatant + RNase | 3.0 ± 0.2 | 7.4 ± 1.5 |
RT-PCR data are shown as the number of copies of the gene of interest per 1,000 copies of the β-actin gene and are representative of two independent experiments (n = 3). Statistical significance was assessed via a two-tailed, unpaired Student t test with the Welch correction.
For Cxcl10: ***, P < 0.001 versus medium. For Tyki: *, P < 0.05 versus medium.
For Cxcl10: **, P < 0.01 versus B. burgdorferi RNA. For Tyki: *, P < 0.05 versus B. burgdorferi RNA.
For Cxcl10: **, P < 0.01 versus BSKII. For Tyki: **, P < 0.01 versus BSKII.
Since we have demonstrated in this report that IFN-responsive gene transcription by BMDMs stimulated with live B. burgdorferi is IRF3 dependent, we next asked whether IRF3 contributes to the inductive effect mediated by B. burgdorferi culture medium or sonicate. Application of B. burgdorferi culture supernatants to IRF3−/− BMDMs resulted in greatly diminished transcription of the IFN-responsive genes Ifit1 (54.9 ± 12.8 copies/1,000 β-actin copies for B6 versus 7.4 ± 2.6 copies/1,000 β-actin copies for IRF3−/− BMDMs) and Cxcl10 (79.8 ± 11.4 copies/1,000 β-actin copies for B6 versus 2.0 ± 0.2 copies/1,000 β-actin copies for IRF3−/− BMDMs) compared with the gene expression levels obtained from comparably stimulated B6 BMDMs. Administration of sonicate to IRF3−/− BMDMs also resulted in reduced Ifit1 (32.2 ± 1.8 copies/1,000 β-actin copies for B6 versus 13.7 ± 1.2 copies/1,000 β-actin copies for IRF3−/− BMDMs) and Cxcl10 (43.5 ± 1.8 copies/1,000 β-actin copies for B6 versus 12.6 ± 2.1 copies/1,000 β-actin copies for IRF3−/− BMDMs) transcript levels. Taken together, these data indicate that a non-nucleic acid ligand(s) generated during B. burgdorferi growth contributes to the IRF3-dependent IFN-responsive gene transcription.
DISCUSSION
We recently uncovered a novel and previously unappreciated role for type I IFN in the development of severe Lyme arthritis in genetically susceptible C3H/HeN mice. In addition, we demonstrated that induction of IFN-responsive gene transcripts in BMDMs was entirely dependent upon a functional type I IFN receptor but independent of TLR2, -4, and -9 and of the adapter molecule MyD88 (30). The current report extends our previous observations by closely examining the nature of the ligand(s) and signal transduction pathways utilized by B. burgdorferi to elicit the downstream production of type I IFN-responsive genes by BMDMs. Here we present data indicating that B. burgdorferi utilizes multiple ligands for IRF3-dependent induction of type I IFN-responsive genes.
Although our previous work determined that IFN profile induction occurred independently of MyD88-dependent TLRs, we had not yet assessed whether this response was TRIF dependent. Similar to what has been reported for the extracellular group B streptococci (11) and for the intracellular bacteria Listeria monocytogenes (16, 23), Francisella tularensis (15), and Mycobacterium tuberculosis (34), we found that IFN-responsive gene transcripts are still highly induced in the absence of a functional TRIF adapter molecule (Fig. 1). These data indicated that initiation of a type I IFN response by B. burgdorferi-stimulated BMDMs does not involve TLR signaling. L. monocytogenes, F. tularensis, and M. tuberculosis each have the ability to escape from the phagosomal compartment of the macrophage and enter the cytosol of the cell. While in the cytosol, recognition of DNA derived from these pathogens, along with MDP-based signaling through NOD2, results in the downstream production of type I IFN by the infected macrophage (16, 23, 34). Our observation that B. burgdorferi does not utilize the cytoplasmic NLR NOD2 for the induction of IFN-responsive gene transcripts (Fig. 1) is not surprising, as it has been reported that this spirochete remains in the phagosome and does not escape to the cytosol (38). In the same vein, our finding that forced cytosolic delivery of B. burgdorferi genomic DNA or sonicate did not result in appreciable IFN-responsive gene transcription suggests that primary cytosolic signaling does not occur as a response to recognition of B. burgdorferi DNA. While we are unable to formally exclude the possibility of NLRP3 (cryopyrin) or other potential NLRs as potential receptors governing IFN profile induction, the failure of cytosolically administered B. burgdorferi to activate IFN-responsive gene transcription argues against this possibility. In addition, Liu and colleagues recently reported that the inflammasome is not utilized for B. burgdorferi-directed host defense (24). Collectively, our data indicate that B. burgdorferi likely utilizes an unidentified, potentially novel BMDM receptor to trigger transcription of IFN-responsive genes. Interestingly, B. burgdorferi also differs from other bacterial pathogens that elicit the production of IFN-responsive genes in that it does not have genes for a type III or IV secretion apparatus, nor does it possess phospholipases.
Type I IFN profile gene transcription by B. burgdorferi-stimulated BMDMs was entirely dependent upon an early STAT1-dependent feedback amplification loop, the effects of which were evident by 1 h poststimulation (Table 1 and data not shown). Interestingly, the induction of all analyzed IFN-responsive transcripts, including Tyki, Ifit1, and Ifit2, was suppressed in B. burgdorferi-stimulated STAT1−/− BMDMs (Table 1 and data not shown). These genes were reported to be among the first IFN-inducible genes transcribed by BMDMs without a requirement for secondary cytokine signaling in response to cytosolic L. monocytogenes (23). These results indicate that the temporal and compartment-specific transcription of these genes differs between B. burgdorferi and L. monocytogenes. The results obtained with B. burgdorferi-stimulated STAT1−/− BMDMs are intriguing but somewhat difficult to interpret, because antibody-mediated blockage of the type I IFN receptor prevents the development of severe Lyme arthritis (30), yet STAT1−/− mice still develop arthritis (9). However, joint tissue is very complex and multiple different types of cells are present, whereas cultured BMDMs are a homogenous single-cell population. Although absent in STAT1−/− BMDMs, compensatory pathways for type I IFN-responsive gene induction may be triggered by other cell types present within, or trafficking to, the joint microenvironment of STAT1−/− mice.
Similar to what has been reported for the majority of bacterial pathogens (11, 14, 15, 23), induction of IFN-responsive genes by B. burgdorferi-stimulated BMDMs was IRF3 dependent (Fig. 2). Previous studies conducted with Listeria monocytogenes (23, 43), Legionella pneumophila (43), and GBS (11) indicated that IRF3-dependent induction of type I IFN is driven by cytosolic recognition of pathogen-derived DNA. However, in stark contrast to the case for other bacterial pathogens, B. burgdorferi-mediated transcription of macrophage IFN-responsive genes was not governed by recognition, cytosolic or otherwise, of its DNA (Fig. 3). This finding was in agreement with our previous work utilizing B. burgdorferi-stimulated TLR9−/− BMDMs to demonstrate that this DNA sensor was not required for type I IFN profile gene transcription by B. burgdorferi-stimulated mouse macrophages (30). However, while this paper was in preparation, Petzke and colleagues reported that B. burgdorferi stimulated the TLR9-dependent production of type I IFN by human peripheral blood mononuclear cells (PBMCs) (35). Potential reasons for the discrepancies in the results obtained by these studies could include differences in experimental methodology and in both the species origin and cell types analyzed. We analyzed a single, pure cell population, the BMDM, from mice, whereas Petzke et al. utilized a mixed human cell population (PBMCs).
It was recently reported that L. pneumophila induces type I IFN production by BMDMs via ligation of the MyD88-independent RIG-I and MDA5 RNA recognition receptors (31). We now report that B. burgdorferi-derived RNA is one ligand that triggers IFN profile induction by BMDMs (Fig. 3A). Live B. burgdorferi stimulates the MyD88-independent transcription of IFN-responsive genes (Fig. 1C) (30) and contains 16S rRNA (Fig. 3C), indicating that RNA likely contributes to the IFN-stimulatory capacity of live B. burgdorferi. Unfortunately, since live B. burgdorferi was resistant to RNase A digestion (Fig. 3C), it is unclear what proportion of the IFN-stimulatory activity can be attributed to RNA. However, in agreement with our results, Petzke and colleagues did find that inhibition of TLR7 on human PBMCs reduced type I IFN responses to live B. burgdorferi (35). IFN-responsive gene transcription was also governed by a second ligand: a non-nucleic acid, cell-associated molecule present in sonicated B. burgdorferi (Fig. 3B and C). The identity of this cell-associated molecule is currently unknown. Although studies with synthetic TLR2 ligands have failed to identify a signal transduction pathway that culminates in the downstream transcription of type I IFN, we demonstrated that BMDM IFN profile gene transcription could be induced by recombinant B. burgdorferi lipidated OspA (Fig. 4). These findings are in agreement with our previous report on the induction of IFN-β transcripts by purified OspA in macrophages from C3H/HeJ mice (27).
Significant induction of IFN-responsive gene transcripts was also mediated by a fourth ligand: a non-nucleic acid molecule(s) present in B. burgdorferi culture supernatant. Intriguingly, we also demonstrated that the inductive capacity mediated by components released during culture of B. burgdorferi was entirely dependent upon functional IRF3. Although LPS stimulation of TRIF can result in IRF3-dependent transcription of type I IFNs, B. burgdorferi lacks LPS (46). This allowed us to control for potential endotoxin contamination of medium components by utilizing the TLR4-defective C3H/HeJ BMDMs, where significant elevation of IFN-responsive gene transcription was stimulated by medium collected from cultured bacteria but not by unused medium (Fig. 4). To our knowledge, this is the first report to describe IRF3-dependent induction of type I IFN-responsive genes by soluble components present in bacterial culture supernatant. While the origin of these soluble factors is currently unknown, potential candidates include, but may not be limited to, proteins, small molecules, or lipids.
In conclusion, we have presented evidence that B. burgdorferi utilizes multiple ligands to trigger IFN-responsive gene transcription by BMDMs. Our data support the existence of at least four ligands: B. burgdorferi-derived RNA; a distinct non-nucleic acid, cell-associated molecule present in sonicated bacteria; the lipoprotein OspA; and a non-nucleic acid factor released by cultured B. burgdorferi. It is also possible that lipoprotein could be one constituent of the cell-associated or released factor. The novelty of these findings is of heightened significance since the production of type I IFN in response to B. burgdorferi assumes a critical role in the development of Lyme arthritis in mice but not in host defense (30). This important distinction may explain why the ligands/pathways utilized by B. burgdorferi-stimulated BMDMs for the transcription of type I IFN-responsive genes likely differ from those triggered as a host defense response to other bacterial pathogens. In addition, the recent findings that human monocytes also produce type I IFN in response to B. burgdorferi indicate that induction of this pathway may also have important clinical consequences in human Lyme disease patients (35, 38). Therefore, elucidation of the signal transduction pathways utilized for type I IFN production by macrophages in response to B. burgdorferi infection could lead to the development of improved therapeutic agents for Lyme disease patients, particularly those afflicted with arthritis.
Acknowledgments
We acknowledge the University of Utah biostatistics core for advice regarding statistical analysis. We thank Charles Brown for C3H Stat1−/− mice and Douglas Golenbock and Katherine Fitzgerald for IRF3−/− femurs. We also thank Lynn Sonderegger for insightful discussions regarding this work.
This work was supported by Public Health Service grants AI-32223 and AI-43521 to J.J.W., AI-24158 to J.H.W., and Training Program in Microbial Pathogenesis 5T32-AI055434 and by an Arthritis Foundation Award to J.C.M.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 19 April 2010.
REFERENCES
- 1.Adachi, O., T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi, and S. Akira. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143-150. [DOI] [PubMed] [Google Scholar]
- 2.Alverson, J., S. F. Bundle, C. D. Sohaskey, M. C. Lybecker, and D. S. Samuels. 2003. Transcriptional regulation of the ospAB and ospC promters from Borrelia burgdorferi. Mol. Microbiol. 48:1665-1677. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. Current protocols in molecular biology, vol. 1. Wiley Press, New York, NY.
- 4.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour, A. G., S. L. Tessier, and W. J. Todd. 1983. Lyme disease spirochetes and ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody. Infect. Immun. 41:795-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barthold, S. W., D. H. Persing, A. L. Armstrong, and R. A. Peeples. 1991. Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am. J. Pathol. 139:263-273. [PMC free article] [PubMed] [Google Scholar]
- 7.Behera, A. K., E. Hildebrand, R. T. Bronson, G. Perides, S. Uematsu, S. Akira, and L. T. Hu. 2006. MyD88 deficiency results in tissue-specific changes in cytokine induction and inflammation in interleukin-18-independent mice infected with Borrelia burgdorferi. Infect. Immun. 74:1462-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolz, D. D., R. S. Sundsbak, Y. Ma, S. Akira, C. J. Kirschning, J. F. Zachary, J. H. Weis, and J. J. Weis. 2004. MyD88 plays a unique role in host defense but not arthritis development in Lyme disease. J. Immunol. 173:2003-2010. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. R., V. A. Blaho, K. L. Fritsche, and C. M. Loiacono. 2006. Stat1 deficiency exacerbates carditis but not arthritis during experimental lyme borreliosis. J. Interferon Cytokine Res. 26:390-399. [DOI] [PubMed] [Google Scholar]
- 10.Brown, J. P., J. F. Zachary, C. Teuscher, J. J. Weis, and R. M. Wooten. 1999. Dual role of interleukin-10 in murine lyme disease: regulation of arthritis severity and host defense. Infect. Immun. 67:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charrel-Dennis, M., E. Latz, K. A. Halman, P. Trieu-Cuot, K. A. Fitzgerald, D. L. Kasper, and D. T. Golenbock. 2008. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe 4:543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, V. A., S. Dixit, S. M. O'Brien, X. Alvarez, B. Pahar, and M. T. Philipp. 2009. Live Borrelia burgdorferi spirochetes elicit inflammatory mediators from human monocytes via the Toll-like receptor signaling pathway. Infect. Immun. 77:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gattorno, M., L. Chicha, A. Gregrio, F. Ferlito, F. Rossi, D. Jarrossay, A. Lanzavecchia, A. Martini, and M. G. Manz. 2007. Distinct expression pattern of IFNa and TNFa in juvenile idiopathic arthritis synovial tissue. Rheumatology 46:657-665. [DOI] [PubMed] [Google Scholar]
- 14.Gratz, N., M. Siller, B. Schaljo, Z. A. Pirzada, I. Gattermeier, I. Vojtek, C. J. Kirschning, H. Wagner, S. Akira, E. Charpentier, and P. Kovarik. 2008. Group A Streptococcus activates type I interferon production and MyD88-dependent signaling without involvement of TLR2, TLR4, and TLR9. J. Biol. Chem. 283:19879-19887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry, T., A. Brotcke, D. S. Weiss, L. J. Thompson, and D. M. Monack. 2007. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 204:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herskovits, A. A., V. Auerbach, and D. A. Portnoy. 2007. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 3:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirschfeld, M., C. J. Kirschning, R. Schwandner, H. Wesche, J. H. Weis, R. M. Wooten, and J. J. Weis. 1999. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by Toll-like receptor 2. J. Immunol. 163:2382-2386. [PubMed] [Google Scholar]
- 18.Hirschfeld, M., Y. Ma, J. H. Weis, S. N. Vogel, and J. J. Weis. 2000. Repurification of lipopolysaccharide eliminates signaling through both human and murine Toll-like receptor 2. J. Immunol. 165:618-622. [DOI] [PubMed] [Google Scholar]
- 19.Jeyaseelan, S., S. K. Young, M. B. Fessler, Y. Liu, K. C. Malcolm, M. Yamamoto, S. Akira, and G. S. Worthen. 2007. Toll/IL-1 receptor domain-containing adaptor inducing IFN-B (TRIF)-mediated signaling contributes to innate immune responses in the lung during Escherichia coli pneumonia. J. Immunol. 178:3153-3160. [DOI] [PubMed] [Google Scholar]
- 20.Koga, R., S. Hamano, H. Kuwata, K. Atarashi, M. Ogawa, H. Hisaeda, M. Yamamoto, S. Akira, K. Himeno, M. Matsumoto, and K. Takeda. 2006. TLR-dependent induction of IFN-B mediates host defense against Trypanosoma cruzi. J. Immunol. 177:7059-7066. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, H., T. Kawai, and S. Akira. 2009. Pathogen recognition in the innate immune response. Biochem. J. 420:1-16. [DOI] [PubMed] [Google Scholar]
- 22.Lazarus, J. J., M. A. Kay, A. L. McCarter, and R. M. Wooten. 2008. Viable Borrelia burgdorferi enhances interleukin-10 production and suppresses activation of murine macrophages. Infect. Immun. 76:1153-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leber, J. H., G. T. Crimmins, S. Raghavan, N. P. Meyer-Morse, J. S. Cox, and D. A. Portnoy. 2008. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 4:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, N., A. A. Belperron, C. J. Booth, and L. K. Bockenstedt. 2009. The caspase 1 inflammasome is not required for control of murine Lyme borreliosis. Infect. Immun. 77:3320-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, N., R. R. Montgomery, S. W. Barthold, and L. K. Bockenstedt. 2004. Myeloid differentiation antigen 88 deficiency impairs pathogen clearance but does not alter inflammation in Borrelia burgdorferi-infected mice. Infect. Immun. 72:3195-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma, Y., J. C. Miller, H. Crandall, E. T. Larsen, D. M. Dunn, R. B. Weiss, M. Subramanian, J. H. Weis, J. F. Zachary, C. Teuscher, and J. J. Weis. 2009. Interval-specific congenic lines reveal QTL with penetrant Lyme arthritis phenotypes on chromosomes 5, 11, and 12. Infect. Immun. 77:3302-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, Y., K. P. Seiler, K. Tai, L. Yang, M. Woods, and J. J. Weis. 1994. Outer surface lipoproteins of Borrelia burgdorferi stimulate nitric oxide production by the cytokine-inducible pathway. Infect. Immun. 62:3663-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathian, A., A. Weinberg, M. Gallegos, J. Banchereau, and S. Koutouzov. 2005. IFN-alpha induces early lethal lupus in preautoimmune (New Zealand Black × New Zealand White) F1 but not in BALB/c mice. J. Immunol. 172:2499-2506. [DOI] [PubMed] [Google Scholar]
- 29.Meerpohl, H. G., M. L. Lohmann-Matthes, and H. Fischer. 1976. Studies on the activation of mouse bone marrow-derived macrophages by the macrophage cytotoxicity factor (MCF). Eur. J. Immunol. 6:213-217. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. C., Y. Ma, J. Bian, K. C. F. Sheehan, J. F. Zachary, J. H. Weis, R. D. Schreiber, and J. J. Weis. 2008. A critical role for type I IFN in arthritis development following Borrelia burgdorferi infection of mice. J. Immunol. 181:8492-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monroe, K. M., S. M. McWhirter, and R. E. Vance. 2009. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog. 5:e1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison, T. B., J. J. Weis, and C. T. Wittwer. 1998. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 24:954-958, 960, 962. [PubMed] [Google Scholar]
- 33.Ornstein, K., and A. G. Barbour. 2006. A reverse transcriptase-polymerase chain reaction assay of Borrelia burgdorferi 16S rRNA for highly sensitive quantification of pathogen load in a vector. Vector-Borne Zoonotic Dis. 6:103-112. [DOI] [PubMed] [Google Scholar]
- 34.Pandey, A. K., Y. Yang, Z. Jiang, S. M. Fortune, F. Coulombe, M. A. Behr, K. A. Fitzgerald, C. M. Sassetti, and M. A. Kelliher. 2009. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 5:e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petzke, M. M., A. Brooks, M. A. Krupna, D. Mordue, and I. Schwartz. 2009. Recognition of Borrelia burgdorferi, the Lyme disease spirochete by TLR7 and TLR9 induces a type I IFN response by human immune cells. J. Immunol. 183:5279-5292. [DOI] [PubMed] [Google Scholar]
- 36.Poltorak, A., X. He, I. Smirnova, M. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in the Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 37.Power, M. R., B. Li, M. Yamamoto, S. Akira, and T. Lin. 2007. A role of Toll-IL-1 receptor domain-containing adaptor-inducing IFNB in the host response to Pseudomonas aeruginosa lung infection in mice. J. Immunol. 178:3170-3176. [DOI] [PubMed] [Google Scholar]
- 38.Salazar, J. C., S. Duhnam-Ems, C. La Vake, A. R. Cruz, M. W. Moore, M. J. Caimano, L. Velez-Climent, J. Shupe, W. Krueger, and J. D. Radolf. 2009. Activation of human monocytes by live Borrelia burgdorferi generates TLR2-dependent and -independent responses which include induction of IFNB. PLoS Pathog. 5:e1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salazar, J. C., C. D. Pope, T. J. Sellati, H. M. Feder, Jr., T. G. Kiely, K. R. Dardick, R. L. Buckman, M. W. Moore, M. J. Caimano, J. G. Pope, P. J. Krause, and J. D. Radolf. 2003. Coevolution of markers of innate and adaptive immunity in skin and peripheral blood of patients with erythema migrans. J. Immunol. 171:2660-2670. [DOI] [PubMed] [Google Scholar]
- 40.Santiago-Raber, M. L., R. Baccala, K. M. Haraldsson, D. Choubey, T. A. Stewart, D. H. Kono, and A. N. Theofilopoulos. 2003. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J. Exp. Med. 197:777-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 42.Schoenfeld, R., B. Araneo, Y. Ma, L. Yang, and J. J. Weis. 1992. Demonstration of a B-lymphocyte mitogen produced by the Lyme disease pathogen, Borrelia burgdorferi. Infect. Immun. 60:455-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stetson, D. B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93-103. [DOI] [PubMed] [Google Scholar]
- 44.Strueby, L., B. Nair, A. Kirk, and R. M. Taylor-Gjevre. 2005. Arthritis and bursitis in multiple sclerosis patients treated with interferon-beta. Scand. J. Rheumatol. 34:485-488. [DOI] [PubMed] [Google Scholar]
- 45.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell. Microbiol. 8:907-922. [DOI] [PubMed] [Google Scholar]
- 46.Takayama, K., R. J. Rothenberg, and A. G. Barbour. 1987. Absence of lipopolysaccharide in the Lyme disease spirochete Borrelia burgdorferi. Infect. Immun. 55:2311-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaena de Avalos, S., I. J. Blader, M. Fisher, J. C. Boothroyd, and B. A. Burleigh. 2002. Immediate/early response to Trypanosoma cruzi infection involves minimal modulation of host cell transcription. J. Biol. Chem. 277:639-644. [DOI] [PubMed] [Google Scholar]
- 48.Vogel, S. N., D. Johnson, P. Perera, A. Medvedev, L. Lariviere, S. T. Qureshi, and D. Malo. 1999. Functional characterization of the effect of the C3H/HeJ defect in mice that lack an Lpsn gene: in vivo evidence for a dominant negative mutation. J. Immunol. 162:5666-5670. [PubMed] [Google Scholar]
- 49.Wang, G., M. M. Petzke, R. Iyer, H. Wu, and I. Schwartz. 2008. Pattern of proinflammatory cytokine inductine in RAW264.7 mouse macrophages is identical for virulent and attenuated Borrelia burgdorferi. J. Immunol. 180:8306-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, X., Y. Ma, A. Yoder, H. Crandall, J. F. Zachary, R. S. Fujinami, J. H. Weis, and J. J. Weis. 2008. T cell infiltration is asoociated with increased Lyme arthritis in TLR2−/− mice. FEMS Immunol. Med. Microbiol. 52:124-133. [DOI] [PubMed] [Google Scholar]
- 51.Weis, J. J., Y. Ma, and L. F. Erdile. 1994. Biological activities of native and recombinant Borrelia burgdorferi outer surface protein A: dependence on lipid modification. Infect. Immun. 62:4632-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, L. E., D. Widman, S. H. Dikman, and P. D. Gorevic. 2002. Autoimmune disease complicating antiviral therapy for hepatitis C virus infection. Semin. Arthritis Rheum. 32:163-173. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, T. Y., and R. A. Daynes. 2007. Glucocorticoid conditioning of myeloid progenitors enhances TLR4 signaling via negative regulation of the phosphatidylinositol 3-kinase-Akt pathway. J. Immunol. 178:2517-2526. [DOI] [PubMed] [Google Scholar]