Abstract
The Yersinia adhesin YadA mediates the adhesion of the human enteropathogen Yersinia enterocolitica to collagens and other components of the extracellular matrix. Though YadA has been proposed to bind to a specific site in collagens, the exact binding determinants for YadA in native collagen have not previously been elucidated. We investigated the binding of YadA to collagen Toolkits, which are libraries of triple-helical peptides spanning the sequences of type II and III human collagens. YadA bound to many of them, in particular to peptides rich in hydroxyproline but with few charged residues. We were able to block the binding of YadA to collagen type IV with the triple-helical peptide (Pro-Hyp-Gly)10, suggesting that the same site in YadA binds to triple-helical regions in network-forming collagens as well. We showed that a single Gly-Pro-Hyp triplet in a triple-helical peptide was sufficient to support YadA binding, but more than six triplets were required to form a tight YadA binding site. This is significantly longer than the case for eukaryotic collagen-binding proteins. YadA-expressing bacteria bound promiscuously to Toolkit peptides. Promiscuous binding could be advantageous for pathogenicity in Y. enterocolitica and, indeed, for other pathogenic bacteria. Many of the tightly binding peptides are also targets for eukaryotic collagen-binding proteins, and YadA was able to inhibit the interaction between selected Toolkit peptides and platelets. This leads to the intriguing possibility that YadA may interfere in vivo with host processes mediated by endogenous collagen-binding proteins.
Adhesion to host tissues is a common and often essential step in the infection processes of bacterial pathogens and is usually among the first stages in colonizing the host. In many cases, the extracellular matrix (ECM) is the site of bacterial adhesion, and the ability to adhere to components of ECM is essential for the virulence of several pathogenic bacteria (reviewed in reference 61).
One such bacterium is Yersinia enterocolitica, an enteropathogen that causes a range of gastrointestinal symptoms, from mild diarrhea to enterocolitis and mesenteric lymphadenitis (7, 10). On occasion, the primary gastric infection is followed by sequelae such as reactive arthritis, glomerulonephritis, and erythema nodosum. After the bacterium crosses the intestinal epithelium, the major adhesin in Y. enterocolitica infections is the Yersinia adhesin A (YadA). YadA is an outer membrane protein that mediates adhesion to ECM molecules and is additionally involved in other virulence-related functions, such as binding to epithelial cells, autoagglutination, and phagocytosis resistance (reviewed in reference 11). YadA also confers serum resistance to Y. enterocolitica (1, 3, 26). Furthermore, YadA promotes the formation of densely packed microcolonies in three-dimensional collagen gels (17). YadA from Y. enterocolitica binds primarily to diverse types of collagen, whereas in Y. pseudotuberculosis, an N-terminal extension of 30 amino acids transforms YadA into a chiefly fibronectin-binding molecule (21). YadA is essential for virulence in Y. enterocolitica, as its loss leads to avirulence in mice (46, 57), but it appears not to be required for virulence in Y. pseudotuberculosis (6, 20, 25). In the third human-pathogenic species of Yersinia, the infamous Y. pestis, YadA is not expressed at all due to a frameshift in the gene (47, 55). However, two chromosomally encoded YadA-like adhesins were recently discovered in Y. pestis (16).
The structure of YadA resembles a lollipop; the C-terminal membrane anchor is followed by an extended coiled-coil stalk capped by the globular head domain at the N terminus (23). The collagen-binding activity of homotrimeric YadA has been mapped to the globular head, whose crystal structure has been solved (39). The head is a left-handed parallel β-roll, and together with the neck region that connects the α-helical stalk to the β-roll domain, it forms a very stable structure (39). The relatively flat surface of YadA does not contain any obvious features that could form the binding site, but mutagenesis studies have pinpointed several critical residues (39, 45). The affinity of YadA for human collagen type I is approximately 0.3 μM, as measured by surface plasmon resonance (39).
Collagens have a distinctive triple-helical structure (reviewed in reference 8). Formation of the collagenous triple helix requires repeats of the sequence triplet G-X-X′, where the glycine at position 1 is absolutely required. The X and X′ positions are often occupied by imino acids; X is commonly proline and X′ is usually 4-hydroxyproline (O), whose presence greatly stabilizes the triple helix. Indeed, 10% of native collagen sequences consist of the GPO triplet (42). Peptides containing GPO or GPP repeats also adopt a collagen-like triple helix (9). Using such model peptides, we showed that YadA binds as tightly to a hydroxyproline-rich collagenous triple helix as to native collagen (30). However, other studies claim that YadA binds to a specific region in the α1 chains of type I and II collagens rather than to the triple-helical collagenous conformation (49, 50).
To further investigate the YadA binding determinants in collagen, we studied the binding of YadA to libraries of triple-helical peptides spanning the entire sequence of the homotrimeric type II and type III human collagens (28, 43). These “Toolkits” are composed of overlapping peptides containing 27 amino acids of the collagen sequence. Each successive peptide advances 18 amino acids along the collagen sequence, so that there are nine unique residues in the center and nine-residue overlaps with both the previous and the following peptide. In addition, the collagen-specific sequence is flanked by the sequence GPC-(GPP)5 at the N terminus and by (GPP)5-GPCG at the C terminus to increase the stability of the triple helix.
YadA bound promiscuously to the Toolkit peptides, with a binding pattern very different from that for eukaryotic collagen-binding proteins. However, YadA bound well to several peptides which contained binding sites for host collagen-binding proteins. Although there was no clear binding site or motif in the high-binding-affinity peptides (high-binding peptides), we did find that YadA binds best to peptides with several hydroxyproline residues and a low net charge. We concluded that the YadA binding determinants in fibrillar collagens are segments with a high hydroxyproline-to-charged-residue ratio. Since these stretches are relatively common in collagens, we suggest that binding promiscuously to such sequences may be advantageous for pathogenic organisms such as Y. enterocolitica. It may allow them to adhere opportunistically to host tissues and thus aid in causing disease.
MATERIALS AND METHODS
Production and purification of YadA.
A fragment of YadA from Y. enterocolitica serotype O:3 containing the collagen-binding head group and stalk (YadA24-378) was produced and purified as described previously (30, 38). Briefly, YadA24-378 was expressed from the expression plasmid pOP-1 in Escherichia coli M15(pREP4) (Qiagen). We lysed the cells by sonication and applied the clarified supernatant to a Ni-Tris-carboxymethyl ethylene diamine (Ni-TED) column (Macherey-Nagel). The column was washed, and YadA24-378 was eluted with imidazole. The eluate was concentrated, and the concentration of YadA24-378 was estimated by measuring the absorbance at 280 nm. The resulting protein was trimeric (see Fig. S1 in the supplemental material).
Collagens and collagen-like peptides.
Bovine collagen type I was from Devro, Stirling, United Kingdom. Bovine collagen type II and type III and human collagen type IV were from Sigma. Fibrous (Ethicon) collagen type I was from Ethicon Corp., Somerville, NJ. The peptides (POG)10 and (POG)5 were purchased from Peptides International Inc., Louisville, KY. The peptide Gly− was from Innovagen AB, Lund, Sweden. The synthesis of the collagen Toolkit peptides has been described before (28, 43). The sequences of the peptides in the Toolkits are given in Table S1 in the supplemental material. Collagen-related peptide (CRP), the peptide VWF-III, and the control peptides GPP10 and GFOGER were synthesized as described previously (27, 31, 36, 43). All other peptides, except for the cyclic peptides, were synthesized as described earlier (36, 56). The synthesis of the cyclic GPO peptides is described in the supplemental material. To ensure that the peptides formed triple helices, their melting temperatures were measured by polarimetry, using a Jasco P1020 polarimeter as described previously (56). The melting temperature (Tm) values for previously unpublished peptides are summarized in Table S2 in the supplemental material.
Solid-phase binding assay (SPBA) with purified YadA.
We coated wells of Immulon 2HB plates (Thermo Scientific) with 100 μl of collagen or collagen-like peptides at 10 μg/ml in 0.01 M acetic acid, either overnight at +4°C or for 1 h at room temperature (RT). The wells were then blocked with 150 μl phosphate-buffered saline (PBS; 20 mM sodium phosphate, pH 7.4, 150 mM NaCl) containing 50 mg/ml bovine serum albumin (BSA; Sigma) and incubated at RT for 30 min. We then added 100 μl of YadA24-378 diluted to 10 μg/ml in adhesion buffer (PBS plus 1 mg/ml BSA) and incubated the plates for 1 h. We washed the wells three times with 150 μl of washing buffer (adhesion buffer plus 0.1% Tween 20) before adding 100 μl of the primary antibody diluted 1:200 in adhesion buffer. This was the monoclonal YadA antibody 3G12 (54), which recognizes a nonlinear epitope at the N-terminal end of the stalk and therefore does not interfere with the YadA-collagen interaction (2). After 1 h of incubation, the wells were washed three times and 100 μl of the secondary antibody solution added. As the secondary antibody, we used a horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (from either Dako or Santa Cruz Biotechnology) diluted 1:5,000 in adhesion buffer. For detection, we added 100 μl of substrate solution, either Immunopure TMB substrate (Thermo Scientific) or SigmaFast OPD (Sigma), and allowed the reaction to proceed until color had developed. The reaction was stopped with an equal amount of 2.5 M sulfuric acid, and the absorbance was read at 450 nm (TMB) or 490 nm (OPD). For plotting of data, the mean absorbances and standard errors of the means calculated for three replicate wells were used. The results are representative of at least two experiments.
SPBA with YadA-expressing cells.
To test the binding of YadA-expressing cells to Toolkit peptides, we used Y. enterocolitica serotype O:3 strain 6471/76 (YeO3) and, as a control, its virulence plasmid-cured derivative, 6471/76-c (YeO3-c), which does not express YadA (51). Overnight cultures were grown in medium E plus 5 mM CaCl2 (MedECa [52]) at +37 °C. For the experiments, the bacteria were diluted to an optical density at 600 nm (OD600) of 0.2 in adhesion buffer (PBS plus 0.1 mg/ml BSA).
We coated wells of Immunol 2HB plates with peptides or collagen type I and blocked them as described above. One hundred microliters of bacterial suspension was added to the wells and incubated for 1 h at RT. The plates were washed three times with 150 μl adhesion buffer, after which 100 μl of primary antibody solution (1:10 dilution in adhesion buffer) was added to the wells. The primary antibody was the monoclonal antibody A6, which reacts with the O antigen of YeO3 lipopolysaccharide (40). After 1 h of incubation at RT, we washed the wells three times as described above and added the secondary antibody solution (goat anti-mouse-HRP [P260; Dako] diluted 1:2,000 in adhesion buffer). We incubated the plates for 30 min, after which we washed the wells four times as described above. One hundred microliters of OPD substrate solution (S-2045; Dako) was added, and color was allowed to develop. The reaction was stopped by adding 100 μl of 0.5 M sulfuric acid, and the absorbance was measured at 485 nm. For plotting of data, the mean absorbances and standard errors of the means calculated for three replicate wells were used. The results are representative of two experiments.
Data analysis.
To analyze the data obtained from the binding experiments, the number of each amino acid type in the specific part of each Toolkit peptide was counted. In addition, the numbers of imino acid residues (P and O), charged residues (D, E, K, and R), and GPO repeats and the net charge of each peptide were calculated. To analyze the effect of peptide hydrophobicity on YadA binding, the mean hydrophobicity of the peptide was calculated, using a predetermined value for each amino acid residue (4), for all of the amino acids in the specific peptide sequence [i.e., not including the flanking GPC-(GPP)5 sequences]. Pearson product moment correlation coefficients were estimated using SAS, version 9.1 (SAS Institute Inc., Cary, NC). A P value of ≤0.05 was taken as statistically significant. The binding data for Toolkits II and III were analyzed separately and in combination. We used a similar approach for G-X, X-X′, and X′-G dipeptides. The frequency of each dipeptide in the Toolkit peptides was calculated, after which the Pearson correlation coefficient was estimated between binding efficiencies and the number of each dipeptide in the Toolkit peptides, again with a P value of 0.05 considered significant.
Whole-blood perfusion experiments and image analysis.
Glass coverslips (thickness 1; Menzel-Glaser, Braunschweig, Germany) were coated with type I collagen or combinations of synthetic collagen-like peptides. The peptides used were CRP [cross-linked GCO-(GPO)10-GCOG peptides], GFOGER, VWF-III [GPC-(GPP)5-GPRGQOGVMGFO-(GPP)5GPC], III-4, II-22, and III-30. Each peptide, dissolved in 0.01 M acetic acid, was used at 0.1 mg/ml in a mixture containing a total of 0.3 mg/ml of peptide. Fibrous Ethicon collagen was used at 0.1 mg/ml. Coverslips were covered with peptide solution and held in a humid chamber overnight at 4°C. After removal of excess fluid, they were blocked for 30 min with 1% BSA in HEPES buffer (36 mM NaCl, 2.7 mM KCl, 5 mM HEPES, 10 mM glucose, 2 mM MgCl2, 2 mM CaCl2, pH 7.4). The blocking buffer was then removed and the coverslip incubated for 1 h with 100 μg/ml of YadA24-378 in binding buffer (20 mM sodium phosphate, pH 8.0, 300 mM NaCl, 25 mM imidazole) or with binding buffer alone. The coverslip, in a 125-μm-deep flow chamber, was mounted on an FV300 laser scanning confocal microscope (Olympus, Southend-on-Sea, United Kingdom) and washed for 1 min with HEPES buffer. Blood from healthy volunteers free from medication for 2 weeks was collected into 40 μM Phe-Pro-Arg-chloromethylketone (PPACK; for anticoagulation) and supplemented hourly with 10 μM PPACK. Before use, the blood was incubated for 15 min with 1 μM 3,3′-dihexyloxacarbocyanine iodide (DIOC6), a lipophilic dye, to stain platelets. For the experiments, the blood was drawn through the chamber for 5 min by use of a syringe pump at a shear rate of 1,000 s−1. Residual blood was then flushed using HEPES buffer before image acquisition.
For imaging, z stacks (vertical sequences of images of a given field of view with step changes [Δz] of 0.69 μm and encompassing the entire thrombus height) were collected using an Olympus UplanFLN 40× (numerical aperture [NA], 1.30) oil-immersion objective, a field size of 360 μm by 360 μm, a confocal aperture of 60 μm, and a 488-nm laser. z stacks were exported to ImageJ 1.35 (NIH) for analysis. Thrombus surface coverage, mean thrombus height, and ZV50 were calculated from the z stacks. Mean thrombus height is a volume measurement derived by taking the total amount of thrombus present on the coverslip and expressing it in relation to the field area (the units are μm3/μm2, giving μm, a height measurement). ZV50 is the height of the center of mass of the thrombus. This gives a measure of the thrombus height and, thus, the degree to which the platelets are activated.
RESULTS
Binding of YadA to Toolkit II and Toolkit III.
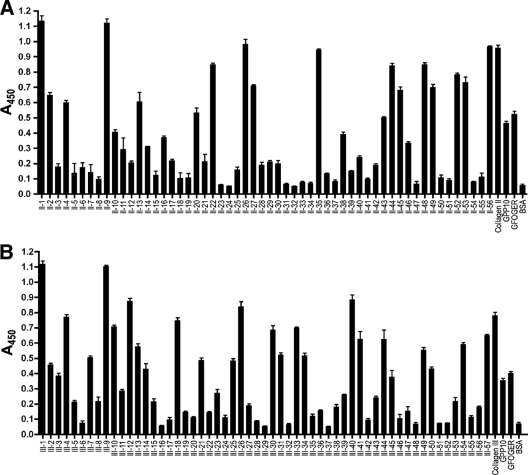
We used SPBA to investigate the binding of YadA to the two Toolkits. As controls, we included the peptides GPP10 [GPC-(GPP)10-GPC] (43) and GFOGER. The latter contains the high-affinity binding site for α2β1 integrin flanked by five GPP repeats (27). We also included appropriate native collagens for comparison, with BSA as a negative control. YadA bound to most peptides in both Toolkits, but with widely varying relative affinities (Fig. 1). This indicates that the GPC-(GPP)5 sequences flanking the Toolkit peptides are not enough alone to promote tight YadA binding, because otherwise YadA would bind to all Toolkit peptides. Presumably, (GPP)5 is not long enough to promote binding, though YadA did bind the control peptide GPP10, but at a significantly lower level than those for the collagens and many Toolkit peptides.
FIG. 1.
Binding of YadA to collagen Toolkits in SPBA. Wells of microtiter plates were coated with Toolkit peptides and probed with YadA. Panel A shows the binding of YadA to Toolkit II, and panel B shows the binding of YadA to Toolkit III. Collagens of the appropriate type, GPP10, and GFOGER were included as controls. A triple-helical conformation is known to be necessary for YadA binding (30). Binding to BSA shows background levels. Toolkit III contains an internal control for the triple-helical conformation, as peptides III-6 and III-52 do not form a collagenous triple helix, unlike the other Toolkit peptides (28, 43). As expected, YadA bound to neither of these peptides. The columns show the mean absorbances at 450 nm for three replicate wells; error bars denote standard errors of the means.
The binding behaviors of YadA with Toolkits II and III were similar. In Toolkit II, five peptides (II-1, II-9, II-26, II-35, and II-56) bound at least as well as native collagen type II (Fig. 1A), while other peptides showed very little or no binding, i.e., giving a signal of 0.1 or below (Fig. 1A). The rest showed intermediate binding levels. Similarly, for Toolkit III, YadA bound strongly to five peptides (III-1, III-9, III-12, III-26, and III-40), and again, several peptides did not bind YadA (Fig. 1B), with most peptides giving an intermediate response. YadA is thus a fairly promiscuous binder; there is clearly no single, unique, high-affinity binding site(s) for it in either collagen type II or collagen type III.
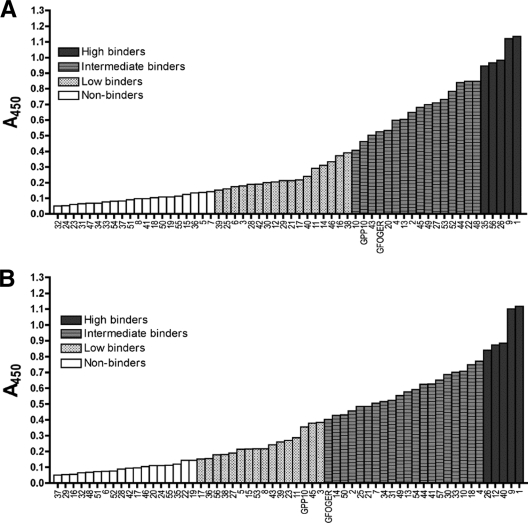
When sorted by absorbance at 450 nm, the peptides did not form a bimodal distribution (binders and nonbinders) but rather revealed a continuum of relative affinities (Fig. 2). We decided that a response that was less than or equal to three times the signal of BSA, i.e., 0.15 absorbance unit (AU), represented background-level binding, so these peptides were classed as nonbinders. Binding between 0.15 and 0.4 AU we considered low binding, whereas binding at or above the level of collagen (0.9 AU for Toolkit II and 0.8 AU for Toolkit III) we considered high binding. The rest of the peptides were classed as intermediate binders. For both Toolkits, the control peptide GFOGER fell into the intermediate category, whereas GPP10 was classed as a low binder in Toolkit III but just fell into the intermediate category in Toolkit II. However, in both Toolkits, the response given by GPP10 lay very close to the boundary between low and intermediate binding (Fig. 2). This was expected, as YadA binds to (PPG)10 300-fold more weakly than to (POG)10 (30), and GPP10 and (PPG)10 have virtually identical sequences.
FIG. 2.
Distributions of Toolkit peptides sorted according to YadA binding response. Toolkit peptides were classed as nonbinders (response of <0.15 AU at 450 nm; white columns), low binders (response between 0.15 and 0.4 AU; striped columns), intermediate binders (response of >0.4, but below that of the corresponding collagen; gray columns), and high binders (response above that of the corresponding collagen; black columns). The peptides were sorted according to mean absorbances at 450 nm for three replicate wells, as shown in Fig. 1. Panel A shows the sorted peptides from Toolkit II and panel B the sorted peptides from Toolkit III.
YadA-expressing bacteria bind to Toolkit peptides.
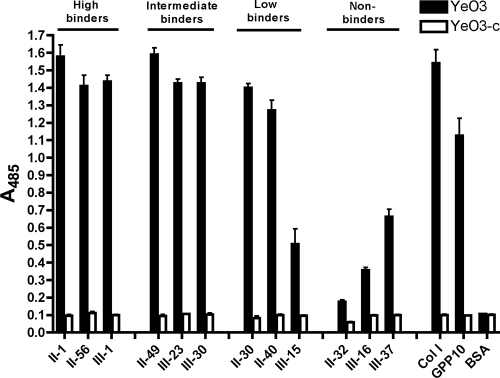
To test whether bacteria expressing YadA on the cell surface behave in a similar manner to the recombinant protein, we performed an adhesion assay with Y. enterocolitica and a subset of the Toolkit peptides. We randomly chose three peptides from each binding category (high, intermediate, low, and nonbinders) so that overall there was an equal representation from both Toolkits. The YadA-expressing strain YeO3 bound well to high- and intermediate-binding Toolkit peptides, giving an almost collagen-level response (Fig. 3). In this setup, low-binding peptides exhibited appreciable binding, and even the nonbinders showed a response above background levels (Fig. 3). The tight binding exhibited by the cells to the peptides with intermediate and low relative affinities can be explained through the avidity imparted by the high density of YadA on the cell surface (23). Intermediate relative affinities of the peptides for recombinant YadA molecules translate into strong binding when numerous adhesins bind cooperatively. Even when the affinity of recombinant YadA for the peptide was at background levels, YadA-expressing cells bound well, showing that residual affinity for the collagen triple helix is enough to promote appreciable binding. The negative-control strain YeO3-c failed to bind any of the Toolkit peptides, demonstrating that the adhesion is YadA mediated.
FIG. 3.
Binding of YadA-expressing bacteria to Toolkit peptides in SPBA. Bacteria were incubated in wells of a microtiter plate coated with a subset of Toolkit peptides. The results show the mean absorbances at 493 nm for three replicate wells; error bars denote standard errors of the means. YeO3 is a YadA-positive strain, whereas YeO3-c does not express YadA. Collagen type I, GPP10, and BSA were included as controls.
Analysis of binding data.
All 10 peptides binding YadA with high relative affinities (Table 1) are rich in imino acids, particularly hydroxyprolines, and most have few charged residues, or the charged residues are clustered so as to provide substantial stretches (at least three G-X-X′ repeats) without charges. The paucity of lysine residues is notable; there is just a single lysine in the high-binding peptides. No consensus sequence or clear binding motif can be discerned from the sequences, though some of the peptides (II-26, II-35, and III-12) contain a short charged motif with the consensus sequence ERGET. Peptide II-56 contains a similar sequence (RSGET), and III-26 contains a motif with similar charge properties to ERGET (DKGDT). However, we do not believe that this is a binding motif, as similar sequences can be found in nonbinding peptides (see Table S3 in the supplemental material), such as II-24, II-55, III-35, and III-48. In the high-binding peptides, imino acids or other hydrophobic residues flank the ERGET motifs, but this is also the case in nonbinders such as II-8 and III-36. ERGET is thus not a binding determinant for YadA.
TABLE 1.
Sequences of high-binding peptides
| Peptide | A450 | Sequencea |
|---|---|---|
| II-1 | 1.136 | GPMGPMGPRGPOGPAGAOGPQGFQGNO |
| II-9 | 1.122 | GNDGQOGPAGPOGPVGPAGGOGFOGAO |
| II-26 | 0.983 | GERGEQGAOGPSGFQGLOGPOGPOGEG |
| II-35 | 0.946 | GSAGARGAOGERGETGPOGPAGFAGPO |
| II-56 | 0.968 | GPRGRSGETGPAGPOGNOGPOGPOGPO |
| III-1 | 1.119 | GLAGYOGPAGPOGPOGPOGTSGHOGSO |
| III-9 | 1.102 | GLOGAAGARGNDGARGSDGQOGPOGPO |
| III-12 | 0.875 | GQRGEOGPQGHAGAQGPOGPOGINGSO |
| III-26 | 0.840 | GPOGPTGPGGDKGDTGPOGPQGLQGLO |
| III-40 | 0.886 | GAAGFOGARGLOGPOGSNGNOGPOGPS |
Charged residues (D, E, K, and R) are shown in bold. Stretches of at least three continuous G-X-X′ repeats containing no charged residues are underlined. The flanking GPC-(GPP)5 sequences are not shown.
Since we were unable to determine any clear binding determinants directly from the sequences, we performed correlation analyses between YadA binding and the number of each amino acid present in each peptide. We examined the data from Toolkits separately and in combination (see Table S4 in the supplemental material). Binding correlated well with hydroxyproline content; the Pearson correlation coefficient was between 0.3 and 0.5, which was at a significant level (P < 0.02) for all three data sets (see Table S4 in the supplemental material). The correlation with proline was not as strong, but though it did not reach significance in the Toolkit II data, the correlation was significant (P < 0.03) for Toolkit III and the combined Toolkit data. Phenylalanine too showed a significant positive correlation in Toolkit II and the combined Toolkits.
We also noted several statistically significant negative correlations for charged residues. Lysine showed a strong negative correlation with binding (P < 0.0001) (see Table S2 in the supplemental material) in all the data sets, and glutamic and aspartic acid had significant negative correlations in the combined Toolkit data (see Table S4 in the supplemental material). Of the charged residues, only arginine failed to show a clear correlation in this analysis. None of the other amino acids had any noticeable effect on binding.
We also examined the distributions of common X-X′ dipeptides in the high- and low-binding peptides (see Table S5 in the supplemental material). PO was highly enriched in the high-binding set, as were certain other hydroxyproline-containing dipeptides, but charged dipeptides occurred less frequently than expected if the distribution were random. Conversely, the low-binding peptides had more charged dipeptides than expected (see Table S5 in the supplemental material). To quantitate these results, we calculated the correlations between dipeptides and binding. When all of the Toolkit data were considered, there were significant positive correlations for the dipeptides PO and FQ in the X-X′ dipeptides but significant negative correlations for the dipeptides PK and EK (see Table S6 in the supplemental material). Among the G-X dipeptides, GF and GP had significant positive correlations, and GE exhibited the only significant negative correlation. Among the X′-G dipeptides, the only two with a significant correlation were OG (positive) and KG (negative). Phenylalanine and proline are thus favored at position X, whereas glutamate is disfavored. Lysine is disfavored in the X′ position, and as expected, hydroxyproline is preferred.
To gain further insight into binding determinants, we tested the correlations between binding, imino acid content, the number of GPO repeats in the peptide, peptide hydrophobicity, positively charged residue content, negatively charged residue content, and charged residue content (Table 2). To calculate hydrophobicities, we used the scale of Black and Mould (4) because it includes a hydrophobicity value for hydroxyproline. All of these correlations were strong and statistically significant (P < 0.0005). However, the net charge of the peptide (number of positively charged residues minus number of negatively charged residues) did not correlate with binding. Interestingly, peptides with a large number of charges that were neutralized, i.e., where the net charge was 0 ± 1, bound YadA better than did peptides with an equal number of charged residues but a larger net charge (see Fig. S2 in the supplemental material). Neutralizing charges can partially compensate for the negative effect of charged residues on YadA binding, explaining why ERGET can occur in high binders. Consistent with this, the correlation between the modulus of the net charge and YadA binding showed a negative trend (Table 2).
TABLE 2.
Correlation of binding with different Toolkit peptide propertiesc
| Property | Correlation | P value |
|---|---|---|
| No. of positively charged residues (K and R)a | −0.47 | <0.0001 |
| No. of negatively charged residues (D and E) | −0.33 | 0.0004 |
| No. of charged residues | −0.48 | <0.0001 |
| Net charge of peptide | −0.08 | 0.41 |
| Modulus of net charge of peptide | −0.12 | 0.19 |
| No. of imino acids | 0.48 | <0.0001 |
| No. of GPO repeats | 0.48 | <0.0001 |
| Mean hydrophobicityb | 0.42 | <0.0001 |
We did not consider histidine to be charged, as 90% of histidine residues are deprotonated at the assay pH (7.4).
Calculated using values from reference 4.
In all cases, 111 peptides were analyzed.
The number and relative positions of GPO repeats do not affect YadA binding.
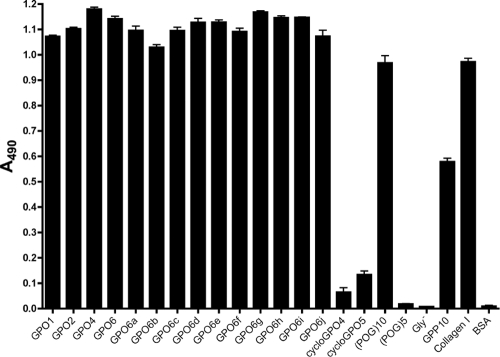
YadA binds strongly to (POG)10 and more weakly to (PPG)10, and our analyses showed that the presence of a large number of GPO repeats in Toolkit peptides promotes binding. To determine how the number of GPO repeats affects the binding of YadA and whether the relative positions of these repeats have an impact on binding, we investigated the binding of YadA to a series of peptides with various numbers of GPO repeats in a GPP background (Table 3). GPO1, GPO2, GPO4, and GPO6 form a series with increasing amounts of GPO repeats (56). GPO6a to -j have equal numbers of GPO repeats, but the spacing between the GPO triplets is different. YadA bound strongly to all peptides containing GPO repeats (Fig. 4). YadA did not bind the non-triple-helical Gly− peptide (32) and also failed to bind (POG)5, which is too short to form a stable triple helix at room temperature (48). Furthermore, YadA did not bind to the cyclic peptides cycloGPO4 and cycloGPO5. However, comparing GPP10 and GPO1, even a single GPO repeat was enough to promote strong YadA binding (Fig. 4), and the GPO6a-to-GPO6j series demonstrates that the relative positions of GPO repeats have no significant effect on the strength of binding. A single GPO triplet was also sufficient to promote YadA aggregation, as assessed by native gel electrophoresis (data not shown).
TABLE 3.
Sequences of GPO and GKO-GPO peptides
| Peptide name | Sequence |
|---|---|
| GPO1 | GCP-(GPP)4-GPO-(GPP)5-GCPG |
| GPO2 | GCP-(GPP)4-(GPO)2-(GPP)4-GCPG |
| GPO4 | GCP-(GPP)3-(GPO)4-(GPP)3-GCPG |
| GPO6 | GCP-(GPP)2-(GPO)6-(GPP)2-GCPG |
| GPO6a | GPC-GPO-(GPP-GPO)5-GPC |
| GPO6b | GPC-GPP-(GPO)2-GPP-(GPO)2-GPP-(GPO)2-(GPP)2-GPC |
| GPO6c | GPC-(GPO)2-(GPP)2-(GPO)2-(GPP)2-(GPO)2-GPP-GPC |
| GPO6d | GPC-(GPO)2-(GPP)2-(GPO)2-(GPP)3-(GPO)2-GPC |
| GPO6e | GPC-(GPP)2-(GPO)6-(GPP)3-GPC |
| GPO6f | GPC-(GPP)2-(GPO)3-GPP-(GPO)3-(GPP)2-GPC |
| GPO6g | GPC-GPP-(GPO)3-(GPP)2-(GPO)3-(GPP)2-GPC |
| GPO6h | GPC-GPP-(GPO)3-(GPP)3-(GPO)3-GPP-GPC |
| GPO6i | GPC-(GPO)3-(GPP)4-(GPO)3-GPP-GPC |
| GPO6j | GPC-(GPO)3-(GPP)5-(GPO)3-GPC |
| GKO-GPO1 | GPC-(GPP)4-GKO-GPO-GKO-(GPP)3-GPC |
| GKO-GPO3 | GPC-(GPP)3-GKO-(GPO)3-GKO-(GPP)2-GPC |
| GKO-GPO4 | GPC-(GPP)2-GKO-(GPO)4-GKO-(GPP)2-GPC |
| GKO-GPO6 | GPC-GPP-GKO-(GPO)6-GKO-GPP-GPC |
| Gly− | (POG)4-PO-(POG)5 |
FIG. 4.
Binding of YadA to GPO peptides in SPBA. Wells of a microtiter plate were coated with peptides with different numbers of GPO repeats (GPO1 to GPO6) or with different spacings between the same number of GPO triplets (GPO6a to -j) and then were probed with YadA. As controls, we included (POG)10, (POG)5, GPP10, and the non-triple-helical peptide Gly−, which is the same as (POG)10 but lacks a central glycine residue (32). Collagen type I was included to assess binding levels. The BSA column shows background levels. The columns show the mean absorbances at 490 nm for three replicate wells; error bars denote standard errors of the means. The difference between GPP10 and GPO1 shows that a single GPO triplet in a GPP background is sufficient for tight binding.
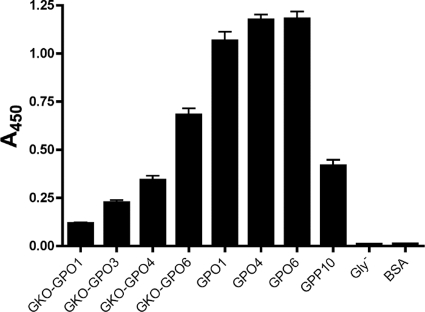
Effect of GKO repeats and their spacing on YadA binding.
To examine how long a minimal YadA binding sequence might be, we synthesized peptides with various numbers of GPO repeats between two GKOs. If the spacing between the GKO triplets was not long enough, YadA should bind more weakly than to the corresponding sequence without lysines. We synthesized four such peptides, GKO-GPO1, GKO-GPO3, GKO-GPO4, and GKO-GPO6 (Table 3), and performed an SPBA experiment to determine the binding of YadA. The GKO triplets did indeed inhibit YadA binding (Fig. 5). The binding of YadA to GKO-GPO1 was only slightly above the background. Increasing the spacing between the residues also increased the relative affinity of YadA for the peptides, in a graded fashion, i.e., YadA bound more tightly to GKO-GPO6 than to GKO-GPO4, which in turn gave a stronger response than that of GKO-GPO3. However, even a spacing of six residues between the GKO repeats was not enough in this setting to give the high-level binding seen with the GPO peptides but, rather, resulted in an intermediate level of binding. YadA thus requires a stretch of more than six uncharged G-X-X′ triplets to achieve strong binding.
FIG. 5.
Binding of YadA to GKO-GPO peptides in SPBA. Wells of a microtiter plate were coated with peptides with different numbers of GPO repeats flanked on either side by GKO and then were probed with YadA. The peptides GPO1, GPO4, GPO6, GPP10, and Gly− were used as controls. The BSA column shows background levels. The columns show the mean absorbances at 450 nm for three replicate wells; error bars denote standard errors of the means.
Binding of YadA to fibrous and network-forming collagens.
YadA is known to bind several fiber-forming (fibrillar) collagens, e.g., types I, II, III, and V (12, 49). However, the present studies were carried out using single-collagen triple helices. We wished to investigate whether YadA would bind to fibrous (i.e., a fibrillar assembly of triple-helical monomers) collagen type I. YadA bound as strongly to fibrous Ethicon collagen as to monomeric type II and type III collagens (see Fig. S3 in the supplemental material). In addition, YadA also bound to CRP, which is composed of cross-linked triple-helical GCO-(GPO)10-GCOG peptides (see Fig. S3 in the supplemental material).
YadA also binds to the network-forming collagen type IV. The binding site for YadA in collagen type IV is reported to reside in the tetrameric 7sL fragment, not in the triple-helical regions (15). However, the head domain of YadA is also responsible for binding to collagen type IV (57). We therefore wished to investigate whether there might be separate binding mechanisms for different classes of collagens. To do this, we studied the effect of blocking the YadA-collagen interaction with the peptides (POG)10 and (POG)5. (POG)10 blocked the binding of YadA to both type I and type IV collagens, whereas (POG)5 failed to do so (see Fig. S4 in the supplemental material). This suggests that YadA binds to both fibrillar and network-forming collagens by the same mechanism, i.e., through triple-helical regions.
Blocking of platelet adhesion and activation by YadA.
YadA binds well to a number of peptides that contain binding sites for eukaryotic collagen-binding proteins. For example, the leukocyte-associated Ig-like receptors (LAIRs) 1 and 2 bind peptides II-56, III-1, and III-30 (29), and discoidin domain receptor 2 (DDR2) binds peptides II-13, II-22, and II-44 (28), which are all strong- or intermediate-affinity ligands for YadA. Platelet deposition upon collagenous substrates depends upon three receptors, namely, integrin α2β1, the activating receptor glycoprotein VI (GpVI), and GpIb, which binds to the von Willebrand factor (VWF) A1 domain, while in turn, the VWF A3 domain binds a single site in collagens (31). Among the integrin α2β1-binding peptides, III-4, III-7, and III-31 (43), as well as the high-affinity ligand GFOGER (27), all bind YadA at an intermediate or stronger level. The best ligand for GpVI from the Toolkits is III-30 (24), an intermediate YadA binder, like II-22 and II-23, which each contain the VWF recognition motif GXRGQOGVMGFO. We recently showed that peptides containing GFOGER, (GPO)10, and GPRGQOGVMGFO will support full thrombus deposition from flowing blood (41).
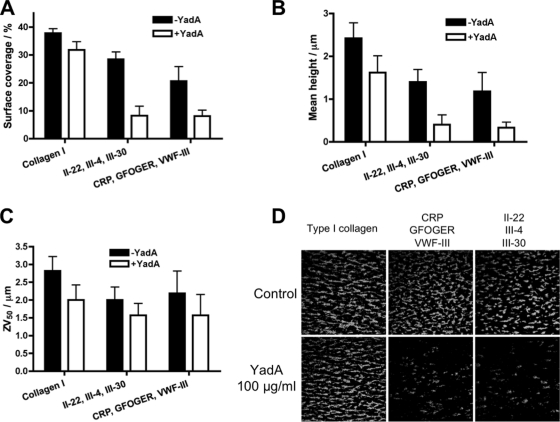
To test whether YadA can interfere with the interaction between human proteins and collagen, we blocked the binding of platelets to immobilized collagen with recombinant YadA. This was performed by drawing blood across coverslips coated with collagen or collagen-like peptides at arterial shear rates. We used our proven set of platelet-binding peptides (CRP, GFOGER, and VWF-III) and a second set, III-4, II-22, and III-30, that were selected as good YadA binders which would also provide platelet-adhesive and -activating properties for thrombus deposition. Blocking with YadA reduced the binding of platelets to both collagen type I and peptides (Fig. 6). In addition, we measured the mean thrombus height and the ZV50 value, which is the height of the center of mass of the thrombus. These values give an indication of the level of platelet activation. Blocking with YadA resulted in a slight decrease in the binding of platelets to collagen type I. However, when each set of collagen-like peptides was blocked with YadA, the decrease was much more pronounced (Fig. 6). YadA was able to inhibit primary (integrin α2β1- and VWF-mediated) adhesion of platelets to these peptides and, as a consequence, to reduce mean thrombus height. Blocking with YadA did not have a large effect on platelet aggregate formation, measured as the ZV50, showing that any adherent platelets became activated and recruited further platelets to the growing thrombus. Platelets express multiple collagen receptors, both directly and indirectly, which possibly explains why YadA did not have as large an effect on the binding of platelets to collagen as to the collagen-like peptides. Collagens contain several binding sites for platelets, and the binding regions for collagen receptors in the heterotrimeric collagen type I have not been investigated directly (see reference 22 for a review). Thus, the overlap in platelet binding sites with YadA is unknown for this collagen type. However, when bound to peptides with an intermediate or high relative affinity for YadA, the bacterial adhesin was clearly able to inhibit the binding of platelets. These data demonstrate that YadA can impede the binding of indigenous proteins to collagen.
FIG. 6.
Inhibition of platelet adhesion to collagen type I or collagen-like peptides by YadA. Coverslips were coated with collagen type I or a mixture or three collagen-like peptides. Blood (incubated with DIOC6 to stain platelets) was drawn across coated coverslips at an arterial shear rate (1,000 s−1), and the adhesion of platelets was determined by confocal microscopy. (A) Surface coverage (%) by platelets. (B) Mean height of thrombi. (C) ZV50 value, i.e., the mean height of the center of mass of the thrombi, a value that measures platelet activation. Results from control experiments without YadA (−YadA) are shown with black columns. White columns show the results from experiments where the coverslips were previously incubated with YadA (+YadA). The columns represent the mean values for four replicates, with standard errors of the means. (D) Representative confocal microscopy images of platelets binding to coverslips coated with collagen type I or a mixture of three collagen-like peptides. The lower row shows the effects of blocking with YadA (100 μg/ml) before running blood over the coverslips; the upper row shows controls.
DISCUSSION
The YadA-collagen interface.
YadA is the first bacterial collagen-binding adhesin to be studied using the collagen Toolkits. The binding determinants for YadA in fibrillar collagens are triple-helical segments rich in imino acids, especially hydroxyprolines, and with several GPO repeats and a small number of charged residues. A triple-helical conformation is required but, contrary to our previous claim (30), is not sufficient for tight binding. However, a specific sequence is not required for YadA binding. The promiscuous nature of YadA binding is highlighted when it is expressed on the cell surface, as YadA molecules cover virtually the entire surface of the cell (23). The binding of Y. enterocolitica cells to collagens is thus multivalent, so cells can adhere even to collagenous sequences with low affinity for YadA.
A minimal high-relative-affinity YadA binding site contains at least six G-X-X′ triplets, as shown by the experiments with the GKO-GPO peptides. However, this is in contrast to the tight binding we observed for certain peptides, such as II-35 and III-19, which have, at most, three uncharged G-X-X′ repeats (Table 1). The uncharged, GPO-rich segments in II-35 and III-19 are, however, located at the edges of the specific sequence. Our explanation is that in these cases, the flanking (GPP)5 sequences participate in forming a binding site. This presumably happens in other peptides as well.
A binding site length of more than six G-X-X′ repeats is consistent with our proposed model of YadA binding to a collagen-mimicking peptide (39). In the model, the peptide lies diagonally across the face of YadA, with 7 of the 10 G-X-X′ triplets contacting the surface of YadA (see Fig. S5 in the supplemental material). The (GPP)5 sequences flanking the Toolkit peptides are also not sufficient for YadA binding, supporting the conclusion that a longer sequence is required. A binding site of more than six G-X-X′ triplets is significantly longer than the target sequences of human collagen-binding proteins. For example, in the crystal structure of integrin α2β1 complexed with GFOGER, the contact region spans only approximately 2.5 triplets (13). Similarly, GpVI appears to interact with only two GPO repeats (56).
The Toolkit peptides represent sequences from fibrillar collagens. We propose that YadA binds to triple-helical regions in the network-forming collagen type IV as well. The binding site for YadA has been reported to reside outside the triple-helical domain of collagen type IV, but we were able to inhibit the binding of YadA to collagen type IV with (POG)10. The blocking effect exhibited by (POG)10 might be due to steric hindrance, if indeed there are distinct mechanisms for binding to fibrillar and network-forming collagens. However, our results suggest that this is not so: the mechanism of binding to different types of collagens is the same, i.e., recognition of and binding to triple-helical segments. But the mechanism by which YadA recognizes a wide variety of triple-helical ligands still remains to be elucidated: how is a single GPO sufficient to elicit tight binding?
Promiscuous binding may be advantageous for pathogens such as Y. enterocolitica.
Promiscuous binding of the kind exhibited by Y. enterocolitica and mediated by YadA may be a commonly used strategy among bacterial pathogens to adhere to host tissues. Indeed, some previous work supports this hypothesis. One example is the collagen-binding domain of Clostridium histolyticum collagenase (ColG). ColG binds (POG)n and (PPG)n when n is large enough to allow triple helix formation (33). Another example is the Staphylococcus aureus collagen adhesin CNA, which binds to multiple sites in collagen and to triple-helical GPO and GPP peptides (63). In the crystal structure of the CNA collagen-binding region complexed with a (GPO)4-GPRGRT-(GPO)4 peptide, the contacts are through the GPO triplets rather than through the more specific sequence in the middle of the peptide (63). Several other bacteria, including Aggregatibacter actinomycetemcomitans, Bartonella henselae, Enterococcus faecalis, and Streptococcus pyogenes, express proteins that bind diverse collagen types, which argues against a specific binding sequence in collagens for these proteins (35, 37, 44, 58, 59). As in the case of YadA, other factors, such as the imino acid content or distribution of charged residues, may be the binding determinants for these proteins as well. Having the ability to bind promiscuously to several sites rather than to a single, high-affinity binding site could be advantageous for pathogenic bacteria, allowing pathogens to gain a foothold in tissues even when presented with only limited collagenous surfaces.
Interaction of YadA and host collagen-binding proteins.
The binding of a number of eukaryotic proteins to the Toolkits has been studied previously, including that of integrin α2β1, von Willebrand factor, DDR2, SPARC, and GpVI (reviewed in reference 14). Recently, the binding of LAIR-1 and -2 to the Toolkits was investigated (29). In all of these cases, the proteins clearly bound to a single or just a few peptides, and the Toolkit peptides could be grouped unambiguously into two populations, binders and nonbinders. This is in marked contrast to the case for YadA, which bound the Toolkit peptides with a continuum of affinities. Similarly, the eukaryotic proteins have a clear binding motif, whereas YadA binds to hydroxyproline-rich stretches with a low net charge, but without a particular sequence preference.
Nevertheless, some of the peptides that bind YadA also bind human proteins. As a test case, we investigated the effect of blocking with YadA on the interaction of platelets with collagen or collagen-like peptides. Our blocking experiments showed that YadA can inhibit the binding of platelets to collagen in vitro. It is tempting to speculate that this may contribute to the bleeding found in severe infection with Y. enterocolitica (62). YadA may also competitively interfere with collagen-mediated regulatory processes in vivo, as hypothesized earlier by Skurnik (53). The spectrum of symptoms caused by Y. enterocolitica includes a number of inflammatory disorders, such as reactive arthritis, erythema nodosum, and Reiter's syndrome (7), all mediated by host immune responses. In a rat model of reactive arthritis, YadA played a major role in eliciting synovial inflammation (18, 19), even though no proliferative bacteria were found in the synovial tissue at the height of arthritis. Our data can explain this: YadA-containing outer membrane fragments or even YadA itself could effectively compete with host proteins for binding sites, thus disturbing host immune responses.
LAIR-1 is a particularly interesting protein in this regard. It is an inhibitory receptor of immune cells that modulates immunological responses, and it is activated upon binding to collagen (34). Inhibition of LAIR-1 signaling through competition with YadA could be partly responsible, in theory, for the inflammation seen in the reactive arthritis or erythema nodosum due to Yersinia infections. Both DDRs and GpVI are also implicated in inflammatory arthritis (5, 60). The possible link between YadA, host proteins, and the sequelae of yersiniosis merits investigation. This and efforts to obtain a crystal structure of YadA in complex with a collagen-like peptide are our future research directions. A crystal of the complex would provide the missing mechanistic information and is clearly the next step in understanding the YadA-collagen interaction.
Supplementary Material
Acknowledgments
This work was funded by the Sigrid Juselius Foundation, the Academy of Finland (grant 114752 to A.G. and grant 114075 to M.S.), The Wellcome Trust, The Medical Research Council, and the British Heart Foundation.
We thank Samir Hamaia (Department of Biochemistry, University of Cambridge) for help with the binding experiments and David Slatter (Department of Biochemistry, University of Cambridge) for doing the polarimetry measurements. We also acknowledge Vesa Elovaara for writing the program with which we calculated the number of each amino acid in the peptides.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 3 May 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Biedzka-Sarek, M., H. Jarva, H. Hyytiäinen, S. Meri, and M. Skurnik. 2008. Characterization of complement factor H binding to Yersinia enterocolitica serotype O:3. Infect. Immun. 76:4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biedzka-Sarek, M., S. Salmenlinna, M. Gruber, A. N. Lupas, S. Meri, and M. Skurnik. 2008. Functional mapping of YadA- and Ail-mediated binding of human factor H to Yersinia enterocolitica serotype O:3. Infect. Immun. 76:5016-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biedzka-Sarek, M., R. Venho, and M. Skurnik. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 73:2232-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, S. D., and D. R. Mould. 1991. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal. Biochem. 193:72-82. [DOI] [PubMed] [Google Scholar]
- 5.Boilard, E., P. A. Nigrovic, K. Larabee, G. F. Watts, J. S. Coblyn, M. E. Weinblatt, E. M. Massarotti, E. Remold-O'Donnell, R. W. Farndale, J. Ware, and D. M. Lee. 2010. Platelets amplify inflammation in arthritis via collagen-dependent microparticle production. Science 327:580-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bölin, I., and H. Wolf-Watz. 1984. Molecular cloning of the temperature-inducible outer membrane protein 1 of Yersinia pseudotuberculosis. Infect. Immun. 43:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodsky, B., and J. A. Ramshaw. 1997. The collagen triple-helix structure. Matrix Biol. 15:545-554. [DOI] [PubMed] [Google Scholar]
- 9.Brodsky, B., G. Thiagarajan, B. Madhan, and K. Kar. 2008. Triple-helical peptides: an approach to collagen conformation, stability, and self-association. Biopolymers 89:345-353. [DOI] [PubMed] [Google Scholar]
- 10.Cover, T. L., and R. C. Aber. 1989. Yersinia enterocolitica. N. Engl. J. Med. 321:16-24. [DOI] [PubMed] [Google Scholar]
- 11.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291:209-218. [DOI] [PubMed] [Google Scholar]
- 12.Emödy, L., J. Heesemann, H. Wolf-Watz, M. Skurnik, G. Kapperud, P. O'Toole, and T. Wadström. 1989. Binding to collagen by Yersinia enterocolitica and Yersinia pseudotuberculosis: evidence for yopA-mediated and chromosomally encoded mechanisms. J. Bacteriol. 171:6674-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emsley, J., C. G. Knight, R. W. Farndale, M. J. Barnes, and R. C. Liddington. 2000. Structural basis of collagen recognition by integrin α2β1. Cell 101:47-56. [DOI] [PubMed] [Google Scholar]
- 14.Farndale, R. W., T. Lisman, D. Bihan, S. Hamaia, C. S. Smerling, N. Pugh, A. Konitsiotis, B. Leitinger, P. G. de Groot, G. E. Jarvis, and N. Raynal. 2008. Cell-collagen interactions: the use of peptide Toolkits to investigate collagen-receptor interactions. Biochem. Soc. Trans. 36:241-250. [DOI] [PubMed] [Google Scholar]
- 15.Flügel, A., H. Schulze-Koops, J. Heesemann, K. Kühn, L. Sorokin, H. Burkhardt, K. von der Mark, and F. Emmrich. 1994. Interaction of enteropathogenic Yersinia enterocolitica with complex basement membranes and the extracellular matrix proteins collagen type IV, laminin-1 and -2, and nidogen/entactin. J. Biol. Chem. 269:29732-29738. [PubMed] [Google Scholar]
- 16.Forman, S., C. R. Wulff, T. Myers-Morales, C. Cowan, R. D. Perry, and S. C. Straley. 2008. yadBC of Yersinia pestis, a new virulence determinant for bubonic plague. Infect. Immun. 76:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freund, S., B. Czech, K. Trülzsch, N. Ackermann, and J. Heesemann. 2008. Unusual, virulence plasmid-dependent growth behavior of Yersinia enterocolitica in three-dimensional collagen gels. J. Bacteriol. 190:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gripenberg-Lerche, C., M. Skurnik, and P. Toivanen. 1995. Role of YadA-mediated collagen binding in arthritogenicity of Yersinia enterocolitica serotype O:8: experimental studies with rats. Infect. Immun. 63:3222-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gripenberg-Lerche, C., M. Skurnik, L. Zhang, K. O. Söderström, and P. Toivanen. 1994. Role of YadA in arthritogenicity of Yersinia enterocolitica serotype O:8: experimental studies with rats. Infect. Immun. 62:5568-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, Y. W., and V. L. Miller. 1997. Reevaluation of the virulence phenotype of the inv yadA double mutants of Yersinia pseudotuberculosis. Infect. Immun. 65:327-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heise, T., and P. Dersch. 2006. Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc. Natl. Acad. Sci. U. S. A. 103:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herr, A. B., and R. W. Farndale. 2009. Structural insights into the interactions between platelet receptors and fibrillar collagen. J. Biol. Chem. 284:19781-19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvis, G. E., N. Raynal, J. P. Langford, D. J. Onley, A. Andrews, P. A. Smethurst, and R. W. Farndale. 2008. Identification of a major GpVI-binding locus in human type III collagen. Blood 111:4986-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapperud, G., E. Namork, M. Skurnik, and T. Nesbakken. 1987. Plasmid-mediated surface fibrillae of Yersinia pseudotuberculosis and Yersinia enterocolitica: relationship to the outer membrane protein YOP1 and possible importance for pathogenesis. Infect. Immun. 55:2247-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirjavainen, V., H. Jarva, M. Biedzka-Sarek, A. M. Blom, M. Skurnik, and S. Meri. 2008. Yersinia enterocolitica serum resistance proteins YadA and Ail bind the complement regulator C4b-binding protein. PLoS Pathog. 4:e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knight, C. G., L. F. Morton, A. R. Peachey, D. S. Tuckwell, R. W. Farndale, and M. J. Barnes. 2000. The collagen-binding A-domains of integrins α1β1 and α2β1 recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J. Biol. Chem. 275:35-40. [DOI] [PubMed] [Google Scholar]
- 28.Konitsiotis, A. D., N. Raynal, D. Bihan, E. Hohenester, R. W. Farndale, and B. Leitinger. 2008. Characterization of high affinity binding motifs for the discoidin domain receptor DDR2 in collagen. J. Biol. Chem. 283:6861-6868. [DOI] [PubMed] [Google Scholar]
- 29.Lebbink, R. J., N. Raynal, T. de Ruiter, D. G. Bihan, R. W. Farndale, and L. Meyaard. 2009. Identification of multiple potent binding sites for human leukocyte associated Ig-like receptor LAIR on collagens II and III. Matrix Biol. 28:202-210. [DOI] [PubMed] [Google Scholar]
- 30.Leo, J. C., H. Elovaara, B. Brodsky, M. Skurnik, and A. Goldman. 2008. The Yersinia adhesin YadA binds to a collagenous triple-helical conformation but without sequence specificity. Protein Eng. Des. Sel. 21:475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisman, T., N. Raynal, D. Groeneveld, B. Maddox, A. R. Peachey, E. G. Huizinga, P. G. de Groot, and R. W. Farndale. 2006. A single high-affinity binding site for von Willebrand factor in collagen III, identified using synthetic triple-helical peptides. Blood 108:3753-3756. [DOI] [PubMed] [Google Scholar]
- 32.Long, C. G., E. Braswell, D. Zhu, J. Apigo, J. Baum, and B. Brodsky. 1993. Characterization of collagen-like peptides containing interruptions in the repeating Gly-X-Y sequence. Biochemistry 32:11688-11695. [DOI] [PubMed] [Google Scholar]
- 33.Matsushita, O., T. Koide, R. Kobayashi, K. Nagata, and A. Okabe. 2001. Substrate recognition by the collagen-binding domain of Clostridium histolyticum class I collagenase. J. Biol. Chem. 276:8761-8770. [DOI] [PubMed] [Google Scholar]
- 34.Meyaard, L. 2008. The inhibitory collagen receptor LAIR-1 (CD305). J. Leukoc. Biol. 83:799-803. [DOI] [PubMed] [Google Scholar]
- 35.Mintz, K. P. 2004. Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen. Microbiology 150:2677-2688. [DOI] [PubMed] [Google Scholar]
- 36.Morton, L. F., P. G. Hargreaves, R. W. Farndale, R. D. Young, and M. J. Barnes. 1995. Integrin α2β1-independent activation of platelets by simple collagen-like peptides: collagen tertiary (triple-helical) and quaternary (polymeric) structures are sufficient alone for α2β1-independent platelet reactivity. Biochem. J. 306:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nallapareddy, S. R., X. Qin, G. M. Weinstock, M. Höök, and B. E. Murray. 2000. Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect. Immun. 68:5218-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nummelin, H., Y. El Tahir, P. Ollikka, M. Skurnik, and A. Goldman. 2002. Expression, purification and crystallization of a collagen-binding fragment of Yersinia adhesin YadA. Acta Crystallogr. D 58:1042-1044. [DOI] [PubMed] [Google Scholar]
- 39.Nummelin, H., M. C. Merckel, J. C. Leo, H. Lankinen, M. Skurnik, and A. Goldman. 2004. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel β-roll. EMBO J. 23:701-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pekkola-Heino, K., M. K. Viljanen, T. H. Stahlberg, K. Granfors, and A. Toivanen. 1987. Monoclonal antibodies reacting selectively with core and O-polysaccharide of Yersinia enterocolitica O:3 lipopolysaccharide. Acta Pathol. Microbiol. Immunol. Scand. C 95:27-34. [DOI] [PubMed] [Google Scholar]
- 41.Pugh, N., A. M. Simpson, P. A. Smethurst, P. G. de Groot, N. Raynal, and R. W. Farndale. 29 March 2010, posting date. Synergism between platelet collagen receptors defined using receptor-specific collagen-mimetic peptide substrata in flowing blood. Blood doi: 10.1182/blood-2010-01-260778. [DOI] [PMC free article] [PubMed]
- 42.Ramshaw, J. A., N. K. Shah, and B. Brodsky. 1998. Gly-X-Y tripeptide frequencies in collagen: a context for host-guest triple-helical peptides. J. Struct. Biol. 122:86-91. [DOI] [PubMed] [Google Scholar]
- 43.Raynal, N., S. W. Hamaia, P. R. Siljander, B. Maddox, A. R. Peachey, R. Fernandez, L. J. Foley, D. A. Slatter, G. E. Jarvis, and R. W. Farndale. 2006. Use of synthetic peptides to locate novel integrin α2β1-binding motifs in human collagen III. J. Biol. Chem. 281:3821-3831. [DOI] [PubMed] [Google Scholar]
- 44.Riess, T., S. G. Andersson, A. Lupas, M. Schaller, A. Schäfer, P. Kyme, J. Martin, J. H. Wälzlein, U. Ehehalt, H. Lindroos, M. Schirle, A. Nordheim, I. B. Autenrieth, and V. A. Kempf. 2004. The Bartonella adhesin A mediates a proangiogenic host cell response. J. Exp. Med. 200:1267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Trülzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185:3735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roggenkamp, A., H. R. Neuberger, A. Flügel, T. Schmoll, and J. Heesemann. 1995. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol. Microbiol. 16:1207-1219. [DOI] [PubMed] [Google Scholar]
- 47.Rosqvist, R., M. Skurnik, and H. Wolf-Watz. 1988. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature 334:522-524. [DOI] [PubMed] [Google Scholar]
- 48.Sakakibara, S., K. Inouye, K. Shudo, Y. Kishida, Y. Kobayashi, and D. J. Prockop. 1973. Synthesis of (Pro-Hyp-Gly)n of defined molecular weights. Evidence for the stabilization of collagen triple helix by hydroxypyroline. Biochim. Biophys. Acta 303:198-202. [DOI] [PubMed] [Google Scholar]
- 49.Schulze-Koops, H., H. Burkhardt, J. Heesemann, K. von der Mark, and F. Emmrich. 1992. Plasmid-encoded outer membrane protein YadA mediates specific binding of enteropathogenic yersiniae to various types of collagen. Infect. Immun. 60:2153-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulze-Koops, H., H. Burkhardt, J. Heesemann, K. von der Mark, and F. Emmrich. 1995. Characterization of the binding region for the Yersinia enterocolitica adhesin YadA on types I and II collagen. Arthritis Rheum. 38:1283-1289. [DOI] [PubMed] [Google Scholar]
- 51.Skurnik, M. 1984. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Appl. Bacteriol. 56:355-363. [DOI] [PubMed] [Google Scholar]
- 52.Skurnik, M. 1985. Expression of antigens encoded by the virulence plasmid of Yersinia enterocolitica under different growth conditions. Infect. Immun. 47:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skurnik, M. 1995. Role of YadA in Yersinia-enterocolitica-induced reactive arthritis: a hypothesis. Trends Microbiol. 3:318-319. [DOI] [PubMed] [Google Scholar]
- 54.Skurnik, M., Y. El Tahir, M. Saarinen, S. Jalkanen, and P. Toivanen. 1994. YadA mediates specific binding of enteropathogenic Yersinia enterocolitica to human intestinal submucosa. Infect. Immun. 62:1252-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3:517-529. [DOI] [PubMed] [Google Scholar]
- 56.Smethurst, P. A., D. J. Onley, G. E. Jarvis, M. N. O'Connor, C. G. Knight, A. B. Herr, W. H. Ouwehand, and R. W. Farndale. 2007. Structural basis for the platelet-collagen interaction: the smallest motif within collagen that recognizes and activates platelet glycoprotein VI contains two glycine-proline-hydroxyproline triplets. J. Biol. Chem. 282:1296-1304. [DOI] [PubMed] [Google Scholar]
- 57.Tamm, A., A. M. Tarkkanen, T. K. Korhonen, P. Kuusela, P. Toivanen, and M. Skurnik. 1993. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10:995-1011. [DOI] [PubMed] [Google Scholar]
- 58.Tang, G., T. Kitten, C. L. Munro, G. C. Wellman, and K. P. Mintz. 2008. EmaA, a potential virulence determinant of Aggregatibacter actinomycetemcomitans in infective endocarditis. Infect. Immun. 76:2316-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Visai, L., S. Bozzini, G. Raucci, A. Toniolo, and P. Speziale. 1995. Isolation and characterization of a novel collagen-binding protein from Streptococcus pyogenes strain 6414. J. Biol. Chem. 270:347-353. [DOI] [PubMed] [Google Scholar]
- 60.Vogel, W. F., R. Abdulhussein, and C. E. Ford. 2006. Sensing extracellular matrix: an update on discoidin domain receptor function. Cell. Signal. 18:1108-1116. [DOI] [PubMed] [Google Scholar]
- 61.Westerlund, B., and T. K. Korhonen. 1993. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9:687-694. [DOI] [PubMed] [Google Scholar]
- 62.Zheng, H., Y. Sun, S. Lin, Z. Mao, and B. Jiang. 2008. Yersinia enterocolitica infection in diarrheal patients. Eur. J. Clin. Microbiol. Infect. Dis. 27:741-752. [DOI] [PubMed] [Google Scholar]
- 63.Zong, Y., Y. Xu, X. Liang, D. R. Keene, A. Höök, S. Gurusiddappa, M. Höök, and S. V. Narayana. 2005. A ‘collagen hug’ model for Staphylococcus aureus CNA binding to collagen. EMBO J. 24:4224-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.