Abstract
Enterotoxigenic Escherichia coli (ETEC) causes considerable morbidity and mortality due to diarrheal illness in developing countries, particularly in young children. Despite the global importance of these heterogeneous pathogens, a broadly protective vaccine is not yet available. While much is known regarding the immunology of well-characterized virulence proteins, in particular the heat-labile toxin (LT) and colonization factors (CFs), to date, evaluation of the immune response to other antigens has been limited. However, the availability of genomic DNA sequences for ETEC strains coupled with proteomics technology affords opportunities to examine novel uncharacterized antigens that might also serve as targets for vaccine development. Analysis of whole or fractionated bacterial proteomes with convalescent-phase sera can potentially accelerate identification of secreted or surface-expressed targets that are recognized during the course of infection. Here we report results of an immunoproteomics approach to antigen discovery with ETEC strain H10407. Immunoblotting of proteins separated by two-dimensional electrophoresis (2DE) with sera from mice infected with strain H10407 or with convalescent human sera obtained following natural ETEC infections demonstrated multiple immunoreactive molecules in culture supernatant, outer membrane, and outer membrane vesicle preparations, suggesting that many antigens are recognized during the course of infection. Proteins identified by this approach included established virulence determinants, more recently identified putative virulence factors, as well as novel secreted and outer membrane proteins. Together, these studies suggest that existing and emerging proteomics technologies can provide a useful complement to ongoing approaches to ETEC vaccine development.
Infectious diarrhea substantially impacts human health in the developing world, where hundreds of millions of infections occur each year. Several pathogens, rotavirus, Shigella, Vibrio cholerae, and enterotoxigenic Escherichia coli (ETEC), each contribute significantly to this disease burden and collectively result in an estimated 2 million deaths due to diarrheal illness annually (52). Therefore, ETEC remains a high priority for vaccine development.
Enterotoxigenic E. coli strains constitute a phenotypically and genetically diverse pathotype that have in common the production of enterotoxin heat-labile toxin (LT) and/or heat-stable toxin (ST). In the classic paradigm for ETEC pathogenesis, organisms must colonize the small intestine via fimbrial colonization factor antigens (CFAs) for effective toxin delivery and subsequent diarrhea (18). Since the early identification of colonization factors (CFs) as important virulence determinants (15), these structures have been a central focus of ETEC vaccine development, and significant inroads have been made into the identification of a broad array of CFs (22, 43), with over 25 antigens identified thus far. ETEC vaccines currently in development are designed to target the most prevalent CFs (56). Moreover, recent elegant structural characterization of the colonization factor antigen I (CFA/I) pilus has provided additional molecular details of pilus tip adhesin molecules that might be exploited (33) as more highly conserved vaccine targets.
However, the remarkable plasticity of E. coli genomes (45) and studies demonstrating that many ETEC strains do not produce an identifiable CF (40, 54) suggest that additional antigens would likely need to be considered to produce a broadly protective vaccine. While much is known about the immunology of the CFs and LT following infection (44, 46, 63), very little is known about the nature of immune responses to ETEC in general, and there is no information regarding immunogenicity of more recently discovered putative virulence factors.
Furthermore, large-scale epidemiologic studies have suggested that additional plasmid or chromosomally encoded factors contribute to the development of an effective protective immune response attributable to prior natural infections with ETEC (55). However, the identity of other antigens that might be involved in the development of protective immune responses to ETEC remains largely unexplored.
The advent of high-throughput sequencing of multiple genomes and advances in proteomics permit avenues for discovery of novel antigens which might be useful in ETEC vaccine development. Two complete ETEC genomes, ETEC H10407 and E24377A (45), and one draft genome sequence, B7A (45), as well as several plasmid sequences (21) are now publicly available. While it is anticipated that dozens if not hundreds of ETEC genome sequences will ultimately be made available, these existing genomes permit some initial antigen discovery and validation efforts that were not previously possible.
Recent studies of mice have demonstrated that mice exposed to ETEC are protected from subsequent intestinal colonization (47). Therefore, these studies were undertaken to characterize the nature of protective immune responses afforded by prior exposures to ETEC in this model and to validate immune responses to selected antigens using sera from patients naturally infected with ETEC.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids employed in these studies are provided in Table 1 . ETEC strain H10407 was originally isolated in Bangladesh from a patient with severe, cholera-like diarrheal illness (13). The H10407 isolate used in the present study was provided by Marcia Wolfe, and it is derived from good manufacturing practice (GMP) lots of H10407 produced at Walter Reed Army Institute of Research. This strain is fully virulent in human volunteer clinical challenge studies (8).
TABLE 1.
Bacterial strains and plasmids used in these studies
| Bacterial strain or plasmid | Relevant genotype or descriptiona | Reference or source |
|---|---|---|
| Bacterial strains | ||
| H10407 | ETEC (Bangladesh) serotype O78:H11, LT+ ST+, CFA/I | 13 |
| ETP98116 | ETEC (ICDDR,B), ST+, CFA/I | This study |
| ETP98118 | ETEC (ICDDR,B), ST+, CFA/I | This study |
| ETP98087 | ETEC (ICDDR,B), LT+ ST+, CS2 CS3 | This study |
| jf1412 | H10407 fliC::Kmr mutant; nonmotile | 49 |
| E. coli Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74recA1araD139 Δ(ara leu)7697galUgalKrpsL (Strr) endA1nupG | Invitrogen |
| Plasmids | ||
| pJL017 | etpBA cloned into pBAD/Myc-His A, with etpA in frame with myc and six-His coding regions | 47 |
| pJL030 | etpC gene cloned into pACYC184; Cmr | 49 |
Abbreviations: ICDDR,B, International Centre for Diarrhoeal Disease Research, Bangladesh; LT+, heat-labile toxin positive; ST+, heat-stable toxin positive; CFA/I, colonization factor antigen I; CS2, coli surface antigen 2; Kmr, kanamycin resistance; Strr, streptomycin resistance; Cmr, chloramphenicol resistance.
Flow cytometry.
To demonstrate recognition of the bacterial surface by immune antisera, suspensions of ETEC H10407 in phosphate-buffered saline (PBS) were fixed with 2% paraformaldehyde for 15 min, washed twice with PBS, and blocked with 1% bovine serum albumin (BSA) in PBS for 30 min. Fixed cells were then incubated with pre- or postimmune mouse sera (diluted 1:50 in blocking buffer) for 1 h at room temperature and then washed three times with PBS. Washed cells were then incubated with Alexa Fluor (488)-labeled anti-mouse IgG, IgM, and IgA antibodies (1:10,000), washed three times, and resuspended in PBS for analysis of cell-bound fluorescence by flow cytometry using a BD FACSCalibur 4-color, dual-laser flow cytometer with FACStation data management system.
Subcellular fractionation of ETEC H10407. (i) Growth conditions.
Prior to processing of bacterial fractions, ETEC strain H10407 was grown under a variety of conditions previously shown to support the production of one or more known virulence factors. Strain H10407 from frozen stocks maintained at −80°C was used to inoculate CFA agar plates (Casamino Acids [1%], yeast extract [0.078%], MgSO4 [0.4 mM], MnCl2 [0.04 mM], agar [2%] [pH 7.4]) (12) to induce production of colonization factors and CYE-G medium (Casamino Acids [30 g/liter], yeast extract [3 g/liter], K2HPO4 [0.5 g/liter], glucose [2 g/liter] [pH 8.0]) to facilitate the secretion of LT as previously described (10, 50).
(ii) OMP.
Outer membranes from ETEC H10407 were prepared by sucrose gradient ultracentrifugation as described previously with slight modifications (19, 53). Briefly, 5 ml of overnight bacterial culture was diluted to 500 ml in either Luria broth or CYE-G medium and incubated for 5 h at 225 rpm and 37°C. Bacterial pellets obtained by centrifugation at 5,000 × g for 10 min were resuspended in 8 ml of cold buffer containing 50 mM Tris HCl and 1 mM EDTA (pH 7.8), and the suspension was lysed twice in a French pressure cell. Unbroken cells were removed by centrifugation at 3,000 × g for 10 min, and the resulting supernatant containing inner and outer membranes was centrifuged at 100,000 × g for 1 h in an SW28 rotor. The pellet was suspended in cold HEPES buffer (10 mM HEPES [pH 7.4]) and loaded as the top layer on a discontinuous gradient prepared from 2.02 M, 1.44 M, and 0.78 M sucrose in HEPES buffer. After 16 h of centrifugation at approximately 80,000 × g and 4°C, material was collected from the 2.02 and 1.44 M interface, diluted 10-fold in HEPES buffer, and recentrifuged at approximately 110,000 × g for 1 h. The supernatant was discarded, and the resulting outer membrane preparation (OMP) was stored at −80°C.
(iii) Concentration of culture supernatants.
Mid-logarithmic-phase cultures of strain H10407 were centrifuged at 10,000 × g for 10 min and 4°C, then clarified supernatants were filtered through a 0.22-μm vacuum filter, and the filtrate was then concentrated (approximately 150 times) through a 10,000-molecular-weight-cutoff (10,000-MWCO) filter (Millipore) in a stirred ultrafiltration cell. Final concentration and desalting were performed at 4°C using a 10,000-MWCO centrifugal filter.
(iv) Preparation of H10407 vesicles.
Vesicles were prepared from H10407 culture supernatants as previously described (29). Briefly, cultures of strain H10407 were centrifuged at 5,000 × g for 10 min; clarified culture supernatants were then filtered through a 0.45-μm vacuum filter, and the filtrate was then centrifuged at 40,000 × g to pellet vesicles. This pellet was then resuspended in sterile water, and a total of 20 μg of protein was separated by SDS-PAGE.
Two-dimensional separation of proteins. (i) Liquid-phase isoelectric focusing.
Protein preparations were suspended in 18 ml of solution containing 8 M urea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 3% ampholyte (Bio-Rad). This suspension was subjected to liquid-phase isoelectric focusing (Rotofor; Bio-Rad) at 12 W for 3 to 5 h on a pH gradient from pH 3 to 10. Individual fractions of approximately 0.9 ml were collected under vacuum and concentrated to a final volume of approximately 50 μl using a 10,000-MWCO centrifugal concentrator (Millipore).
(ii) SDS-PAGE and immunoblotting.
A 10-μl volume of sample was loaded onto two identical 4 to 15% gradient SDS-PAGE gels. Following the second dimension electrophoretic separation, one gel was stained with Sypro Ruby (Molecular Probes), and then imaged using a Typhoon variable-mode imager (GE Healthcare). Proteins separated on the remaining gel were transferred to nitrocellulose for immunoblotting using pooled sera from mice repeatedly challenged with wild-type H10407 bacteria or pooled human convalescent-phase sera from patients with ETEC diarrheal illness obtained at the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B) in Dhaka, Bangladesh.
Identification of immunogenic proteins. (i) In-gel tryptic digestion of proteins.
Protein bands corresponding to immunoreactive species on the Western blot were excised from the Sypro Ruby-stained gel and taken to the University of Tennessee Health Sciences Center (UTHSC) Mass Spectrometry Core laboratory for identification. Here gel bands were minced and destained in acetonitrile-100 mM ammonium bicarbonate solution (50%, vol/vol), dried under vacuum, and digested for approximately 18 h at 37°C with trypsin (0.0004%, wt/vol). The digestion supernatant was transferred to another centrifuge tube, and 50 μl of acetonitrile-trifluoroacetic acid (TFA) solution (5% acetonitrile and 60% TFA [vol/vol]) was added to the remaining gel fragments. These gel fragments were sonicated twice to extract the remaining tryptic peptides. Supernatants were combined and dried. The dried residue was reconstituted in 15 μl TFA (0.1%).
Pipette tip chromatography columns (ZipTipC18; Millipore) were used to desalt and preferentially enrich the tryptic peptides according to the manufacturer's manual. Briefly, the columns were prewet with 50% acetonitrile and equilibrated in 0.1% TFA. The reconstituted solution was loaded onto a column, followed by washing with 0.1% TFA. The tryptic peptides were eluted with 10 μl of a 50% acetonitrile solution, the eluate was dried in a vacuum centrifuge, and the residue was reconstituted in 4 μl of 0.1% formic acid solution. Two microliters of that solution was then used for liquid chromatography (LC)-tandem mass spectometry (MS-MS) analysis.
(ii) Liquid chromatography-mass spectrometry and database searches.
High-performance liquid chromatography (HPLC) was performed at a flow rate of 0.5 μl/min in solvent A (0.1% formic acid solution) and solvent B (0.1% formic acid in a solution of 90% methanol) with a gradient elution time profile of 100% solvent A (0 to 5 min), followed by 100% solvent A to 20% solvent A-80% solvent B (5 to 60 min). Peptide elution was coupled to mass spectrometry analysis using a nano electrospray ionization source mass spectrometer (Q-TOF II; Micromass, Manchester, United Kingdom) in positive-ion mode with a source temperature of 90°C and capillary voltage of 3.0 kV. MS-MS scans (5 s) were followed by an MS scan (1 s); double- and triple-charged precursor ions with a count of ≥20 were selected to perform MS-MS. Masslynx software (version 3.5) was used to process the chromatographic and mass spectrometric data, and the MS-MS data were transferred into pkl format files. These mass spectra were then used to query protein databases using MASCOT (39). Alternatively, mass spectra were used to query predicted peptides encoded by the H10407 genome available via the Welcome Trust Sanger Institute website (http://www.sanger.ac.uk/Projects/E_coli_H10407/) using customized BioPerl (http://www.bioperl.org) scripts and FindPept (24). A complete description of these scripts, the algorithms used in their execution, and instructions for implementation are provided in the supplemental material. Proteins identified in strain H10407 were localized to coding sequences within the genome using rapid annotation subsystem technology (RAST) (http://rast.nmpdr.org/) (3).
Expression and purification of recombinant proteins.
Recombinant EtpA was produced as previously described (17, 49). Briefly, E. coli strain Top10, which does not produce native flagellin (27), was used as a host for the recombinant etpBAC locus two-partner secretion plasmids pJL017 and pJL030. These two plasmids, pJL017 and pJL030, carry the etpBA and etpC genes, respectively, permitting expression and secretion of glycosylated, myc-polyhistidine-tagged recombinant EtpA (rEtpA), which was purified by metal affinity chromatography using recently described protocols (17).
Intestinal infection of mice with ETEC H10407.
Mice were infected enterally with ETEC strain H10407 as previously described (1). Briefly, strain H10407 was grown to mid-logarithmic phase in Luria broth, pH 7.4, and resuspended in sterile PBS such that the final concentration of bacteria was approximately 1 × 107 CFU in a final volume of 300 μl. This amount was then administered by gavage to 20 ETEC-naïve ICR mice that had been pretreated with streptomycin to eliminate native flora and cimetidine to reduce stomach acidity prior to challenge. This procedure was repeated on days 14 and 28, and mice were subsequently sacrificed on day 35 (47).
Hyperimmune mouse and convalescent human sera used in immunoassays.
To obtain preimmune sera from mice, venipuncture of facial vein was conducted prior to challenge; postimmune sera were obtained by cardiocentesis following euthanasia. Existing deidentified human convalescent-phase sera previously collected from patients hospitalized with ETEC infections were obtained from ICDDR,B. These sera were used under established protocols approved by the Institution Review Boards of ICDDR,B and the Memphis VA Medical Center. Control age-matched (<5 years) sera from ETEC-naïve children were obtained from Le Bonheur Children's Medical Center, Memphis, TN.
Existing human convalescent-phase serum samples from patients previously hospitalized at ICDDR,B were first tested against recombinant ETEC proteins by screening kinetic enzyme-linked immunosorbent assays (ELISAs) using a 1:50 dilution of sera. Positive sera were subjected to additional testing by immunoblotting and kinetic ELISAs as previously described (49). Briefly, recombinant proteins (4 μg/ml in 0.1 M NaHCO3 buffer [pH 8.6]) were used to coat ELISA plates (overnight, 4°C). After the plates were washed with Tris-buffered saline (TBS) containing 0.05% Tween 20 (TBS-T), they were blocked (1 h, 37°C) with 1% BSA in TBS-T (Blocker; Thermo Scientific). Dilutions of sera prepared in blocking solution were added to blocked ELISA plates and incubated for 1 h at 37°C. After the plates were washed with TBS-T, horseradish peroxidase (HRP)-labeled goat anti-mouse secondary antibody (IgA, IgM, and IgG) was added (1:10,000). After 1 h at 37°C, the plates were washed and developed with 3,3′,5,5′-tetramethylbenzidine (TMB) peroxidase substrate, and kinetic absorbance was measured (58) (620 nm, 30-s acquisition intervals) on a Molecular Devices Spectramax 340PC microplate reader. SoftMax Pro software (v5.0.1) was used to record and report data (Vmax, expressed as milliunits per minute; Vmax represents the maximum slope of the kinetic display of optical density versus time).
Caco-2 in vitro bacterial adherence assays.
Caco-2 adherence assays for ETEC were performed as previously described (20). Briefly, Caco-2 cells, propagated in accordance with ATCC protocols were split 1:2 on the day prior to infection and used to seed 96-well tissue culture plates. EtpA-producing ETEC strains were grown overnight (37°C, 225 rpm) in 2 ml of Luria broth inoculated from frozen stocks maintained at −80°C. Following dilution and growth to mid-logarithmic phase the following morning, bacteria were introduced into semiconfluent Caco-2 monolayers at a multiplicity of infection (MOI) of approximately 10:1. Antibodies were added immediately prior to introduction of bacteria. After 1 h of incubation at 37°C and 5% CO2, monolayers were washed four times with RPMI 1640, and cell-associated bacteria were recovered by plating dilutions of Triton X-100 (0.1%) lysates onto Luria agar.
RESULTS
Multiple ETEC surface proteins are recognized during infection.
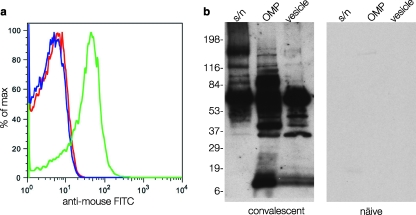
To demonstrate first that the development of a protective immune response in mice correlated with production of antibodies that recognized the surface of ETEC, we performed flow cytometry on whole ETEC H10407 bacteria incubated with sera obtained before and after intestinal challenge of mice with this strain. These studies demonstrated that following exposure to ETEC H10407, mice did mount demonstrable antibody responses to the surface of ETEC as shown by flow cytometry (Fig. 1a).
FIG. 1.
Recognition of ETEC surface antigens following experimental infection. (a) Bacterial surface recognition flow cytometry data obtained following incubation of ETEC strain H10407 with sera from mice obtained before (red peak) and after repeated enteric challenge with strain H10407 (green peak). The blue peak shows unlabeled organisms. FITC, fluorescein isothiocyanate; max, maximum. (b) Immunoblots demonstrating that multiple surface-expressed ETEC proteins are recognized by convalescent mouse sera relative to controls. Different fractions are shown (supernatant [s/n], outer membrane proteins [OMP], and outer membrane vesicles [vesicle]). The positions of molecular mass markers (in kilodaltons) are shown to the left of the immunoblot.
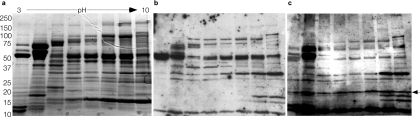
Therefore, additional studies were conducted to examine the nature of the immune responses following infection with ETEC. In these studies, ETEC H10407 was first grown under conditions known to support the production of virulence factors, including the heat-labile toxin and known colonization factors. Preliminary examination of subcellular protein fractions, including outer membrane preparations, (OMPs), concentrated culture supernatants, and outer membrane vesicles (OMVs) by immunoblotting with sera obtained from animals following infection with strain H10407 demonstrated multiple immunoreactive species that were not identified by preimmune sera, suggesting that many proteins are recognized during the course of infection with ETEC (Fig. 1b). Interestingly, using either pooled sera from mice infected experimentally with H10407 or pooled sera from children infected with ETEC, we obtained remarkably similar results (Fig. 2a to c). Sera from naïve mice or humans demonstrated little or no immunoreactivity (not shown), while multiple highly immunoreactive bands were observed with convalescent-phase sera.
FIG. 2.
Human and murine responses to ETEC infection are comparable. (a) Stained SDS-PAGE gel following two-dimensional (2-D) separation of OMPs from ETEC strain H10407. (b) Immunoblot of same proteins using sera from mice obtained following repeated oral challenge with H10407. (c) Immunoblot performed using pooled convalescent-phase sera from ETEC-infected Bangladeshi patients. The arrow to the right of the gel indicates protein recognized exclusively following human ETEC infection, subsequently identified as the surface antigen 2 superfamily protein (CDD accession no. cl01155), OmpW. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel.
On the basis of these preliminary results, additional studies were carried out to identify proteins that were recognized following infection. Among the proteins identified using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) were known immunogenic virulence proteins recognized following natural infection or experimental challenge in humans, including the heat-labile toxin (16, 60), and CfaB, the major structural protein of the CFA/I pilus (8), as well as putative usher proteins and chaperones required for fimbrial biogenenesis (Table 2).
TABLE 2.
Immunoreactive proteins identified by MALDI-TOF MS
| Group | Band no.a | Sourceb | Accession no.c | Characterization of protein |
Peptide matches |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Descriptiond | Mass (kDa) | pI | No. matched | Coverage (%) | MOWSE scoree/database | ||||
| Virulence | 01 | S, O, V | YP_003229794 | FliC | Flagellin H11 monomer | 50.92 | 4.70 | 181 | 85 | 8,628/NCBInr |
| 02 | S | AAO17297 | EatA | Autotransporter | 147.61 | 6.23 | 9 | 9 | 272/NCBInr | |
| 03 | S | YP_001464349.1 | Antigen 43 | Autotransporter | 97.27 | 5.92 | 12 | 10 | Sangerf | |
| 50 | S, O, V | AAX13509.2 | EtpA | TpsA adhesin | 176.92 | 9.77 | 19 | 8 | 375/NCBInr | |
| 08 | O | AAC41414.1 | CfaA | Periplasmic chaperone | 27.2 | 9.79 | 9 | 43 | Sanger | |
| 09 | O | A7ZK44 | FimD2 | Fimbrial usher | 95.2 | 5.42 | 20 | 19 | Sanger | |
| 17, 36, 54 | O, W, S | P02971 | CfaB | Pilin subunit | 17.45 | 7.93 | 5 | 48 | 252/NCBInr | |
| 49 | O | ABV16486 | Antigen 43 | Autotransporter | 97.29 | 5.56 | 4 | 5 | 75/NCBInr | |
| 43 | V | AAB02982 | LT-B | Heat-labile toxin b subunit | 14.06 | 8.76 | 5 | 30 | 145/NCBInr | |
| 43 | V | ABM92275 | CexE | Secreted ETEC protein, unknown function | 12.63 | 9.32 | 3 | 26 | 154/NCBInr | |
| 51 | V | Q9XD84 | TibA | Autotransporter/adhesin | 101.05 | 5.58 | 9 | 10 | 294/NCBInr | |
| Molecular chaperone | 22 | S | AAR21890 | GroEL | Chaperonin | 57.28 | 4.84 | 19 | 61 | 1,386/NCBInr |
| 21 | S | NP_308041 | DnaK (Hsp70) | Chaperonin | 69.07 | 4.83 | 32 | 60 | 1,587/NCBInr | |
| 06 | O | NP_285857 | (DegP/HtrA) | Membrane-associated serine protease | 46.80 | 7.88 | 17 | 52 | 548/NCBInr | |
| Outer membrane protein | 07, 20, 12 | O | NP_309068.1 | OmpA | Porin | 37.18 | 5.99 | 17 | 51 | 695/NCBInr |
| 39, 11 | V, O | AAB40749 | NmpC | Porin | 41.5 | 4.94 | 13 | 37 | 997/NCBInr | |
| 10 | O | YP_405685.1 | FecA homologue | Fe transport membrane receptor | 85.16 | 5.99 | 17 | 16 | Sanger | |
| 14, 42 | O, V | NP_752830 | OmpX | Ail-like adhesin? | 18.59 | 6.56 | 7 | 57 | 333/NCBInr | |
| 19 | O | NP_414692.1 | FhuA-like protein | Ferrichrome outer membrane transporter | 82.2 | 5.35 | 17 | 19 | Sanger | |
| 53 | O | NP_417795.1 | Bfr | Bacterioferrin | 18.48 | 4.69 | 8 | 67 | 472/NCBInr | |
| 59, 60 | O | YP_540467.1 | OmpW | Surface antigen 2 superfamily protein | 25.856 | 6.28 | 5 | 32 | 229/NCBInr | |
| Hypothetical | 34 | O | NP_288109.1 | Hypothetical | 7.89 | 9.6 | 2 | 75 | Sanger | |
| 45 | O | ZP_04533914.1 | Hypothetical | 75.53 | 8.91 | 20 | 27 | Sanger | ||
| 23 | S | NP_755083.1 | Hypothetical | 30.42 | 4.97 | 23 | 67 | 789/NCBInr | ||
| 28 | S | NP_311861.1 | Hypothetical | 26.41 | 9.35 | 10 | 53 | Sanger | ||
| 27 | S | YP_853622 | Hypothetical | Similar to APEC01 protein | 36.42 | 5.53 | 5 | 25 | 138/NCBInr | |
| 31 | S | YP_002408614 | Hypothetical | Conserved, hypothetical (phage origin) | 12.76 | 5.82 | 3 | 56 | 263/NCBInr | |
| 35 | W | NP_286070 | Hypothetical | Conserved hypothetical in EHEC strains | 9.92 | 5.36 | 3 | 49 | 72/NCBInr | |
| 15 | O | NP_288111 | Hypothetical | 11.58 | 4.83 | 4 | 45.7 | Sanger | ||
| 33 | ZP_04533918.1 | Hypothetical | Homologue in Crohn's isolate E. coli 3_2_53FAA | 16.65 | 10.10 | 9 | 44 | Sanger | ||
| 37 | V | YP_002388453.1 | YghJ | Predicted membrane lipoprotein | 168.65 | 4.92 | 13 | 13 | 624/NCBInr | |
| 41 | V | NP_288218 | Hypothetical | MipA domain; envelope biogenesis | 27.78 | 5.50 | 4 | 22 | 90/NCBInr | |
| 40 | V | YP_541993 | Hypothetical | Homologue in UPEC (UTI89) | 30.42 | 4.97 | 7 | 42 | 205/NCBInr | |
| Transcription/translation | 24 | S | NP_312218.1 | Elongation factor G | 77.53 | 5.24 | 22 | 37 | 791/NCBInr | |
| 30 | S | CAA47740.1 | H-NS | 15.53 | 5.43 | 5 | 45 | 363/NCBInr | ||
| Housekeeping/metabolic | 26 | S | H85928 | Phosphopyruvate hydratase | 45.57 | 5.32 | 19 | 54 | 644/NCBInr | |
| 44 | O | ZP_03072175.1 | Pyruvate dehydrogenase | 99.61 | 5.46 | 8 | 8 | 214/NCBInr | ||
| 29 | S | AAC43517 | Thiol peroxidase | 17.88 | 4.75 | 4 | 37 | 400/NCBInr | ||
| 32 | S | AAA24696 | Thioredoxin | 11.89 | 4.67 | 4 | 47 | 210/NCBInr | ||
| 25 | S | NP_285812.1 | Dihydrolipoamide dehydrogenase | 50.66 | 5.79 | 18 | 43 | 590/NCBInr | ||
The band number refers to the band from the stained gel. The bands were numbered chronologically in order of submission for MALDI-TOF MS identification.
Source: S, supernatant; O, outer membrane; V, vesicle; W, unfractionated whole-cell lysate.
The accession number refers to the closest homologue identified by BLAST(P) search.
TpsA, two-partner secretion exoprotein family; APEC, avian pathogenic Escherichia coli; EHEC, enterohemorrhagic Escherichia coli; UPEC, uropathogenic Escherichia coli.
MOWSE score generated using the MASCOT algorithm to query the NCBInr database.
Data for peptide lacking matches in the NCBInr database were matched to predicted proteins based on the Sanger sequence for strain H10407 using custom Perl scripts and FindPept.
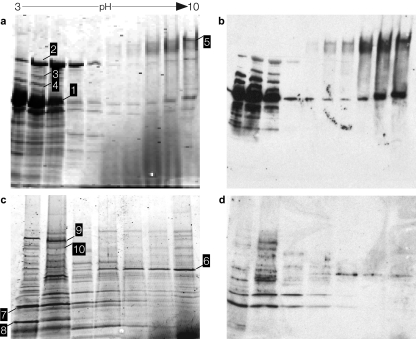
In addition, we identified a number of immunoreactive proteins that have more recently been discovered during the course of molecular pathogenesis studies of ETEC. These proteins included the TibA (34) and EatA (38) autotransporter proteins, the EtpA two-partner secretion protein (20), and the flagellin (FliC) molecule (49) corresponding to the H11 serotype (H10407 is serotype O78:H11) (Table 2 and Fig. 3).
FIG. 3.
Identification of immunoreactive proteins. (a) Supernatant proteins from ETEC strain H10407 separated by liquid-phase isoelectric focusing (pH gradient 3 to 10) followed by SDS-PAGE. (b) Immunoblot of panel a. (c) Gel image of outer membrane preparation from strain jf1412 (fliC mutant), separated in 2 dimensions. (d) Immunoblot of panel c. (Blots were developed with pooled sera from mice repeatedly infected with H10407.) The white numbers in black boxes on each panel correspond to selected individual immunoreactive protein bands chosen for identification. The protein band numbers (shown first) and proteins identified were as follows: 1, flagellin (E. coli, serotype, H11); 2, EatA autotransporter; 3, antigen 43 autotransporter protein; 4, DnaK (Hsp70) chaperone; 5, EtpA two-partner secretion exoprotein; 6, DegP membrane-associated serine protease; 7, OmpA; 8, CfaA periplasmic chaperone; 9, fimbrial usher; 10, outer membrane iron transport receptor.
Interestingly, both flagellin and EtpA were identified in each subcellular fraction (supernatant, OMP, and OMV) examined, likely relating to the interaction of these two proteins (49). Because of the relative abundance of flagellin associated with outer membrane fractions, we also examined OMPs prepared from jf1412, a fliC mutant of H10407 to facilitate the identification of other immunoreactive proteins (Fig. 3c).
Also identified using this approach were a number of “hypothetical” proteins that are not currently annotated as virulence proteins or for which no function has been assigned. These “hypothetical” proteins included a number of proteins that share significant homology with hypothetical proteins identified in other E. coli pathovars, including uropathogenic E. coli (UPEC) (36, 62), avian pathogenic E. coli (APEC) (30), and isolates linked to Crohn's disease, suggesting that these proteins may play as yet undetermined roles in pathogenesis.
Conversely, these studies also identified a number of proteins that are not directly connected to virulence but that have been identified as immunogenic proteins in similar analyses of other pathogens. These include the molecular chaperones DnaK, GroEL, and DegP (HtrA) which have previously been shown to be recognized as part of the humoral immune response to a wide variety of pathogenic bacteria, including V. cholerae (51), and group A streptococci (7).
Outer membrane vesicles are enriched with virulence proteins.
Much of the heat-labile toxin secreted by ETEC has been shown to be packaged into outer membrane vesicles (29). Therefore, we looked specifically in OMV preparations for an immunoreactive band corresponding to the heat-labile toxin. In addition to the B subunit of the heat-labile toxin, we identified multiple immunoreactive species in vesicle preparations corresponding to a number of established or putative virulence proteins, including flagellin, EtpA, the TibA autotransporter protein, and CexE (41), an extracytoplasmic ETEC protein of unknown function for which expression is regulated by CfaD transcriptional activator that also regulates expression of the CFA/I pilus. Likewise, a number of previously hypothetical proteins and putative porin proteins were identified exclusively in these subfractions of strain H10407.
Validation of immunoproteomics results with rEtpA glycoprotein.
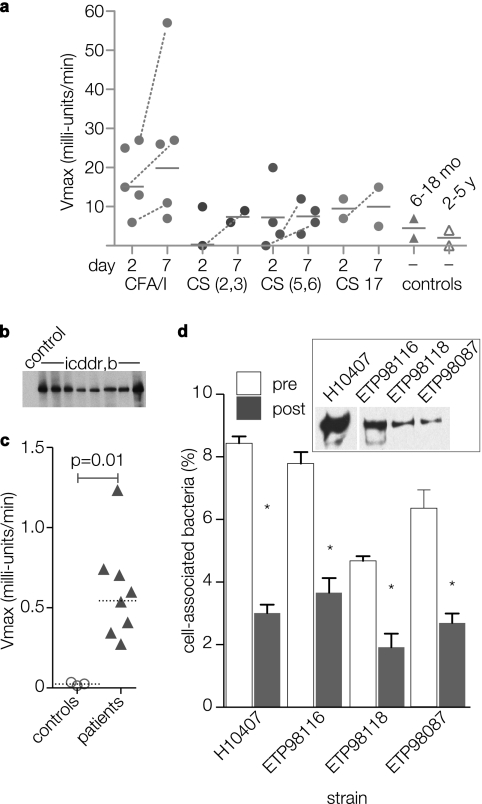
Earlier studies have shown that sera from mice infected with ETEC H10407 recognize recombinant versions of both EtpA and its two-partner secretion pore, EtpB (47). The present studies demonstrate that the sera of mice infected with strain H10407 also recognize the native fully glycosylated EtpA protein. To examine whether the sera from individual patients infected with ETEC also recognized EtpA, we examined existing serum specimens maintained at ICDDR,B against the full-length recombinant EtpA glycoprotein (rEtpAgp) by kinetic ELISAs using sera diluted 1:50. Sera from 13 ETEC-infected patients were compared with 2 uninfected control patients aged 6 to 18 months and 2 uninfected controls aged 2 to 5 years. In general, patients acutely infected with ETEC had significantly higher IgA responses to rEtpA than the controls did (P = 0.035), but particularly when samples obtained at day 7 following infection were compared to controls (P = 0.023). Seven of 13 patients had demonstrable increases in kinetic IgA responses during convalescence, and in five patients, this difference in measured kinetic rates was substantial (between 2- and 90-fold increase from day 2 to day 7) (Fig. 4a). Conversely, we found no difference in IgG responses to EtpA between patients and ICDDR,B controls (not shown). Sera from eight of these ICDDR,B patients along with corresponding ETEC isolates were subsequently subjected to additional evaluation. Both immunoblotting (Fig. 4b) and kinetic ELISA (Fig. 4c) against rEtpAgp confirmed the presence of anti-EtpA antibody (total IgG, IgA, and IgM) at titers of ≥1:1,028 compared to age-matched (<5 years of age) control sera obtained from children hospitalized in the United States where ETEC is not endemic (P = 0.01).
FIG. 4.
Human immune responses to EtpA. (a) Screening kinetic ELISA detection of anti-rEtpAgp (IgA) antibodies performed on early convalescent-phase sera (1:50 dilution) from Bangladeshi patients infected with ETEC. Each symbol represents a value for an individual patient obtained on either day 2 or 7 following initial hospitalization. Horizontal lines represent geometric mean values; dashed lines connect only patient sample values corresponding to demonstrable increases in value from day 2 to day 7 of hospitalization. The CF type for isolated strains is shown on the x axis (CS, coli surface antigen). The controls were age-matched children without ETEC infections (6- to 18-month-old children or 2- to 5-year-old children). The y axis shows absorbance readings at Vmax (in milliunits per minute). (b and c) Immunoblots and kinetic ELISA comparison of sera from patients in Dhaka, Bangladesh, and age-matched control sera from patients in the United States. Values represent total anti-EtpAgp antibody in day 7 samples (diluted 1:1,028) relative to controls. (d) Caco-2 cell in vitro adherence assays with EtpA-producing ETEC strains isolated from patients in Dhaka, Bangladesh (inset immunoblot shows EtpA produced by three isolates from ICDDR,B and ETEC H10407 positive-control strain). Data are shown for preimmune sera (pre) and anti-EtpA postimmune polyclonal antisera (post) (1:100 dilution). Bars represent means plus standard deviations (error bars) for four replicates. Values that were significantly different (P ≤ 0.05) for preimmune sera and postimmune antisera by Mann-Whitney analysis are indicated by an asterisk.
In subsequent analysis of ETEC strains isolated from these patients, we were able to confirm the presence of both the etpA gene by PCR (not shown) and production of the EtpA protein by immunoblotting of culture supernatants in three isolates (Fig. 4d, inset). As in earlier studies (49), we demonstrated that antibodies against EtpA were able to inhibit adherence of these ETEC strains to intestinal epithelial cell monolayers in vitro, suggesting that these strains make immunologically and functionally similar EtpA molecules.
DISCUSSION
Considerable effort has been devoted to the development of a safe and broadly protective vaccine for enterotoxigenic Escherichia coli (61). While there is not currently a vaccine for ETEC that can offer sustained, broad-based protection (5), multiple lines of evidence suggest that an ETEC vaccine is feasible. First, in areas where ETEC is not endemic, the incidence of ETEC infections is highest in early childhood (42), and longitudinal studies have demonstrated that prior exposure to ETEC ultimately protects against subsequent infection (55). Humans (32) as well as experimental animals (47) exposed to ETEC are protected on subsequent challenge, and active (14) or passive (57) immunization affords protection against challenge in experimental human challenge models of ETEC.
Prior to the advent of whole-genome sequencing projects, much of the effort expended toward development of an ETEC vaccine necessarily focused intensively on a limited subset of antigens, namely, the fimbrial colonization factors and toxins (56). Although there are considerable data regarding immune responses to these established ETEC virulence factors, there is little or no information regarding either the expression or immune recognition of other proteins during the course of infection. The availability of DNA sequence from multiple ETEC strains provides new avenues for antigen discovery, including immunoproteomics approaches (6, 37, 59) similar to those employed here.
The present studies, undertaken as an initial investigation of protective immune responses that develop with repeated exposure to ETEC, demonstrate that multiple antigens in addition to CFs and LT are recognized during both experimental infection in mice or natural infections in humans. These antigens are in part comprised by a variety of putative virulence proteins discovered in the course of more recent molecular pathogenesis investigations of ETEC strain H10407, including the exoprotein adhesin EtpA (20, 49), flagellin (49), and the TibA (11, 34) and EatA (38) autotransporter proteins.
A number of immunogenic outer membrane proteins were identified in our studies of H10407. These proteins include Ail adhesin-like OmpX, porins OmpA and NmpC, OmpW, and the Ag43 autotransporter protein. Interestingly, each of these proteins was also identified in a search for antigenic proteins of UPEC following urinary tract infection (25). Also, among outer membrane proteins identified were putative iron receptor proteins. Similar proteins have recently been identified in studies of uropathogenic E. coli, where they were shown to be immunogenic, induced in vivo, and act as protective antigens in murine models of urinary tract infection (2).
While our studies demonstrate that multiple ETEC antigens are recognized during infection, newly available genomic data also highlight potential challenges inherent in finding highly conserved protective antigens. Recent sequencing of multiple E. coli pathovars indicate that there may be relatively few genes that are truly specific to any one set of E. coli pathogens (45), including ETEC. A number of the proteins identified in the present studies correspond to (previously) hypothetical coding sequences that have close homologues in other E. coli pathovars. These previously unexplored antigens could be of particular value in future vaccine development efforts. Comparisons of the genomes of seven different UPEC isolates identified 173 genes that were shared by all of these strains but not with commensal strains of E. coli (36). However, many of these genes encoded hypothetical or predicted proteins of unknown function, while none of the known virulence factors was universally represented in uropathogenic strains. Similarly, other than the heat-labile and/or heat-stable toxins, none of the other established or putative virulence factors of ETEC has been identified in every isolate.
Recent studies do suggest however that one or more novel virulence proteins could complement existing vaccine strategies. Both EtpA and flagellin, identified here as immunogenic proteins, have also been shown to be protective antigens in a murine model of ETEC infection (48). Although most human strains of ETEC are flagellated (64), significant work is needed to investigate the molecular epidemiology of ETEC with respect to EtpA. Nonetheless, these data provide additional evidence that EtpA is expressed during both experimental and human infection and support the idea that novel antigens in addition to those currently targeted in present iterations of vaccines (61) could be useful in future development.
Interestingly, both of these proteins were identified in each of the fractions analyzed. As each flagellum is comprised of as many as 20,000 flagellin, and ETEC, like other E. coli bacteria, make peritrichous flagella with multiple structures, this protein subunit is easily the most abundant protein exported by ETEC, as is true for other flagellated Gram-negative pathogens (31). The identification of EtpA in each of these subcellular fractions is not surprising, given the recently identified interaction of this exoprotein adhesin with flagellin (49).
Many of the known or putative virulence proteins in these studies were found in analysis of subcellular fractions obtained from outer membrane vesicles (OMVs). The results of these studies would suggest that ETEC OMVs, similar to vesicles from other pathogens investigated thus far (4, 35), are enriched for a number of immunogenic virulence proteins in addition to the heat-labile toxin (23, 29).
A potential limitation of the present studies is that laboratory culture conditions may not completely mimic those encountered by these pathogens in vivo. In addition, current separation techniques, relative antigen abundance, and proteomics technology will limit identification of less abundant antigens. These studies were also limited to one particular ETEC strain. We also identified several housekeeping or metabolic genes which have no known link to virulence but are highly immunogenic (28), have been noted in similar studies of other pathogens (59), or like thiol peroxidase are associated with the bacterial membrane and involved in envelope remodeling in the context of the host (26).
Emerging proteomics technology, including the availability of pathogen-specific proteome microarrays (9), coupled with comparative genomic hybridization (36) studies to identify highly conserved surface-expressed antigens can overcome many of these limitations. Nevertheless, the present data, combined with recent discovery (20) and characterization (49) of novel ETEC virulence proteins, should engender enthusiasm for investigating additional avenues to protect against these important pathogens.
Supplementary Material
Acknowledgments
We thank Haibao Hwan of the UTHSC Mass Spectrometry Core laboratory for assistance with the LC-MS studies, David Brand and Ed Rosloniec for assistance with flow cytometry studies, Phil Hardwidge for review of the manuscript, and Ramy Aziz for aid using National Microbial Pathogen Database Resources, including RAST.
These studies were funded by a Merit Review Award (J.M.F.) from the Department of Veterans Affairs. The findings in these studies do not represent the views of the Department of Veterans Affairs or the U.S. Government.
Editor: S. M. Payne
Footnotes
Published ahead of print on 10 May 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Allen, K. P., M. M. Randolph, and J. M. Fleckenstein. 2006. Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect. Immun. 74:869-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alteri, C. J., E. C. Hagan, K. E. Sivick, S. N. Smith, and H. L. Mobley. 2009. Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathog. 5:e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, and O. Zagnitko. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlanda Scorza, F., F. Doro, M. J. Rodriguez-Ortega, M. Stella, S. Liberatori, A. R. Taddei, L. Serino, D. Gomes Moriel, B. Nesta, M. R. Fontana, A. Spagnuolo, M. Pizza, N. Norais, and G. Grandi. 2008. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DeltatolR IHE3034 mutant. Mol. Cell Proteomics 7:473-485. [DOI] [PubMed] [Google Scholar]
- 5.Boedeker, E. C. 2005. Vaccines for enterotoxigenic Escherichia coli: current status. Curr. Opin. Gastroenterol. 21:15-19. [PubMed] [Google Scholar]
- 6.Chitlaru, T., O. Gat, H. Grosfeld, I. Inbar, Y. Gozlan, and A. Shafferman. 2007. Identification of in vivo-expressed immunogenic proteins by serological proteome analysis of the Bacillus anthracis secretome. Infect. Immun. 75:2841-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, J. N., R. D. Ramirez, B. J. Currie, S. J. Cordwell, S. P. Djordjevic, and M. J. Walker. 2005. Surface analyses and immune reactivities of major cell wall-associated proteins of group A streptococcus. Infect. Immun. 73:3137-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coster, T. S., M. K. Wolf, E. R. Hall, F. J. Cassels, D. N. Taylor, C. T. Liu, F. C. Trespalacios, A. DeLorimier, D. R. Angleberger, and C. E. McQueen. 2007. Immune response, ciprofloxacin activity, and gender differences after human experimental challenge by two strains of enterotoxigenic Escherichia coli. Infect. Immun. 75:252-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, D. H., X. Liang, J. E. Hernandez, A. Randall, S. Hirst, Y. Mu, K. M. Romero, T. T. Nguyen, M. Kalantari-Dehaghi, S. Crotty, P. Baldi, L. P. Villarreal, and P. L. Felgner. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. U. S. A. 102:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorsey, F. C., J. F. Fischer, and J. M. Fleckenstein. 2006. Directed delivery of heat-labile enterotoxin by enterotoxigenic Escherichia coli. Cell. Microbiol. 8:1516-1527. [DOI] [PubMed] [Google Scholar]
- 11.Elsinghorst, E. A., and J. A. Weitz. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, D. G., and D. J. Evans, Jr. 1978. New surface-associated heat-labile colonization factor antigen (CFA/II) produced by enterotoxigenic Escherichia coli of serogroups O6 and O8. Infect. Immun. 21:638-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, D. G., D. J. Evans, Jr., and H. L. DuPont. 1977. Virulence factors of enterotoxigenic Escherichia coli. J. Infect. Dis. 136(Suppl.):S118-S123. [DOI] [PubMed] [Google Scholar]
- 14.Evans, D. G., D. J. Evans, Jr., A. R. Opekun, and D. Y. Graham. 1988. Non-replicating oral whole cell vaccine protective against enterotoxigenic Escherichia coli (ETEC) diarrhea: stimulation of anti-CFA (CFA/I) and anti-enterotoxin (anti-LT) intestinal IgA and protection against challenge with ETEC belonging to heterologous serotypes. FEMS Microbiol. Immunol. 1:117-125. [DOI] [PubMed] [Google Scholar]
- 15.Evans, D. G., R. P. Silver, D. J. Evans, Jr., D. G. Chase, and S. L. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12:656-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, D. J., Jr., G. Ruiz-Palacios, D. E. Evans, H. L. DuPont, L. K. Pickering, and J. Olarte. 1977. Humoral immune response to the heat-labile enterotoxin of Escherichia coli in naturally acquired diarrhea and antitoxin determination by passive immune hemolysis. Infect. Immun. 16:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleckenstein, J., and K. Roy. 2009. Purification of recombinant high molecular weight two-partner secretion proteins from Escherichia coli. Nat. Protoc. 4:1083-1092. [DOI] [PubMed] [Google Scholar]
- 18.Fleckenstein, J. M., P. R. Hardwidge, G. P. Munson, D. A. Rasko, H. Sommerfelt, and H. Steinsland. 2010. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 12:89-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleckenstein, J. M., D. J. Kopecko, R. L. Warren, and E. A. Elsinghorst. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64:2256-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleckenstein, J. M., K. Roy, J. F. Fischer, and M. Burkitt. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74:2245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Froehlich, B., J. Parkhill, M. Sanders, M. A. Quail, and J. R. Scott. 2005. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J. Bacteriol. 187:6509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 23.Gankema, H., J. Wensink, P. A. Guinee, W. H. Jansen, and B. Witholt. 1980. Some characteristics of the outer membrane material released by growing enterotoxigenic Escherichia coli. Infect. Immun. 29:704-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gattiker, A., W. V. Bienvenut, A. Bairoch, and E. Gasteiger. 2002. FindPept, a tool to identify unmatched masses in peptide mass fingerprinting protein identification. Proteomics 2:1435-1444. [DOI] [PubMed] [Google Scholar]
- 25.Hagan, E. C., and H. L. Mobley. 2007. Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect. Immun. 75:3941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haugen, B. J., S. Pellett, P. Redford, H. L. Hamilton, P. L. Roesch, and R. A. Welch. 2007. In vivo gene expression analysis identifies genes required for enhanced colonization of the mouse urinary tract by uropathogenic Escherichia coli strain CFT073 dsdA. Infect. Immun. 75:278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 28.Holmgren, A., and B. M. Sjoberg. 1972. Immunochemistry of thioredoxin. I. Preparation and cross-reactivity of antibodies against thioredoxin from Escherichia coli and bacteriophage T4. J. Biol. Chem. 247:4160-4164. [PubMed] [Google Scholar]
- 29.Horstman, A. L., and M. J. Kuehn. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489-12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komoriya, K., N. Shibano, T. Higano, N. Azuma, S. Yamaguchi, and S. I. Aizawa. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34:767-779. [DOI] [PubMed] [Google Scholar]
- 32.Levine, M. M., D. R. Nalin, D. L. Hoover, E. J. Bergquist, R. B. Hornick, and C. R. Young. 1979. Immunity to enterotoxigenic Escherichia coli. Infect. Immun. 23:729-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Y. F., S. Poole, K. Nishio, K. Jang, F. Rasulova, A. McVeigh, S. J. Savarino, D. Xia, and E. Bullitt. 2009. Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:10793-10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindmark, B., P. K. Rompikuntal, K. Vaitkevicius, T. Song, Y. Mizunoe, B. E. Uhlin, P. Guerry, and S. N. Wai. 2009. Outer membrane vesicle-mediated release of cytolethal distending toxin (CDT) from Campylobacter jejuni. BMC Microbiol. 9:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lloyd, A. L., D. A. Rasko, and H. L. Mobley. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 189:3532-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowalk, A. J., R. D. Gilmore, Jr., and J. A. Carroll. 2006. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect. Immun. 74:3864-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel, S. K., J. Dotson, K. P. Allen, and J. M. Fleckenstein. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 40.Peruski, L. F., Jr., B. A. Kay, R. A. El-Yazeed, S. H. El-Etr, A. Cravioto, T. F. Wierzba, M. Rao, N. El-Ghorab, H. Shaheen, S. B. Khalil, K. Kamal, M. O. Wasfy, A.-M. Svennerholm, J. D. Clemens, and S. J. Savarino. 1999. Phenotypic diversity of enterotoxigenic Escherichia coli strains from a community-based study of pediatric diarrhea in periurban Egypt. J. Clin. Microbiol. 37:2974-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pilonieta, M. C., M. D. Bodero, and G. P. Munson. 2007. CfaD-dependent expression of a novel extracytoplasmic protein from enterotoxigenic Escherichia coli. J. Bacteriol. 189:5060-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qadri, F., A. Saha, T. Ahmed, A. Al Tarique, Y. A. Begum, and A. M. Svennerholm. 2007. Disease burden due to enterotoxigenic Escherichia coli in the first 2 years of life in an urban community in Bangladesh. Infect. Immun. 75:3961-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qadri, F., A. M. Svennerholm, A. S. Faruque, and R. B. Sack. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao, M. R., T. F. Wierzba, S. J. Savarino, R. Abu-Elyazeed, N. El-Ghoreb, E. R. Hall, A. Naficy, I. Abdel-Messih, R. W. Frenck, Jr., A. M. Svennerholm, and J. D. Clemens. 2005. Serologic correlates of protection against enterotoxigenic Escherichia coli diarrhea. J. Infect. Dis. 191:562-570. [DOI] [PubMed] [Google Scholar]
- 45.Rasko, D. A., M. J. Rosovitz, G. S. Myers, E. F. Mongodin, W. F. Fricke, P. Gajer, J. Crabtree, V. Sperandio, and J. Ravel. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rivas, M., N. Binsztein, G. Basanta, M. Vergara, M. Quiroga, R. Cinto, and A. M. Svennerholm. 1995. Antibody responses against Escherichia coli heat-labile toxin and colonization factor antigens I and II in Argentinian children. J. Infect. Dis. 171:1045-1049. [DOI] [PubMed] [Google Scholar]
- 47.Roy, K., D. Hamilton, K. P. Allen, M. P. Randolph, and J. M. Fleckenstein. 2008. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect. Immun. 76:2106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy, K., D. Hamilton, M. M. Ostmann, and J. M. Fleckenstein. 2009. Vaccination with EtpA glycoprotein or flagellin protects against colonization with enterotoxigenic Escherichia coli in a murine model. Vaccine 27:4601-4608. [DOI] [PubMed] [Google Scholar]
- 49.Roy, K., G. M. Hilliard, D. J. Hamilton, J. Luo, M. M. Ostmann, and J. M. Fleckenstein. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sack, D., S. Huda, P. Neogi, R. Daniel, and W. Spira. 1980. Microtiter ganglioside enzyme-linked immunosorbent assay for Vibrio and Escherichia coli heat-labile enterotoxins and antitoxin. J. Clin. Microbiol. 11:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahu, G. K., R. Chowdhury, and J. Das. 1994. Heat shock response and heat shock protein antigens of Vibrio cholerae. Infect. Immun. 62:5624-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaetti, C. 2009. Vaccines for enteric diseases: update on recent developments. Expert Rev. Vaccines 8:1653-1655. [DOI] [PubMed] [Google Scholar]
- 53.Schnaitman, C. A. 1970. Examination of the protein composition of the cell envelope of Escherichia coli by polyacrylamide gel electrophoresis. J. Bacteriol. 104:882-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaheen, H. I., I. A. Abdel Messih, J. D. Klena, A. Mansour, Z. El-Wakkeel, T. F. Wierzba, J. W. Sanders, S. B. Khalil, D. M. Rockabrand, M. R. Monteville, P. J. Rozmajzl, A. M. Svennerholm, and R. W. Frenck. 2009. Phenotypic and genotypic analysis of enterotoxigenic Escherichia coli in samples obtained from Egyptian children presenting to referral hospitals. J. Clin. Microbiol. 47:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinsland, H., P. Valentiner-Branth, H. K. Gjessing, P. Aaby, K. Molbak, and H. Sommerfelt. 2003. Protection from natural infections with enterotoxigenic Escherichia coli: longitudinal study. Lancet 362:286-291. [DOI] [PubMed] [Google Scholar]
- 56.Svennerholm, A. M., and J. Tobias. 2008. Vaccines against enterotoxigenic Escherichia coli. Expert Rev. Vaccines 7:795-804. [DOI] [PubMed] [Google Scholar]
- 57.Tacket, C. O., G. Losonsky, H. Link, Y. Hoang, P. Guesry, H. Hilpert, and M. M. Levine. 1988. Protection by milk immunoglobulin concentrate against oral challenge with enterotoxigenic Escherichia coli. N. Engl. J. Med. 318:1240-1243. [DOI] [PubMed] [Google Scholar]
- 58.Tsang, V. C., B. C. Wilson, and S. E. Maddison. 1980. Kinetic studies of a quantitative single-tube enzyme-linked immunosorbent assay. Clin. Chem. 26:1255-1260. [PubMed] [Google Scholar]
- 59.Vytvytska, O., E. Nagy, M. Bluggel, H. E. Meyer, R. Kurzbauer, L. A. Huber, and C. S. Klade. 2002. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. Proteomics 2:580-590. [DOI] [PubMed] [Google Scholar]
- 60.Wachsmuth, I. K., J. G. Wells, and R. W. Ryder. 1977. Escherichia coli heat-labile enterotoxin: comparison of antitoxin assays and serum antitoxin levels. Infect. Immun. 18:348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker, R. I., D. Steele, and T. Aguado. 2007. Analysis of strategies to successfully vaccinate infants in developing countries against enterotoxigenic E. coli (ETEC) disease. Vaccine 25:2545-2566. [DOI] [PubMed] [Google Scholar]
- 62.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wenneras, C., F. Qadri, P. K. Bardhan, R. B. Sack, and A. M. Svennerholm. 1999. Intestinal immune responses in patients infected with enterotoxigenic Escherichia coli and in vaccinees. Infect. Immun. 67:6234-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolf, M. K. 1997. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10:569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.