Abstract
Biofilm formation by the periodontal pathogen Aggregatibacter actinomycetemcomitans is dependent upon autoinducer-2 (AI-2)-mediated quorum sensing. However, the components that link the detection of the AI-2 signal to downstream gene expression have not been determined. One potential regulator is the QseBC two-component system, which is part of the AI-2-dependent response pathway that controls biofilm formation in Escherichia coli. Here we show that the expression of QseBC in A. actinomycetemcomitans is induced by AI-2 and that induction requires the AI-2 receptors, LsrB and/or RbsB. Additionally, inactivation of qseC resulted in reduced biofilm growth. Since the ability to grow in biofilms is essential for A. actinomycetemcomitans virulence, strains that were deficient in QseC or the AI-2 receptors were examined in an in vivo mouse model of periodontitis. The ΔqseC mutant induced significantly less alveolar bone resorption than the wild-type strain (P < 0.02). Bone loss in animals infected with the ΔqseC strain was similar to that in sham-infected animals. The ΔlsrB, ΔrbsB, and ΔlsrB ΔrbsB strains also induced significantly less alveolar bone resorption than the wild type (P < 0.03, P < 0.02, and P < 0.01, respectively). However, bone loss induced by a ΔluxS strain was indistinguishable from that induced by the wild type, suggesting that AI-2 produced by indigenous microflora in the murine oral cavity may complement the ΔluxS mutation. Together, these results suggest that the QseBC two-component system is part of the AI-2 regulon and may link the detection of AI-2 to the regulation of downstream cellular processes that are involved in biofilm formation and virulence of A. actinomycetemcomitans.
Dental plaque is a complex and dynamic microbial community that forms as a biofilm on the surfaces of teeth and oral tissues (20, 22, 31, 50). It is comprised of over 700 species of bacteria (1, 20-22, 31, 50) and is the prime etiological agent of three common human oral diseases: dental caries, gingivitis, and periodontal disease (25, 26, 43). Major shifts in microbial populations within the oral biofilm have been associated with the progression of disease, as diseased sites often have increased populations of pathogenic species relative to healthy sites in the oral cavity (24-26). The host and/or microbial signals that contribute to the population shifts associated with disease are still unknown. The oral cavity is subject to continual environmental flux, including changes in pH, temperature, osmolarity, and nutrient supply, and it is possible that these stresses contribute in part to microbial population shifts in the biofilm. However, it is clear that oral bacteria rapidly detect environmental fluctuations and respond appropriately, allowing them to successfully coexist and thrive in the oral cavity (2, 18, 26).
Both intra- and interspecies communication is known to occur between bacteria, and these signaling processes potentially enable the organisms to coordinate their behavior and function by regulating gene expression as a community. One mechanism of communication, termed quorum sensing, is a cell density-dependent response (21, 28, 39, 55) which, in Gram-negative bacteria, is mediated by the production, release, and detection of soluble signal molecules called autoinducers. A variety of chemical species function as autoinducers, including acylated homoserine lactones (15, 30), quinolone derivatives (32), and furan derivatives (e.g., autoinducer-2 [AI-2]) (28, 39, 49, 52). As a population of bacteria expands, the external concentration of the autoinducer increases until a threshold is attained, at which point a signal transduction cascade is initiated that alters gene expression and behavior of the microbial community. Quorum sensing has been shown to control cell density-dependent behaviors, such as the expression of virulence factors, biofilm formation, and iron acquisition, in a variety of organisms (4, 15, 49, 51). Thus, it has been suggested that quorum sensing may allow bacteria in a biofilm to react coordinately as a multicellular organism to changes in the external environment.
The dental pathogen Aggregatibacter actinomycetemcomitans, a Gram-negative organism associated with aggressive forms of periodontitis and other systemic infections (6, 29, 42, 54), possesses an AI-2-dependent quorum-sensing system (12). AI-2 produced by A. actinomycetemcomitans regulates expression of virulence factors, biofilm formation, and iron uptake and also influences the planktonic growth of the organism under conditions of iron limitation (13, 38, 41). A. actinomycetemcomitans possesses two periplasmic AI-2 receptors, LsrB and RbsB, both of which are linked to ABC transporters (19, 40), suggesting that A. actinomycetemcomitans may import AI-2. However, exactly how the detection and/or importation of AI-2 is linked to downstream gene regulation remains to be determined. In Escherichia coli, the QseBC two-component signal transduction system has been suggested to regulate biofilm formation (14), and A. actinomycetemcomitans contains an operon that displays 70 to 80% sequence similarity to the QseBC genes of E. coli. QseBC is also similar to the FeuPQ two-component system that regulates iron uptake in Rhizobium leguminosarum, Sinorhizobium meliloti, and Brucella suis (11), and iron acquisition is known to be regulated by AI-2 (13) and dramatically influences biofilm formation (35, 36). In this study, we show that qseBC is part of the AI-2 regulon and that its induction requires a functional AI-2 receptor. The qseBC operon also contributes to biofilm formation and virulence of A. actinomycetemcomitans, since inactivation of qseC results in reduced biofilm growth and attenuates virulence in vivo. These results suggest that QseBC may couple the detection and/or importation of AI-2 to the downstream regulation of gene expression that controls these processes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains employed in this study are listed in Table 1. A. actinomycetemcomitans JP2 and 652 are afimbriated, smooth-colony-morphotype strains and were grown at 37°C under microaerophilic conditions in brain heart infusion broth (BHI; Becton Dickinson and Company [BD], Sparks, MD) supplemented with 40 mg of NaHCO3 (Sigma-Aldrich, St. Louis, MO) per liter. The luxS-deficient strain of A. actinomycetemcomitans was grown as described above, but in medium supplemented with kanamycin (25 μg/ml; Sigma-Aldrich). The ΔluxS mutant complemented with a plasmid-borne copy of luxS was grown in BHI supplemented with kanamycin (25 μg/ml) and streptomycin (50 μg/ml; Sigma-Aldrich). The A. actinomycetemcomitans ΔlsrB and ΔrbsB mutant strains were cultured in BHI supplemented with spectinomycin (50 μg/ml; Sigma-Aldrich) and kanamycin (25 μg/ml), respectively. The A. actinomycetemcomitans ΔlsrB ΔrbsB double mutant was cultured in BHI supplemented with kanamycin (25 μg/ml) and spectinomycin (50 μg/ml). The ΔqseC mutant was grown in BHI supplemented with spectinomycin (50 μg/ml), and the qseC-complemented strain was cultured in BHI supplemented with kanamycin (25 μg/ml).
TABLE 1.
Bacterial strains used in this study
| Bacterial strain | Description | Source or reference |
|---|---|---|
| A. actinomycetemcomitans strains | ||
| 652 | Wild type, serotype c, minimally leukotoxic strain | 7 |
| 652 ΔlsrB | lsrB::spec single-receptor mutant | 40 |
| 652 ΔrbsB | rbsB::kan single-receptor mutant | 19 |
| 652 ΔlsrB ΔrbsB | lsrB::spec rbsB::kan double-receptor mutant | 40 |
| 652 ΔqseC | qseC::spec mutant | This study |
| 652 ΔqseC comp | qseC mutant complemented with pYGKqseC | This study |
| JP2 | Wild type, serotype b, highly leukotoxic strain | 7 |
| JP2-12 | luxS::kan mutant | 12 |
| JP2-12/750 | luxS mutant complemented with pJRD215luxS | 13 |
| E. coli strains | ||
| DH5α TqseC | DH5α carrying pGEMTqseC | This study |
| DH5α TqseCcomp | DH5α carrying pBSKQseC-spec | This study |
| pVT1461 | Contains Specr cassette | K. Mintz |
| P. gingivalis strains | ||
| 33277 | Wild type | ATCC |
| ΔluxS | 33277 luxS::ermF mutant | 10 |
| V. harveyi BB170 | Sensor 1− sensor 2+ | 5 |
Porphyromonas gingivalis strains were grown in reduced Trypticase soy broth (TSB; BD) supplemented with yeast extract (1 g per liter; BD), menadione (1 μg per ml; Sigma-Aldrich), and hemin (5 μg per ml; Sigma-Aldrich). The medium was reduced for 24 h under anaerobic conditions by equilibration in an atmosphere consisting of 10% CO2, 10% H2, and 80% N2. The P. gingivalis ΔluxS mutant (kindly supplied by R. Lamont, University of Florida, Gainesville, FL) was grown as described above, but the medium was supplemented with erythromycin (10 μg/ml; Sigma-Aldrich) immediately before inoculation.
Vibrio harveyi BB170 (sensor 1− sensor 2+) was a gift from B. Bassler (Princeton University) and was grown overnight in AB medium with aeration at 30°C (5). AB medium consists of 0.3 M NaCl, 50 mM MgSO4, 0.2% Casamino Acids, 10 mM potassium phosphate (pH 7.0), 1 mM l-arginine, 2% glycerol, 1 μg per ml thiamine, and 10 ng per ml riboflavin. E. coli strains were grown in Luria-Bertani (LB) medium (BD) with aeration at 37°C. E. coli strains containing plasmid pGEM-T or pYGK were cultured as described above, using LB supplemented with 100 μg per ml ampicillin or 25 μg per ml kanamycin, respectively.
Construction of mutant strains.
The A. actinomycetemcomitans qseBC operon was identified from the genomic sequence of strain HK1651 (Los Alamos National Laboratory [http://www.oralgen.lanl.gov/]) and was annotated as the ygiX and qseC genes. To construct the fragment for inactivation of qseC, parts of the ygiX and qseC genes were amplified using genomic DNA of strain 652 as the template, with primers P1 and P2 (Table 2). The following PCR program was used: 94°C for 10 min for 1 cycle and then 94°C for 30 s, 60°C for 1 min, and 72°C for 2 min for 30 cycles. The PCR products were then ligated with pGEM-T Easy (Promega, Madison, WI) and transformed into E. coli DH5α. The resulting plasmid, pGEMTQseC, was purified from E. coli, cleaved by digestion with KpnI/BamHI, and ligated into pBSK. The resulting plasmid, pBSKQseC, was then cleaved with BamHI and treated with alkaline phosphatase for insertion of a spectinomycin resistance cassette. The spectinomycin resistance cassette was obtained by PCR amplification using plasmid pVT1461 (kindly supplied by K. Mintz, University of Vermont) as the template, with primers P3 and P4, and then ligated into pBSKQseC to create pBSKQseC-spec. This plasmid was then transformed into E. coli DH5α, and recombinant clones were confirmed by PCR using primers P1 and P4. Purified pBSKQseC-spec plasmid was introduced into A. actinomycetemcomitans by electroporation, with ampicillin resistance and spectinomycin resistance selection. The lack of a QseC transcript in the mutant strain was confirmed by reverse transcription-PCR (RT-PCR) using primers P6 and P7.
TABLE 2.
Primers used for PCR
| Primer | Sequence (5′-3′)a | Target gene | Product size (kbp) |
|---|---|---|---|
| P1 | GGTACCTCGCCGTGGATTGGTTTACCGAC | ygix (5′ primer) | 1.030 |
| P2 | GGATCCGGCGTTTATGCGACGGTTTG | qseC (3′ primer) | 1.030 |
| P3 | GGATCCATCGATTTTCGTTCGTG | Specr cassette (5′ primer) | 1.141 |
| P4 | GGATCCCATATGCAAGGGTTTAT | Specr cassette (3′ primer) | 1.141 |
| P6 | TAAGTGGAATAATTACAGCCTGCG | qseC (5′ primer) | 0.139 |
| P7 | TTGTTGTGCGTCAAACACTTGGTTC | qseC (3′ primer) | 0.139 |
| 5′ qseC-comp-HindIII | AAGCTTATGAAACTGAGTAAGTGG | qseC | 1.374 |
| 3′ qseC-comp-BamHI | GGATCCCAACTGAATCTCTGCC | qseC | 1.374 |
| 5′ ltx-pro-KpnI | GGTACCAATGAAAAAAAACAAAGCG | Leukotoxin promoter | 0.308 |
| 3′ ltx-pro-HindIII | AAGCTTACTCGTTTTCCTTTTTCATTAG | Leukotoxin promoter | 0.308 |
| 5′ QseBC induction | TCGCCGTGGATTGGTTTACCGAC | qseB | 0.159 |
| 3′ QseBC induction | GAATCAGCACCGGCACATCCTGC | qseB | 0.159 |
| 5′ 5S rRNA | GCGGGGATCCTGGCGGTGACCTACT | 5S-2 rRNA | 0.089 |
| 3′ 5S rRNA | GCGATCTAGACCACCTGAAACCATACC | 5S-2 rRNA | 0.089 |
Bold sequences indicate restriction enzyme sites used to facilitate cloning into the appropriate shuttle vectors.
To make the qseC-complemented strain, the entire qseC gene was amplified using 652 genomic DNA as the template, with primers 5′ qseC-comp-HindIII and 3′ qseC-comp-BamHI. The leukotoxin promoter (ltx-pro) was also amplified from 652 genomic DNA, using the 5′ ltx-pro-KpnI and 3′ ltx-pro-HindIII primers. The following PCR program was used: 94°C for 10 min for 1 cycle and then 94°C for 30 s, 60°C for 1 min, and 72°C for 2 min for 30 cycles. Both the qseC and leukotoxin PCR products were then ligated into pGEM-T Easy (Promega, Madison, WI) and transformed into E. coli DH5α. The resulting plasmids, pGEMTQseCcomp and pGEMTltx-pro, were purified from E. coli. The pGEMTQseCcomp plasmid was cleaved by digestion with HindIII and BamHI, and the pGEMTltx-pro plasmid was cleaved by digestion with KpnI and HindIII. Both digested fragments—the qseC gene and ltx-pro—were then ligated into pYGK. The resulting plasmid, pYGKqseC, was transformed into E. coli. The plasmid was then purified from an overnight culture of E. coli and transformed into the ΔqseC mutant for complementation.
Partial purification of A. actinomycetemcomitans AI-2.
An enriched fraction containing AI-2 from A. actinomycetemcomitans was produced as described by Sperandio et al. (44). Briefly, an overnight culture of A. actinomycetemcomitans cells was diluted 1:20 in fresh medium, cultured to mid-exponential phase (optical density, 0.3) at 37°C, and then harvested by centrifugation. AI-2 was obtained from a 7.2-ml aliquot of the conditioned medium. The culture supernatant was first filtered through a 0.22-μm-pore-size filter and then through a Centricon YM-3 3-kDa exclusion filter (Millipore, Bedford, MA). The resulting filtrate was lyophilized, suspended in 1 ml of cold 5 mM sodium phosphate buffer, pH 6.2, and chromatographed on a C18 Sep-Pak reverse-phase column (Waters Company, Milford, MA) according to the manufacturer's instructions. Induction of bioluminescence of Vibrio harveyi BB170 was monitored in order to follow the AI-2 activity in the column fractions. Active fractions were lyophilized and stored at 4°C.
RNA isolation and real-time PCR.
Overnight cultures of the appropriate A. actinomycetemcomitans strains were diluted 1:20 in fresh BHI medium, with or without partially purified AI-2, and were incubated at 37°C until the mid-exponential growth phase for RNA isolation. Total RNA was isolated from A. actinomycetemcomitans cells by use of a 5 Prime PerfectPure RNA Cell & Tissue kit (5 Prime Inc., Gaithersburg, MD) according to the manufacturer's instructions. To ensure that the samples were free of contaminating genomic DNA, the RNA preparation was digested with RQ RNase-free DNase I (Promega Corporation, Madison, WI). The concentration and purity of each RNA sample were measured via spectrophotometry (ND-1000 spectrophotometer; NanoDrop Technologies, Inc., Wilmington, DE) and were also assessed by gel electrophoresis. Samples were checked for contamination of genomic DNA by real-time PCR, using A. actinomycetemcomitans 5S rRNA primers (Table 2). RNA samples were considered free of significant genomic DNA if no amplification product was detected by real-time PCR after at least 30 cycles of amplification. RNA that was not immediately utilized for a reverse transcription reaction was aliquoted into different tubes and stored at −80°C until future use.
First-strand cDNA was prepared by using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The resulting cDNA was amplified using a Smart Cycler system (Cepheid, Sunnyvale, CA), with a final reaction volume of 25 μl that contained 100 ng of cDNA, primers for qseC (5′ QseBC induction and 3′ QseBC induction; ∼71 μM [final concentration of each]), and 1× FastStart SYBR green master mix (Roche, Indianapolis, IN). The amplification conditions for real-time PCRs were as follows: 35 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and elongation at 72°C for 30 s. The threshold cycle for each real-time PCR was determined from a second derivative plot of total fluorescence as a function of cycle number by using the software package supplied with the Smart Cycler system. All gene-specific threshold values were normalized against threshold values for primers specific for the A. actinomycetemcomitans 5S rRNA gene (∼60 μM [final concentration]). Real-time PCRs were carried out in triplicate, with consistent results. Each real-time PCR end-point amplification product was visualized by electrophoresis on 2% agarose gels.
Biofilm formation and analysis.
A. actinomycetemcomitans biofilms were grown on a saliva-coated cover glass in a polycarbonate flow chamber (model FC81; Biosurface Technologies Corp, Bozeman, MT) (chamber dimensions are 50.5 mm by 12.7 mm by 2.54 mm) at a flow rate of 5.8 ml per hour at 25°C, essentially as described by Shao et al. (41). Briefly, saliva was self-collected, filter sterilized (pore size, 0.22 μm), and incubated on the cover glass (60 mm by 24 mm) for 30 min at 37°C. The saliva-coated cover glass was then fixed in the flow chamber and washed with phosphate-buffered saline (PBS; 100 mM NaH2PO4, 150 mM NaCl) for 10 min at a flow rate of 60 ml per hour by use of a peristaltic pump (Manostat Sarah cassette; Fisher Scientific, Pittsburgh, PA). Overnight cultures of A. actinomycetemcomitans were resuspended in PBS at an optical density at 600 nm of 0.5, inoculated for 1 h into a polycarbonate flow chamber, and then washed with PBS for 30 min. Bound cells were fed BHI medium and allowed to grow for 60 h at a flow rate of 5.8 ml per hour. The resulting biofilm was stained with 0.2 mg/ml fluorescein isothiocyanate (FITC; Sigma-Aldrich) for 1 h in the dark and then washed with PBS for 2 h.
Biofilms were visualized using an Olympus Fluoview FV500 confocal scanning laser microscope (Olympus, Pittsburgh, PA) at a magnification of ×600, using an argon laser. Confocal images were captured from 9 randomly chosen frames from each flow chamber. Biofilm depth was determined by performing z-plane scans from 0 to 100 μm above the cover glass surface. The total biofilm biomass was determined by integrating fluorescence intensity across the z stack, using the Fluoview FV500 software provided by Olympus. Biofilm depth, total biomass, and biofilm topology were also quantified utilizing the COMSTAT image-processing software package (16). Biomass data were analyzed using a pairwise t test (Graphpad Software, Inc.) and are expressed as means ± standard deviations calculated for all the frames obtained for a given biofilm. Experiments were carried out in triplicate.
Mouse periodontitis model.
To assess A. actinomycetemcomitans virulence, the Baker mouse model of periodontitis was utilized (3). This model measures alveolar bone resorption, the clinical presentation of periodontitis in humans, as an outcome, and we used the model previously to assess the virulence of P. gingivalis, another periodontal pathogen (33, 48). All animal procedures received Institutional Animal Care and Use Committee approval and were in accordance with federal guidelines for the care and use of laboratory animals. Specific-pathogen-free female BALB/cByJ mice (10 weeks old; Jackson Laboratory, Bar Harbor, ME) were orally infected with wild-type or mutant strains of A. actinomycetemcomitans or, in some experiments, with wild-type or mutant strains of P. gingivalis. Briefly, prior to infection, each group of mice was given the antibiotics sulfamethoxazole (final concentration of 800 μg/ml; Sigma-Aldrich) and trimethoprim (final concentration of 400 μg/ml; Sigma-Aldrich) in their water bottles ad libitum for 10 days to suppress the normal flora of the mouse oral cavity. After the 10-day antibiotic period, the mice were given regular drinking water without antibiotics for 4 days and then were orally infected a total of five times at 2-day intervals with 1 × 109 CFU/ml of bacteria resuspended in 2% carboxymethylcellulose (CMC; MP Biomedicals, Solon, OH) via a blunt-tipped syringe (Fisher Scientific, Pittsburgh, PA). Animals that served as the negative-control group (sham-infected mice) received only 2% CMC without bacteria. Mice were euthanized by CO2 inhalation and cervical dislocation 47 days after the last infection (for a total experiment time of 70 days).
The maxillae were collected at the end of the experiment and were immersed in 3% H2O2 overnight. Maxillae were then sonicated in 1% bleach, cleaned, rinsed with water, and then stained for 30 s with 1% methylene blue (Ricca Chemical Company, Arlington, TX). To measure the loss of alveolar bone, the distance from the cemento-enamel junction (CEJ) to the alveolar bone crest (ABC) was measured at 7 sites on the buccal side of the right and left maxillary molars (for a total of 14 measurements per animal), utilizing a dissecting microscope fitted with a video image marker measurement system (model VIA-170K; Fryer, Huntley, IL) standardized to give measurements in millimeters. Alveolar bone loss was calculated by subtracting the 14-site mean sum total CEJ-ABC distance for each experimental group from the mean sum total CEJ-ABC distance for the sham-infected group. Results are expressed as changes in bone size, in millimeters, and negative values indicate bone loss compared with sham-infected controls.
Statistical analysis.
Data were evaluated by analysis of variance (ANOVA) and the Dunnette multiple-comparison test, using the InStat program (Graphpad). Two-tailed t tests were also performed where appropriate (comparison of two groups only). Statistical differences were considered significant with P values of <0.05.
RESULTS
AI-2 is required for induction of QseBC in A. actinomycetemcomitans.
In some E. coli strains, the QseBC two-component system is induced by AI-2 and, furthermore, is part of the AI-2-dependent response circuit that controls biofilm growth (14). The genome of A. actinomycetemcomitans encodes a two-component system that is related to QseBC in E. coli. The genes are listed as ygiX and qseC and are annotated as genes for a sensor histidine kinase (qseC) and a response regulator (ygiX) of a two-component signal transduction system (www.oralgen.lanl.gov). In strain 652, the ygiX and qseC genes overlap by 11 bp (data not shown), and RT-PCR indicates that they are coexpressed in an operon (data not shown). The deduced amino acid sequences for ygiX and qseC from strain 652 exhibit significant sequence identity with QseB and QseC from E. coli (data not shown).
To determine if AI-2 regulates the putative QseBC two-component system in A. actinomycetemcomitans, the expression of qseC was examined using real-time PCR with RNA isolated from overnight cultures of the ΔluxS mutant grown in medium alone or in medium supplemented with partially purified AI-2. As shown in Table 3, the presence of AI-2 in the growth medium resulted in a 14.5-fold induction of qseC. As a control, cultures were also grown in medium supplemented with conditioned culture fluid obtained from a ΔluxS strain that was subjected to the same purification scheme used to obtain the AI-2 samples. No increase in qseC expression was observed under these conditions. In addition, the induction of qseC did not occur in the ΔlsrB ΔrbsB strain of A. actinomycetemcomitans, which lacks the AI-2 receptors but can still produce AI-2. This suggests that the QseBC two-component system in A. actinomycetemcomitans is regulated by AI-2 and may be linked to the detection and/or importation of AI-2 by the AI-2 receptors.
TABLE 3.
QseBC two-component system is influenced by AI-2
| Strain and signal | ΔTa | ΔΔT | Fold induction |
|---|---|---|---|
| JP2-12 | −3.1 ± 0.6 | 0.11 ± 0.04 | 1.0 |
| JP2-12 + AI-2 | −0.66 ± 0.2 | 1.52 ± 0.3 | 14.5 ± 4.3 |
| JP2-12 + mockb | 3.6 ± 0.05 | 0.08 ± 0.01 | 0.8 ± 0.3 |
| ΔlsrB ΔrbsB mutantc | 3.2 ± 0.35 | 0.09 ± 0.13 | 0.95 ± 0.2 |
ΔT was calculated by subtracting the CT determined for the qseC reaction from the CT for the 5S rRNA control.
The mock control represents culture fluid from the ΔluxS strain subjected to the purification scheme used to partially purify AI-2.
This strain contains a functional copy of luxS and produces but cannot respond to AI-2.
Inactivation of qseC influences biofilm formation in A. actinomycetemcomitans.
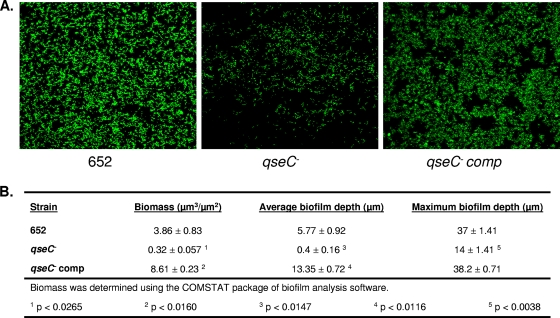
Previous studies demonstrated that biofilm formation by A. actinomycetemcomitans was dependent on AI-2-mediated quorum sensing (41), but how the initial detection of the AI-2 signal leads to downstream regulation of gene expression that influences biofilm growth was not determined. To determine if QseBC plays a role in regulating biofilm growth, wild-type, ΔqseC, and qseC-complemented strains of A. actinomycetemcomitans were cultured in flow cells as described in Materials and Methods. Representative images of the biofilms formed by each of these strains are shown in Fig. 1 A and illustrate that inactivation of qseC resulted in a significant reduction in biofilm growth. Complementation of the mutant with a plasmid-borne copy of qseC restored biofilm growth to wild-type levels. Analysis of the biofilms by use of COMSTAT is shown in Fig. 1B. The total biomass (P < 0.03), average depth (P < 0.02), and maximum depth (P < 0.004) of biofilms formed by the qseC mutant were significantly reduced relative to those of the wild type (Fig. 1B). All biofilm growth parameters were restored to wild-type levels in the complemented strain. Indeed, biomass and average biofilm depth for the complemented strain were greater than those for the wild type, which may reflect gene dosage, as qseC is expressed from a multicopy plasmid in the complemented organism. These results suggest that QseC controls biofilm formation by A. actinomycetemcomitans.
FIG. 1.
qseC is necessary for biofilm growth in A. actinomycetemcomitans. (A) Representative confocal images (in the x-y plane) of 60-hour biofilms formed by wild-type A. actinomycetemcomitans (strain 652) and ΔqseC and qseC-complemented strains. The resulting biofilms were quantified using the COMSTAT program (16). (B) Values for microbial biomass, average biofilm depth, and maximum biofilm depth were determined using COMSTAT image-processing software as described in Materials and Methods. At least 9 individual microscopic frames were analyzed for each biofilm experiment, and three independent biofilm experiments were carried out for each strain.
The A. actinomycetemcomitans ΔqseC mutant and AI-2 receptor mutants are less virulent in a murine model of periodontitis.
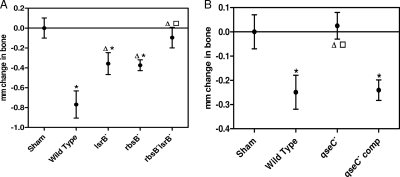
The ability of A. actinomycetemcomitans to grow in a biofilm is essential for its survival in the oral cavity and is associated with virulence. Our previous studies showed that AI-2-dependent quorum sensing (41) and qseC (described above) are essential components that regulate biofilm formation. To determine if AI-2 and qseC influence A. actinomycetemcomitans virulence, we utilized a murine model of periodontitis (3) in which mice were orally infected with 1 × 109 CFU/ml of the wild-type strain or strains lacking either QseC or the AI-2 receptors. As a control, sham-infected mice were infected only with CMC, the vehicle used to deliver the bacterial suspensions. Alveolar bone loss, one of the clinical outcomes of periodontal disease in humans, was then measured 47 days after the last infection. As shown in Fig. 2 A, mice that were infected with the wild type (P = 0.0002), ΔlsrB (P = 0.0261), or ΔrbsB (P = 0.0036) strain of A. actinomycetemcomitans exhibited significantly more alveolar bone resorption than did sham-infected mice. However, animals infected with either the ΔlsrB or ΔrbsB mutant strain exhibited a significant reduction in bone resorption relative to that of the wild type (for the ΔlsrB strain, P = 0.0285; for the ΔrbsB strain, P = 0.0134), suggesting that the detection of AI-2 by either receptor contributes to virulence. Consistent with this, mice that were infected with the ΔlsrB ΔrbsB double-receptor mutant did not display significant levels of bone resorption compared to the sham-infected controls.
FIG. 2.
qseC is necessary for in vivo virulence of A. actinomycetemcomitans. (A) AI-2 receptor mutants caused significantly less alveolar bone resorption than the wild type did. Infection with either the ΔlsrB or ΔrbsB mutant induced more alveolar bone loss than that in sham-infected mice but significantly less than that induced by the wild-type strain. The ΔlsrB ΔrbsB mutant induced bone loss comparable to that observed in sham-infected animals. The asterisks indicate statistically significant differences relative to sham-infected animals, triangles indicate significant differences relative to the wild type, and the square indicates a significant difference relative to the ΔrbsB strain. Values are means ± standard errors of the means (SEM). (B) Infection of mice with the ΔqseC mutant resulted in significantly less bone loss than that induced by the wild-type and complemented strains. The virulence of the mutant was restored to wild-type levels when the qseC mutation was complemented with a plasmid-borne copy of the qseC gene. Asterisks indicate statistical significance relative to sham-infected animals, the triangle indicates statistical significance relative to the wild type, and the square indicates statistical significance relative to the complemented strain. Values are means ± SEM.
As shown in Fig. 2B, animals that were infected with the ΔqseC mutant exhibited significantly less bone resorption than those infected with the wild type (P < 0.02) and were indistinguishable from the sham-infected controls. The virulence of the ΔqseC mutant was restored to wild-type levels when it was genetically complemented with a plasmid-borne copy of qseC. These results suggest that both QseC and the ability to detect AI-2 contribute to A. actinomycetemcomitans virulence and that a defective AI-2-dependent quorum-sensing pathway negatively impacts the ability of A. actinomycetemcomitans to induce alveolar bone resorption.
Inactivation of luxS does not influence A. actinomycetemcomitans virulence.
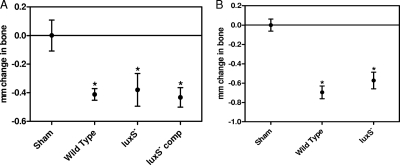
Previous studies showed that a ΔluxS mutant of A. actinomycetemcomitans formed biofilms that exhibited reduced total biomass and biofilm depth relative to those of wild-type biofilms (41). Since the luxS gene is responsible for the production of AI-2 and AI-2 is essential for the formation of biofilms, we next examined the importance of luxS in the in vivo virulence of A. actinomycetemcomitans. As shown in Fig. 3 A, the extent of bone resorption induced by the A. actinomycetemcomitans ΔluxS mutant was indistinguishable from that induced by the wild-type or luxS-complemented strain, but all strains induced alveolar bone resorption over that in the sham-infected controls (for the wild type, P = 0.0052; for the ΔluxS mutant, P = 0.0365; and for the luxS-complemented strain, P = 0.007). Similar results were obtained when animals were infected with wild-type and ΔluxS mutant strains of another periodontal pathogen, P. gingivalis (Fig. 3B). Thus, inactivation of the AI-2 receptors, but not the AI-2 synthase, affected A. actinomycetemcomitans virulence. This suggests that AI-2 produced by indigenous bacteria in the mouse oral cavity may complement the luxS mutation in A. actinomycetemcomitans.
FIG. 3.
The ΔluxS mutation is complemented in vivo. The ΔluxS mutants of A. actinomycetemcomitans (A) and P. gingivalis (B) induced alveolar bone resorption at levels that were comparable to those with the wild type or the luxS-complemented strain. Asterisks indicate statistically significant differences relative to sham-infected animals. Values are means ± SEM.
DISCUSSION
Dental plaque is a complex oral biofilm comprised of a diverse microbial community consisting of more than 700 different bacterial species (1, 20-22, 31, 50) that must coexist and adapt to the fluctuating environment of the oral cavity. It is likely that both intra- and interspecies communication occurs between these organisms and that these processes may enable oral organisms to coordinate their behavior and function based on their local environment and the other organisms that occupy the same niche. For A. actinomycetemcomitans, this intimate cell-to-cell communication potentially occurs via the quorum-sensing circuit dependent upon the soluble signaling molecule AI-2 (12). Indeed, previous studies showed that AI-2 regulates both biofilm formation and various iron acquisition pathways in A. actinomycetemcomitans (13, 41). However, it is not known how the initial detection of AI-2 is linked to downstream regulation of gene expression that controls these complex phenotypes. A. actinomycetemcomitans does not possess the dedicated two-component system that controls the cell density-dependent response of Vibrio spp. to AI-2 (i.e., the LuxQ sensor kinase, LuxU phosphorelay protein, LuxO response regulator, and LuxR master regulator encoded by the lux operon). Instead, the genome of A. actinomycetemcomitans encodes two periplasmic proteins, LsrB and RbsB, which function as receptors for AI-2, and each is linked to a putative ABC transporter that may import the signal (19, 40). Thus, the AI-2 response circuit in A. actinomycetemcomitans may be similar to that described by Li et al. for E. coli, in which the LsrR regulator controls the expression of genes involved in biofilm growth as well as regulating the expression of the lsr operon bound to AI-2 that has been phosphorylated by the LsrK kinase (23). AI-2 has also been suggested to regulate motility and biofilm formation in E. coli through the QseBC two-component system (14, 45). The A. actinomycetemcomitans genome also encodes the QseBC two-component system, and our results here show that qseBC is part of the AI-2 regulon in A. actinomycetemcomitans, since qseBC is induced by the AI-2 signal itself and its induction requires a functional AI-2 receptor. Furthermore, AI-2-dependent induction of QseBC in some E. coli strains requires the MqsR regulator (14). MqsR has been shown to be part of a toxin/antitoxin system and adopts an α/β fold similar to that of the RelE family of bacterial RNase toxins (8). Consistent with this, Yamaguchi et al. have shown that MqsR functions as a GCU-specific mRNA interferase (53). MqsR exhibits sequence similarity to open reading frame AA00673 in the A. actinomycetemcomitans genome (www.oralgen.lanl.gov), which is coexpressed with AA00672. AA00673 and AA00672 also exhibit approximately 65% sequence identity to the HigBA toxin/antitoxin system of Vibrio cholerae, and the HigB toxin is known to function as an mRNase (9). This suggests that QseBC may participate in the AI-2 response circuit in A. actinomycetemcomitans and that AA00673 may represent a paralog of MqsR and contribute to qseBC regulation.
Consistent with this, inactivation of the QseC sensor influenced the formation of biofilms by A. actinomycetemcomitans in that the ΔqseC mutant formed biofilms that were reduced in total biomass, average biofilm depth, and maximum biofilm depth relative to those for biofilms formed by the wild-type strain. The biofilm growth phenotype of the ΔqseC strain closely resembled the growth phenotype of the A. actinomycetemcomitans ΔluxS mutant previously reported by Shao et al. (41). Complementation of the ΔqseC mutation restored biofilm formation, and indeed, the complemented strain formed biofilms that exhibited greater average depth and biomass than those of the wild type. This may have been due to the gene dosage of qseC in the complemented strain, since qseC was expressed from a multicopy plasmid in this organism. These data suggest that the QseC sensor, and presumably the QseB response regulator, may be part of the quorum-sensing circuit mediated by AI-2 in A. actinomycetemcomitans.
However, although the QseC sensor may be common to the AI-2 signal transduction pathways of both A. actinomycetemcomitans and some E. coli strains, other aspects of these AI-2 response circuits appear to differ. Sperandio et al. have shown that E. coli O157:H7 QseC responds to stress hormones (epinephrine/norepinephrine) as well as the microbial signal AI-3 and that QseC may function in a one-to-many-branched signal transduction pathway that activates the QseB, QseF, and KdpE response regulators (17, 34). In addition, the QseE sensor kinase was shown to reside downstream of QseC in the adrenergic signal transduction cascade (17). At present, the signal that the A. actinomycetemcomitans QseC kinase senses is not known, and we have been unsuccessful in identifying AI-3 in A. actinomycetemcomitans extracts (D. R. Demuth, unpublished data). Furthermore, paralogs of the QseE, QseF, and KdpE polypeptides could not be identified in the A. actinomycetemcomitans genome at a search stringency that readily detected QseC with the E. coli QseC sequence as the query. This suggests that the components that reside downstream of QseC in A. actinomycetemcomitans may differ from those in some E. coli strains. Interestingly, E. coli O157:H7 lacks MqsR, and transcription of qseBC is independent of AI-2 in this organism (44).
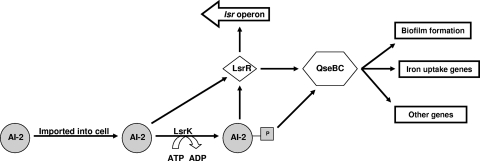
Biofilm growth in A. actinomycetemcomitans is dependent upon AI-2-mediated quorum sensing (41), and formation of biofilms is essential to the in vivo survival and virulence of dental pathogens in the oral cavity. Our results show that AI-2-mediated quorum sensing is important for A. actinomycetemcomitans virulence in vivo, since mutants that were deficient in either of the AI-2 receptors (LsrB or RbsB) induced lower levels of alveolar bone resorption than the wild-type strain and since a double-receptor mutant was essentially avirulent. These phenotypes are also consistent with the biofilm phenotypes exhibited by these mutant strains (41). The ΔqseC mutant was also avirulent and did not induce bone loss over that in the sham-infected control group. This is consistent with QseC participating in and residing downstream of the AI-2 receptors in the AI-2 response circuit, as shown in the model presented in Fig. 4. Nonetheless, the exact mechanism(s) through which QseBC contributes to the intracellular response of AI-2 as well as to biofilm growth in A. actinomycetemcomitans is not known. A. actinomycetemcomitans possesses the protein LsrR, which is the master regulator of the lsr operon in Salmonella enterica (46, 47), and Gonzalez-Barrios et al. suggested that AI-2-dependent biofilm growth of E. coli requires internalization of AI-2 (14). In A. actinomycetemcomitans, the QseBC two-component system may couple the detection/importation of AI-2, either alone or in cooperation with LsrR, to downstream regulation of biofilm formation and other cellular processes.
FIG. 4.
AI-2 quorum-sensing circuit in A. actinomycetemcomitans. For A. actinomycetemcomitans, we hypothesize that AI-2-dependent biofilm formation and other potential AI-2-mediated cellular processes may result from one or two possible pathways. AI-2 is initially bound by its periplasmic receptors and imported into the cell by ABC-type transporters associated with the lsr and rbs operons. Once inside the cell, AI-2 is presumed to be phosphorylated by LsrK, and phosphorylated AI-2 is thought to interact directly with LsrR, resulting in derepression of the lsr operon. However, LsrR may also interact with unphosphorylated AI-2, as suggested by Li et al. (21). Regulation of the QseBC two-component system may occur through direct interaction with AI-2, or its expression may be induced by LsrR. The QseBC two-component system in turn alters the expression of genes that mediate biofilm formation, and potentially other genes, including those important for iron acquisition.
At present, the sets of genes that are regulated by the QseBC two-component system in A. actinomycetemcomitans are not known, but it is interesting that qseBC also exhibits sequence similarity to the feuPQ two-component system that regulates iron acquisition in several other bacterial species. Several operons that encode iron acquisition pathways have been shown to be regulated by AI-2 (13), and iron availability is also known to dramatically influence biofilm formation by A. actinomycetemcomitans (35, 36). Thus, it is possible that the biofilm defect that arises from inactivation of qseC occurs in part from the dysregulation of iron acquisition mechanisms. We are currently determining if the QseBC two-component system coregulates iron acquisition genes, and RT-PCR experiments suggest that at least one ferric uptake transporter is under QseBC control (Demuth, unpublished data).
Given that luxS was previously shown to influence biofilm growth, it was surprising that neither A. actinomycetemcomitans nor P. gingivalis ΔluxS mutants exhibited attenuated virulence in the murine model. A possible explanation for this result is that the ΔluxS mutation was complemented by AI-2 produced by organisms that are indigenous to the murine oral cavity. Consistent with this, McNab et al. have shown that AI-2 cross talk occurs in dual-species biofilms of Streptococcus gordonii and P. gingivalis (27). In this system, dual-species biofilms formed efficiently even if one of the strains harbored a luxS mutation, but no biofilms formed if both strains were LuxS deficient. Our previous studies also showed that AI-2-mediated cross talk is possible between A. actinomycetemcomitans and P. gingivalis and that the AI-2 signal of A. actinomycetemcomitans is capable of modulating the expression of luxS-regulated genes in P. gingivalis (12). In addition, Rickard et al. showed that the ability of the oral microbes Actinomyces naeslundii and Streptococcus oralis to form dual-species biofilms in saliva was dependent on AI-2 produced by S. oralis (37). The inability to detect or respond to AI-2 prevents the stimulation of the AI-2 response circuit and results in reduced biofilm growth and attenuated virulence, whereas the inability to produce AI-2 is likely overcome by the presence of exogenous signals produced by other bacteria in the murine oral cavity.
In summary, we have shown that the QseBC two-component system is induced by AI-2 and that QseC is important for biofilm formation and virulence of A. actinomycetemcomitans. Our results suggest that QseC is part of the AI-2 response circuit and resides downstream of the AI-2 receptor proteins LsrB and RbsB. Further definition of the genes regulated by the QseBC two-component system may identify new targets for therapeutic intervention of aggressive periodontitis and other systemic infections associated with A. actinomycetemcomitans.
Acknowledgments
We thank Janice Ditslear, Kathy Laster, and the rest of the University of Louisville Research Resources Center staff for their help with our mouse studies.
This study was supported by Public Health Service grant RO1-DE14605 from the National Institute of Dental and Craniofacial Research.
Editor: F. C. Fang
Footnotes
Published ahead of print on 19 April 2010.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Actis, L. A., E. R. Rhodes, and A. P. Tomaras. 2003. Genetic and molecular characterization of a dental pathogen using genome-wide approaches. Adv. Dent. Res. 17:95-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, P. J., R. T. Evans, and D. C. Roopenian. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39:1035-1040. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 6.Block, P. J., C. Yoran, A. C. Fox, and A. J. Kaltman. 1973. Actinobacillus actinomycetemcomitans endocarditis: report of a case and review of the literature. Am. J. Med. Sci. 266:387-392. [DOI] [PubMed] [Google Scholar]
- 7.Brogan, J. M., E. T. Lally, K. Poulsen, M. Kilian, and D. R. Demuth. 1994. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect. Immun. 62:501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, B. L., S. Grigoriu, Y. Kim, J. M. Arruda, A. Davenport, T. K. Wood, W. Peti, and R. Page. 2009. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 5:e1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen-Dalsgaard, M., and K. Gerdes. 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62:397-411. [DOI] [PubMed] [Google Scholar]
- 10.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 12.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong, K. P., L. Gao, and D. R. Demuth. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henke, J. M., and B. L. Bassler. 2004. Bacterial social engagements. Trends Cell Biol. 14:648-656. [DOI] [PubMed] [Google Scholar]
- 16.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 17.Hughes, D. T., M. B. Clarke, K. Yamamoto, D. A. Rasko, and V. Sperandio. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog. 5:e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James, C. E., Y. Hasegawa, Y. Park, V. Yeung, G. D. Tribble, M. Kuboniwa, D. R. Demuth, and R. J. Lamont. 2006. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect. Immun. 74:3834-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James, D., H. Shao, R. J. Lamont, and D. R. Demuth. 2006. The Actinobacillus actinomycetemcomitans ribose binding protein RbsB interacts with cognate and heterologous autoinducer 2 signals. Infect. Immun. 74:4021-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolenbrander, P. E., and J. London. 1993. Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 175:3247-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuramitsu, H. K., X. He, R. Lux, M. H. Anderson, and W. Shi. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J., C. Attila, L. Wang, T. K. Wood, J. J. Valdes, and W. E. Bentley. 2007. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J. Bacteriol. 189:6011-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh, P. D. 2003. Are dental diseases examples of ecological catastrophes? Microbiology 149:279-294. [DOI] [PubMed] [Google Scholar]
- 25.Marsh, P. D. 2006. Dental plaque as a biofilm and a microbial community—implications for health and disease. BMC Oral Health 6(Suppl. 1):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsh, P. D., and D. J. Bradshaw. 1997. Physiological approaches to the control of oral biofilms. Adv. Dent. Res. 11:176-185. [DOI] [PubMed] [Google Scholar]
- 27.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNab, R., and R. J. Lamont. 2003. Microbial dinner-party conversations: the role of LuxS in interspecies communication. J. Med. Microbiol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 29.Page, M. I., and E. O. King. 1966. Infection due to Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. N. Engl. J. Med. 275:181-188. [DOI] [PubMed] [Google Scholar]
- 30.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. U. S. A. 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paster, B. J., I. Olsen, J. A. Aas, and F. E. Dewhirst. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 42:80-87. [DOI] [PubMed] [Google Scholar]
- 32.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce, D. L., S. Nishiyama, S. Liang, M. Wang, M. Triantafilou, K. Triantafilou, F. Yoshimura, D. R. Demuth, and G. Hajishengallis. 2009. Host adhesive activities and virulence of novel fimbrial proteins of Porphyromonas gingivalis. Infect. Immun. 77:3294-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reading, N. C., D. A. Rasko, A. G. Torres, and V. Sperandio. 2009. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 106:5889-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhodes, E. R., S. Menke, C. Shoemaker, A. P. Tomaras, G. McGillivary, and L. A. Actis. 2007. Iron acquisition in the dental pathogen Actinobacillus actinomycetemcomitans: what does it use as a source and how does it get this essential metal? Biometals 20:365-377. [DOI] [PubMed] [Google Scholar]
- 36.Rhodes, E. R., C. J. Shoemaker, S. M. Menke, R. E. Edelmann, and L. A. Actis. 2007. Evaluation of different iron sources and their influence in biofilm formation by the dental pathogen Actinobacillus actinomycetemcomitans. J. Med. Microbiol. 56:119-128. [DOI] [PubMed] [Google Scholar]
- 37.Rickard, A. H., R. J. Palmer, Jr., D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60:1446-1456. [DOI] [PubMed] [Google Scholar]
- 38.Schaeffer, L. M., M. L. Schmidt, and D. R. Demuth. 2008. Induction of Aggregatibacter actinomycetemcomitans leukotoxin expression by IS1301 and orfA. Microbiology 154:528-538. [DOI] [PubMed] [Google Scholar]
- 39.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 40.Shao, H., D. James, R. J. Lamont, and D. R. Demuth. 2007. Differential interaction of Aggregatibacter (Actinobacillus) actinomycetemcomitans LsrB and RbsB proteins with autoinducer 2. J. Bacteriol. 189:5559-5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao, H., R. J. Lamont, and D. R. Demuth. 2007. Autoinducer 2 is required for biofilm growth of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect. Immun. 75:4211-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slots, J., H. S. Reynolds, and R. J. Genco. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect. Immun. 29:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Socransky, S. S., and A. D. Haffajee. 1992. The bacterial etiology of destructive periodontal disease: current concepts. J. Periodontol. 63:322-331. [DOI] [PubMed] [Google Scholar]
- 44.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U. S. A. 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 46.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 47.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 48.Wang, M., M. A. Shakhatreh, D. James, S. Liang, S. Nishiyama, F. Yoshimura, D. R. Demuth, and G. Hajishengallis. 2007. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J. Immunol. 179:2349-2358. [DOI] [PubMed] [Google Scholar]
- 49.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 50.Whittaker, C. J., C. M. Klier, and P. E. Kolenbrander. 1996. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50:513-552. [DOI] [PubMed] [Google Scholar]
- 51.Winans, S. C., and B. L. Bassler. 2002. Mob psychology. J. Bacteriol. 184:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 53.Yamaguchi, Y., J. H. Park, and M. Inouye. 2009. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284:28746-28753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zambon, J. J., C. DeLuca, J. Slots, and R. J. Genco. 1983. Studies of leukotoxin from Actinobacillus actinomycetemcomitans using the promyelocytic HL-60 cell line. Infect. Immun. 40:205-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, G., W. Wan, S. Mansouri, J. F. Alfaro, B. L. Bassler, K. A. Cornell, and Z. S. Zhou. 2003. Chemical synthesis of S-ribosyl-l-homocysteine and activity assay as a LuxS substrate. Bioorg. Med. Chem. Lett. 13:3897-3900. [DOI] [PubMed] [Google Scholar]