Abstract
Probiotics are viable microorganisms that are increasingly used for treatment of a variety of diseases. Occasionally, however, probiotics may have adverse clinical effects, including septicemia. Here we examined the role of the intestinal microbiota and the adaptive immune system in preventing translocation of probiotics (e.g., Escherichia coli Nissle). We challenged C57BL/6J mice raised under germfree conditions (GF-raised C57BL/6J mice) and Rag1−/− mice raised under germfree conditions (GF-raised Rag1−/− mice) and under specific-pathogen-free conditions (SPF-raised Rag1−/− mice) with probiotic E. coli strain Nissle 1917, strain Nissle 1917 mutants, the commensal strain E. coli mpk, or Bacteroides vulgatus mpk. Additionally, we reconstituted Rag1−/− mice with CD4+ T cells. E. coli translocation and dissemination and the mortality of mice were assessed. In GF-raised Rag1−/− mice, but not in SPF-raised Rag1−/− mice or GF-raised C57BL/6J mice, oral challenge with E. coli strain Nissle 1917, but not oral challenge with E. coli mpk, resulted in translocation and dissemination. The mortality rate was significantly higher for E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice (100%; P < 0.001) than for E. coli strain Nissle 1917-challenged SPF-raised Rag1−/− mice (0%) and GF-raised C57BL/6J mice (0%). Translocation of and mortality due to strain E. coli Nissle 1917 in GF-raised Rag1−/− mice were prevented when mice were reconstituted with T cells prior to strain E. coli Nissle 1917 challenge, but not when mice were reconstituted with T cells after E. coli strain Nissle 1917 challenge. Cocolonization experiments revealed that E. coli mpk could not prevent translocation of strain E. coli Nissle 1917. Moreover, we demonstrated that neither lipopolysaccharide structure nor flagella play a role in E. coli strain Nissle 1917 translocation and dissemination. Our results suggest that if both the microbiota and adaptive immunity are defective, translocation across the intestinal epithelium and dissemination of the probiotic E. coli strain Nissle 1917 may occur and have potentially severe adverse effects. Future work should define the possibly related molecular factors that promote probiotic functions, fitness, and facultative pathogenicity.
The human gastrointestinal tract contains a complex symbiotic microbiota that is estimated to comprise more than 40,000 species and in some regions more than 1011 organisms (14) and that helps maintain immune homeostasis in the gut-associated lymphoid tissues (20, 29), optimize nutritional uptake (13), and support development of the gut (40). The thick mucus layer that overlies the entire intestinal epithelium and an effective immune system keep this enormous bacterial load strictly sequestered on the luminal side of the gut, preventing penetration across the epithelial barrier (20).
The importance of the cross talk between the microbiota, intestinal epithelial cells, and the innate and adaptive portions of the immune system is indicated by a variety of intestinal pathological conditions, including Crohn's disease, ulcerative colitis, pouchitis, irritable bowel syndrome, and necrotizing enterocolitis (NEC). Immature or genetically compromised immunity (25) results in exaggerated intestinal inflammation (26) or disruption or altered composition of the intestinal mucosa, which in turn disturbs the homeostasis between the human host and its intestinal symbionts. Pathological events change the relative balance between beneficial and aggressive enteric symbionts, turn beneficial bacteria into pathogens (36), or select for novel opportunistic pathogens (2, 28). A qualitatively and quantitatively changed gastrointestinal microbiota, often described as small bowel bacterial overgrowth (SIBO) (14, 33) or dysbiosis (26), may contribute substantially to local chronic inflammation in a vicious cycle and provoke bacterial translocation that leads to fatal sepsis.
This concept provides the rationale for selective therapeutic manipulation of the abnormal microbiota by probiotics for the intestinal diseases that have been described; probiotics are defined as viable microorganisms with beneficial physiological or therapeutic activities (36). Various in vitro and animal studies with probiotics, including Escherichia coli strain Nissle 1917, have demonstrated the capacity of probiotics to reduce intestinal inflammation (29, 42), to strengthen the intestinal barrier against pathogens (15, 46), to increase the host innate immune functions (37), or to prevent adherent and invasive E. coli strains from adhering to and invading human intestinal epithelial cells (9). Indeed, limited clinical trials using E. coli strain Nissle 1917 or other microorganisms have suggested that this therapeutic strategy is efficacious in patients with chronic idiopathic inflammatory bowel diseases (IBD) (23, 34, 35, 36), irritable bowel syndrome (32), and NEC (25).
However, all probiotic bacterial species are not equally beneficial, and each species may have individual mechanisms of action due to specific metabolic activities and cellular structures (10). Some case reports even seem to indicate that probiotics, including E. coli strain Nissle 1917, might promote sepsis (6, 24), and severe adverse effects of probiotics have been observed in patients suffering from acute pancreatitis (3, 5).
Host characteristics, specifically characteristics of the existing microbiota and the intestinal immune status, are often not considered when probiotics are used as therapeutic agents, particularly in genetically or therapeutically immunocompromised patients, including very-low-birth-weight preterm infants (25) and severely ill IBD patients receiving anti-inflammatory therapy (11).
The fact that living bacteria that may turn out to be opportunistic pathogens are therapeutically administered to patients with impaired immune functions and an altered intestinal microbiota raises two important basic questions. First, how important is a functional adaptive immune system for preventing adverse clinical effects of probiotics, such as translocation, bacteremia, and death? Lymphocytes of the gut-associated lymphoid tissue play an essential role in controlling proliferation and differentiation of intestinal epithelial cells (22) and in the maintenance of gut integrity (12). Second, how important is the microbiota in this context? Symbionts have been shown to activate epithelial innate immune signaling pathways that are needed to fight microbial pathogens (20).
To answer the questions mentioned above, we employed mice having a targeted disruption of recombinase-activating gene 1 (Rag1−/−), in which T- and B-lymphocyte development is arrested at the CD4− CD8− double-negative thymocyte or B220+ CD43+ pro-B-cell stage (30). To study the role of the physiological microbiota, these mice were raised under germfree (GF) or specific-pathogen-free (SPF) conditions and were challenged with E. coli strain Nissle 1917. Here we present evidence indicating that when there is a profound deficiency in the adaptive immune system in the presence of both functional innate immunity and the intestinal microbiota, translocation and dissemination of E. coli strain Nissle 1917 do not occur; however, in the absence of the intestinal microbiota, substantial translocation of E. coli strain Nissle 1917 causes high rates of mortality in Rag1−/− mice. We also show that adoptive transfer of naïve CD4+ T cells to Rag1−/− mice raised under GF conditions (GF-raised Rag1−/− mice) after E. coli strain Nissle 1917 challenge increases the mortality rate significantly. Our data might explain why immunocompromised patients that have an immature or disrupted intestinal microbiota and are treated with probiotics have an enhanced risk for severe side effects due to bacterial translocation. Furthermore, we describe a T-cell-mediated pathogenic mechanism that is involved in a fatal outcome for immunocompromised mice fed the probiotic E. coli strain Nissle 1917.
MATERIALS AND METHODS
Mice.
C57BL/6J-Rag1tm1Mom (Rag1−/−) (30) mice and C57BL/6J (B6) mice were used in this study. Mice were bred either under SPF conditions in a barrier-sustained facility or under gnotobiotic conditions in isolators at the University of Ulm, Ulm, Germany. Gnotobiotic mice were maintained in a germfree (GF) environment or were colonized with only one bacterial strain, either E. coli strain Nissle 1917 (kindly provided by Sonnenborn, Ardeypharm, Germany), the mouse intestinal strain E. coli mpk, or the mouse intestinal strain Bacteroides vulgatus mpk (45). E. coli mpk (O not typeable:H8) has been assigned to phylogenic group B1 (45). Initial colonization of GF mice was accomplished by feeding the animals a suspension containing the appropriate bacterial strain(s). Successful colonization was controlled and monitored as described below. The gnotobiotic state was controlled weekly and at the time of necropsy, and this involved culturing for aerobic and anaerobic bacteria, Gram stain examination of feces and intestinal contents, and performing broad-range eubacterial 16S rRNA gene PCR analyses of stool samples from mice, as described previously (45). The presence of Helicobacter spp. was controlled by routine veterinary monitoring. Tumor necrosis factor alpha (TNF-α)-deficient mice (TNF-α−/− mice), backcrossed with B6 mice for 10 generations (31), were kindly provided by R. Mocikat, Helmholtz Zentrum, Munich, Germany.
Bacterial challenge and determination of translocation by culture methods and PCR.
We challenged groups of at least four GF-raised wild-type mice, GF-raised Rag1−/− mice, or Rag1−/− mice raised under SPF conditions (SPF-raised Rag1−/− mice) on day 0 with the probiotic E. coli strain Nissle 1917, the nonpathogenic strain E. coli mpk, or B. vulgatus mpk as described previously (45). In brief, mice were challenged with 1 × 108 CFU of viable E. coli or B. vulgatus by oral gavage. E. coli strain Nissle 1917 expresses flagella, has a semirough lipopolysaccharide (LPS) phenotype, and does not produce known extracellular protein toxins (7, 17). E. coli strain Nissle 1917, an E. coli strain Nissle 1917 ΔfliC mutant deficient in the flagellum filament protein (37), an E. coli strain Nissle 1917 ΔflgE mutant deficient in the flagellum hook gene (37), and an E. coli strain Nissle 1917 strain complemented with a plasmid containing a functional copy of wzy from E. coli strain 536 [E. coli strain Nissle 1917(pBWB536) (17)] (designated the E. coli Nissle 1917 wzy strain), which provides a smooth LPS phenotype, were used in mouse colonization experiments. All E. coli strains were grown in Luria-Bertani broth at 37°C overnight, and B. vulgatus was grown in brain heart infusion (BHI) medium at 37°C under anaerobic conditions. Where appropriate, ampicillin was added to the growth medium at a concentration of 100 μg/ml.
The numbers of bacterial CFU in the mesenteric lymph nodes (MLN), liver, spleen, and lungs and in the feces (intestinal colonization) were determined by homogenization and plating of serial dilutions of the homogenates on blood and Endo agar (aerobic), as well as brain heart infusion (BHI) agar (anaerobic) at different time points after challenge. The numbers of CFU per plate were determined and expressed as log10CFU/g organ (limit of detection, 50 CFU). For identification of E. coli strain Nissle 1917, PCR was performed using primers specific for the pMUT1 plasmid and for the pMUT2 plasmid as described previously (8).
T-cell transfer before and after bacterial challenge.
Lymphocytes were isolated by homogenizing spleens of B6 or TNF-α−/− mice. Erythrocytes were eliminated by incubation of the homogenates in lysis buffer (160 mM NH4Cl, 170 mM Tris; pH 7.4). Splenic naïve CD4+ T cells were purified with a MACS negative selection kit (Miltenyi, Bergisch Gladbach, Germany) by following the manufacturer's instructions. The purity of the CD4+ T-cell population obtained was >90%, and over 80% of the cells were CD62L+ CD4+ T cells. E. coli strain Nissle 1917-challenged GF- or SPF-raised Rag1−/− mice were reconstituted with 5 × 105 CD62L+ CD4+ naïve T cells intraperitoneally 3 days before bacterial challenge or at day 6 after bacterial challenge.
Histology and assessment of cytokine levels in serum.
Lung tissues were fixed in neutral buffered 4% formalin. Formalin-fixed tissues were embedded in paraffin and cut into 2-μm sections. Samples were stained with hematoxylin and eosin (H&E) (Merck, Darmstadt, Germany). Sections were analyzed in a blinded fashion by one pathologist. Serum samples were stored at −80°C. Levels of TNF-α were quantified by an enzyme-linked immunosorbent assay (ELISA) (BD Biosciences, Heidelberg, Germany) performed according to the manufacturer's instructions.
Statistics.
Statistical analysis was performed using Student's t test, analysis of variance (ANOVA), or the Kaplan-Meier log rank test, as indicated below.
RESULTS
The presence of a microbiota prevents translocation of E. coli strain Nissle 1917 in immunodeficient Rag1−/− mice and ensures survival after challenge with E. coli strain Nissle 1917.
To assess the impact of the intestinal microbiota on protection of mice with severely compromised adaptive immunity (30) from bacterial translocation, we challenged GF- or SPF-raised Rag1−/− mice orally with E. coli strain Nissle 1917, a probiotic E. coli strain used to treat IBD and other intestinal diseases (36), or with E. coli mpk, a commensal fecal mouse E. coli strain (45).
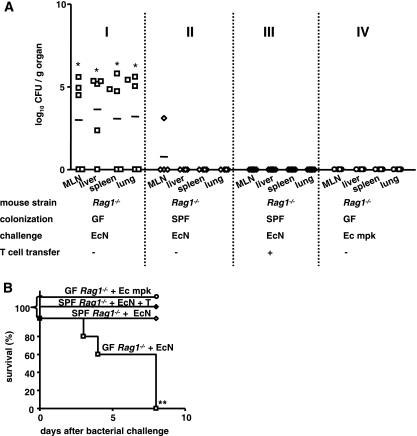
Rag1−/− mice raised under GF conditions were challenged with E. coli strain Nissle 1917 or E. coli mpk, and bacterial translocation and dissemination were assessed by determining the numbers of bacteria in the MLN, liver, spleen, and lungs 7 days later. GF-raised Rag1−/− mice devoid of the intestinal microbiota were found to be highly susceptible to E. coli strain Nissle 1917 challenge; high numbers of bacteria, identified by PCR as E. coli strain Nissle 1917, were detected in the MLN, liver, spleen, or lungs of the animals (Fig. 1A, panel I), and all GF-raised Rag1−/− mice succumbed to the bacteria within 7 days (Fig. 1B). In contrast, for SPF-raised Rag1−/− mice, which had a physiologically highly diverse microbiota, no translocation of E. coli strain Nissle 1917 across the intestinal barrier was observed, whether T cells were present (Fig. 1A, panel III) or not present (Fig. 1A, panel II), and all of these mice survived the E. coli strain Nissle 1917 challenge (Fig. 1B). To examine whether other E. coli strains translocated in GF-raised Rag1−/− mice, we tested the commensal strain E. coli mpk under the same experimental conditions. In contrast to the results for E. coli strain Nissle 1917, translocation of E. coli mpk to the MLN, liver, spleen, and lungs was not observed, and none of the animals in this group died for 7 days after bacterial challenge (Fig. 1B). These results indicate that in the absence of a microbiota, E. coli strain Nissle 1917, but not E. coli mpk, translocated and disseminated in GF-raised Rag1−/− hosts. A microbiota is required to prevent translocation of E. coli strain Nissle 1917 in immunodeficient mice, and translocation of E. coli strain Nissle 1917 across the intestinal barrier is a strain-specific phenomenon and therefore may be related to E. coli strain Nissle 1917-specific fitness or virulence factors.
FIG. 1.
In Rag1−/− mice the microbiota prevents bacterial translocation across the intestinal epithelium and ensures survival after challenge with E. coli Nissle 1917. (A) Groups of at least four GF-raised (panel I) or SPF-raised (panel II) Rag1−/− mice were colonized with 1 × 108 CFU of E. coli Nissle 1917 (EcN) on day 0. Additionally, one group of SPF-raised Rag1−/− mice was reconstituted with T cells 6 days after E. coli strain Nissle 1917 challenge (panel III), and GF-raised Rag1−/− mice were also challenged with 1 × 108 CFU of the commensal E. coli mpk (Ec mpk) (panel IV). At day 7, the numbers of bacterial CFU in the MLN, liver, spleen, and lungs of the animals were determined. Each symbol indicates the data for a single animal. *, P < 0.05 for a comparison with all other groups (one-way ANOVA, Bonferroni multiple-comparison post test). (B) Kaplan-Meier survival curves for E. coli strain Nissle 1917-challenged GF-raised (□) or SPF-raised (⋄) Rag1−/− mice, E. coli strain Nissle 1917-challenged T-cell-reconstituted SPF-raised Rag1−/− mice (⧫), and GF-raised Rag1−/− mice which were challenged with the commensal E. coli mpk (○). **, P < 0.01 (Kaplan-Meier log rank test).
T cells are required to prevent dissemination of E. coli strain Nissle 1917 in GF mice.
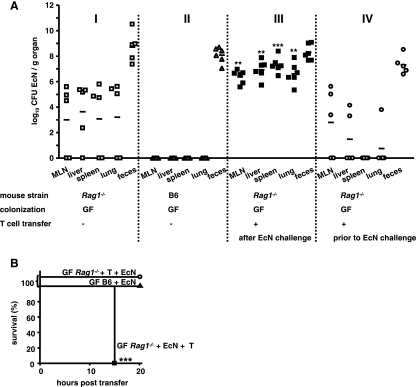
To assess the impact of the adaptive immune system on protection of mice devoid of an intestinal microbiota from bacterial translocation and dissemination, we challenged GF-raised Rag1−/− or B6 mice orally with E. coli strain Nissle 1917 and determined the bacterial translocation and dissemination and the survival of mice. Figure 2A shows that GF-raised Rag1−/− mice (Fig. 2A, panel I), but not GF-raised B6 mice (Fig. 2A, panel II), are susceptible to translocation and dissemination of E. coli strain Nissle 1917, which indicates that T cells are required to prevent dissemination of E. coli strain Nissle 1917 in mice devoid of a microbiota.
FIG. 2.
T cells are required to prevent translocation and dissemination of E. coli strain Nissle 1917 in GF mice. (A) Numbers of CFU in the MLN, liver, spleen, lungs, and feces of groups of at least five E. coli strain Nissle 1917 (EcN)-challenged GF-raised Rag1−/− mice (panel I), E. coli strain Nissle 1917-challenged GF-raised B6 mice (panel II), E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice (panel III) which were reconstituted with naive T cells (panel IV), and GF-raised Rag1−/− mice which received naïve T cells 3 days before challenge with E. coli strain Nissle 1917. At day 7, the numbers of bacterial CFU were determined. Each symbol indicates the data for a single animal. **, P < 0.01 for a comparison with all other groups; ***, P < 0.001 for a comparison with all other groups (one-way ANOVA, Bonferroni multiple-comparison post test). (B) Kaplan-Meier survival curves for E. coli strain Nissle 1917-challenged GF-raised B6 mice (▴), E. coli strain Nissle 1917-challenged T-cell-reconstituted GF-raised Rag1−/− mice (▪), and GF-raised Rag1−/− mice which received T cells 3 days before bacterial challenge (○). ***, P < 0.001 (Kaplan-Meier log rank test).
To test whether adoptive transfer of T cells after E. coli strain Nissle 1917 challenge rescued Rag1−/− mice from E. coli strain Nissle 1917 dissemination and mortality, we intraperitoneally reconstituted GF-raised Rag1−/− mice with 5 × 105 CD62L+ CD4+ naïve T cells 6 days after E. coli strain Nissle 1917 challenge (Fig. 2A, panel III). Strikingly, T-cell reconstitution after E. coli strain Nissle 1917 challenge led to accelerated mortality of GF-raised Rag1−/− mice (Fig. 2B) compared to the mortality observed for non-T-cell-reconstituted mice. In fact, high bacterial counts were obtained for the MLN, liver, spleen, and lungs of all GF-raised Rag1−/− mice (Fig. 2A, panel III) compared to the results for non-T-cell-reconstituted mice (Fig. 1A, panel I, and Fig. 2A, panel I), and the mortality rate was high 15 h after T-cell transfer (Fig. 2B). T-cell reconstitution of Rag1−/− mice 2 days after E. coli strain Nissle 1917 challenge resulted in mild to severe exacerbation of the disease, and the severity of the disease correlated with the mortality of mice (data not shown).
To investigate whether T-cell reconstitution prior to E. coli strain Nissle 1917 challenge rescued Rag1−/− mice without an intestinal microbiota from subsequent translocation of E. coli strain Nissle 1917, we next reconstituted GF-raised Rag1−/− mice with T cells 3 days before E. coli strain Nissle 1917 challenge. This treatment resulted in significantly less bacterial translocation and dissemination and a reduced bacterial burden in all organs investigated (Fig. 2A, panel IV), and all of the mice survived for the whole observation period (Fig. 2B). These results were corroborated by the results of experiments performed with fully immunocompetent B6 mice without an intestinal microbiota, in which E. coli strain Nissle 1917 translocation and dissemination were not observed in any of the organs investigated (Fig. 2A, panel I). Consequently, all B6 mice survived the bacterial challenge (Fig. 2B).
Targeting of TNF-α production in T cells does not reduce T-cell-mediated mortality of E. coli strain Nissle 1917-challenged, T-cell-reconstituted GF-raised Rag1−/− mice.
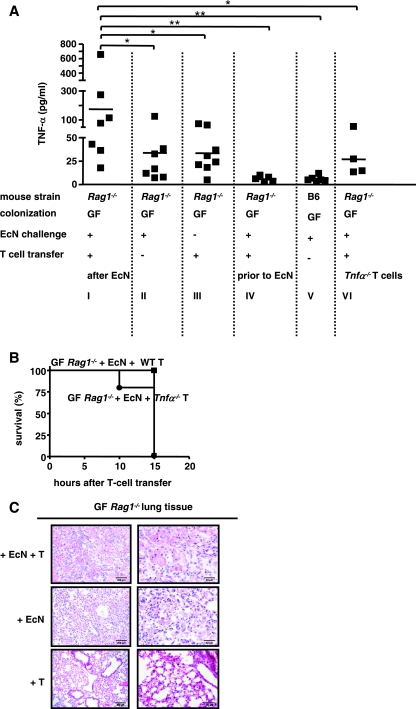
In GF-raised mice challenged with E. coli strain Nissle 1917 and reconstituted with T cells 6 days later enhanced mortality was associated with significantly increased levels of TNF-α in the serum (Fig. 3A I) compared to the levels in non-T-cell-reconstituted mice (Fig. 3A, panel II), suggesting that there were systemic inflammatory events. Increased levels of TNF-α were not observed in nonchallenged T-cell-reconstituted GF-raised Rag1−/− mice (Fig. 3A, panel III), in E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice that were T cell reconstituted before bacterial challenge (Fig. 3A, panel IV), or in E. coli strain Nissle 1917-challenged GF-raised B6 mice (Fig. 3A, panel V). These results suggest that TNF-α production was increased when translocated bacteria directly or indirectly stimulated T cells.
FIG. 3.
Reconstitution of E. coli strain Nissle 1917-challenged Rag1−/− mice with Tnf-α−/− T cells does not reduce T-cell-meditated mortality. (A) TNF-α cytokine concentrations in serum of E. coli strain Nissle 1917 (EcN)-challenged T-cell-reconstituted GF-raised Rag1−/− mice (panel I), E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice (panel II), T-cell-reconstituted GF-raised Rag1−/− mice (panel III), GF-raised Rag1−/− mice which received T cells of wild-type mice 3 days before E. coli strain Nissle 1917 challenge (panel IV), E. coli strain Nissle 1917-challenged GF-raised B6 mice (panel V), and E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice which received Tnf-α−/− naïve T cells (panel VI), as measured by ELISA. *, P < 0.05; **, P < 0.01 (paired Student t test). (B) Kaplan-Meier survival curves for E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice reconstituted with Tnf-α−/− cells (•) or wild-type T cells (WT) (▪). (C) Histology of lung tissues of E. coli strain Nissle 1917-challenged T-cell-reconstituted GF-raised Rag1−/− mice, E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice, and T-cell-reconstituted GF-raised uninfected control mice. All sections were stained with H&E.
To examine whether T-cell-derived TNF-α may contribute to mortality in this model, GF-raised Rag1−/− mice were reconstituted with T cells from Tnf-α−/− mice (31). Figure 3B shows that the attempts to reduce mortality by adoptive transfer of Tnf-α−/− T cells were unsuccessful, although the levels of TNF-α in the serum were reduced in these mice (Fig. 3A, panel VI) compared to the levels in controls with T cells transferred from wild-type mice (Fig. 3A, panel I). These findings suggest that the enhanced mortality was mediated largely by other, TNF-α-independent mechanisms.
To examine the possible mechanisms involved in mortality of E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice, histomorphological analyses were carried out (Fig. 3C). The substantially increased mortality of E. coli strain Nissle 1917-challenged T-cell-reconstituted GF-raised Rag1−/− mice was particularly supported by the histology of the lungs; fibrin deposits, accumulation of alveolar macrophages, scattered neutrophils, and signs of pleuritis were observed in these mice. In some respects, these findings resembled the diffuse alveolar damage that is the morphological manifestation of acute respiratory distress syndrome. E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice without T-cell reconstitution had fewer histopathological changes and only moderate interstitial infiltration, whereas GF-raised Rag1−/− mice showed no significant pathology (Fig. 3C).
Neither semirough lipopolysaccharide nor flagella promote dissemination of E. coli strain Nissle 1917 in and mortality of E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice.
To examine the virulence factors of E. coli strain Nissle 1917 which cause translocation and death in E. coli strain Nissle 1917-challenged GF-raised Rag1−/− mice, we used E. coli strain Nissle 1917 mutants in our animal model. First, we examined the role of flagella, which play an important role in cell adhesion and bacterial motility (39). As E. coli strain Nissle 1917 is flagellated, we hypothesized that the E. coli strain Nissle 1917 ΔfliC and ΔflgE flagellum mutants would not cross the intestinal barrier and would not cause increased mortality in GF-raised Rag1−/− mice reconstituted with T cells after bacterial challenge.
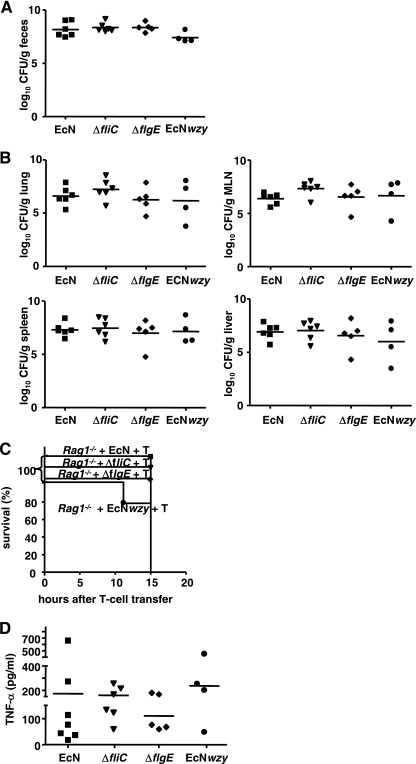
The E. coli strain Nissle 1917 ΔfliC and E. coli strain Nissle 1917 ΔflgE mutants colonized the intestine of GF-raised Rag1−/− mice like E. coli wild-type strain Nissle 1917 (Fig. 4A). Determination of the bacterial counts in the MLN, liver, spleen, and lungs revealed that the flagellum mutants translocated and disseminated as effectively as E. coli wild-type strain Nissle 1917. Thus, the mutant strains did not differ from the wild-type strain in terms of the bacterial burdens in the various organs of the mice 7 days after challenge (Fig. 4B) or in terms of the mortality that they caused (Fig. 4C). Moreover, colonization with E. coli wild-type strain Nissle 1917 and colonization with the mutants resulted in similarly enhanced levels of TNF-α in the serum (Fig. 4D). From these results we concluded that flagella are not required for pathogenicity of E. coli strain Nissle 1917 in GF-raised Rag1−/− mice.
FIG. 4.
Translocation in germfree Rag1−/− mice is independent of LPS- or flagellum-mediated signals. Groups of at least five GF-raised Rag1−/− mice were challenged with 1 × 108 CFU of the ΔfliC mutant, the ΔflgE mutant, the E. coli strain Nissle 1917 wzy mutant (EcNwzy), or E. coli wild-type strain Nissle 1917 (EcN). Six days after challenge Rag1−/− mice were reconstituted with naïve T cells. (A) Numbers of CFU in feces at 1 day after transfer. Each symbol indicates the data for one animal. (B) Numbers of CFU in organs at 1 day after transfer. Each symbol indicates the data for one animal. (C) Kaplan-Meier survival curves for Rag1−/− mice challenged with either E. coli strain Nissle 1917 (▪), the ΔfliC mutant (▾), the ΔflgE mutant (⧫), or the E. coli strain Nissle 1917 wzy mutant (•) after T cell reconstitution. (D) TNF-α concentrations in sera of T-cell-reconstituted GF-raised Rag1−/− mice challenged with E. coli strain Nissle 1917, the ΔfliC mutant, the ΔflgE mutant, or the E. coli strain Nissle 1917 wzy mutant. TNF-α concentrations were determined by ELISA.
Rough-type LPS, which is present in E. coli wild-type strain Nissle 1917, is known to be 100 times more effective for stimulation of epithelial cells than smooth-type LPS, which is present in, e.g., E. coli mpk (1, 4, 19, 45). Therefore, we tested whether a change in the LPS structure of E. coli strain Nissle 1917 resulted in differences in translocation, dissemination, and mortality rates in Rag1−/− mice. To do this, E. coli strain Nissle 1917 transfected with a plasmid containing a functional copy of the E. coli strain 536 wzy gene (pBWB536 [17]) (E. coli strain Nissle 1917 wzy mutant), which resulted in a smooth LPS phenotype for the bacterium, was used for intestinal colonization. Challenge of mice with the E. coli strain Nissle 1917 wzy mutant resulted in colonization of the mouse intestine that was the same as the colonization observed for E. coli wild-type strain Nissle 1917 (Fig. 4A) and in similar bacterial counts in the MLN, liver, spleen, and lungs (Fig. 4B). In line with this, a reduction in mortality (Fig. 4C) or in the TNF-α level in the serum (Fig. 4D) was not observed.
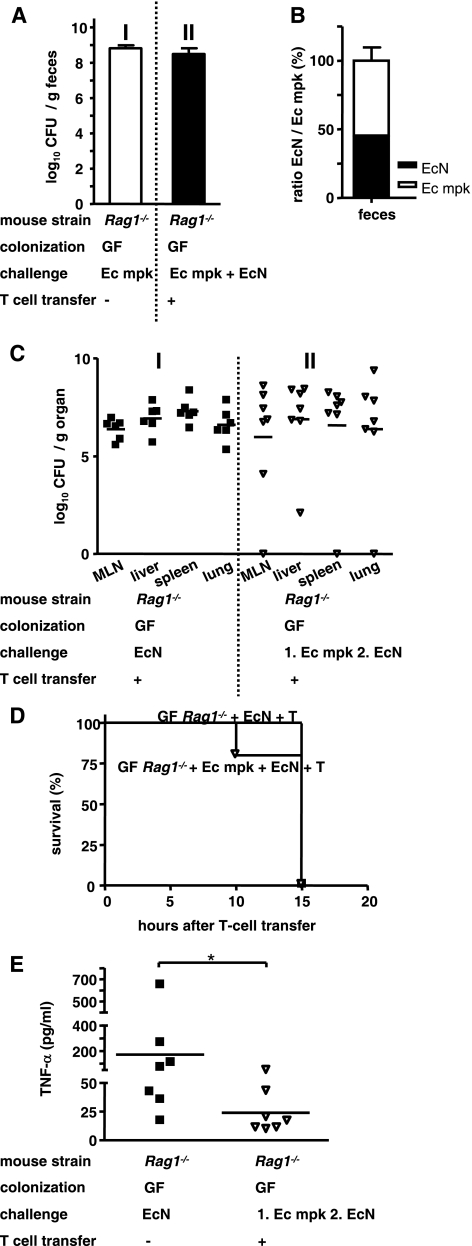
E. coli mpk cannot prevent translocation of E. coli strain Nissle 1917.
In order to test whether competition between E. coli strain Nissle 1917 and the intestinal mouse strain E. coli mpk protected the mice from translocation and dissemination of E. coli strain Nissle 1917 and increased mortality, GF-raised Rag1−/− mice were first challenged with E. coli mpk and then after 3 days were challenged with E. coli strain Nissle 1917 and reconstituted with T cells on day 6 after the E. coli strain Nissle 1917 challenge.
As shown in Fig. 5, E. coli mpk colonized the intestine of GF-raised Rag1−/− mice (Fig. 5A, panel I) as effectively as E. coli strain Nissle 1917. Subsequent challenge with E. coli strain Nissle 1917 did not increase the total number of intestinal CFU (Fig. 5A, panel II). The ratio of E. coli mpk to E. coli strain Nissle 1917, determined by PCR-based detection of the E. coli strain Nissle 1917-specific plasmids pMUT1 and pMUT2, showed that the ratio of E. coli mpk to E. coli strain Nissle 1917 was almost 1:1 (Fig. 5B). This might indicate that E. coli strain Nissle 1917 supersedes E. coli mpk during competition for similar biological niches. As shown in Fig. 5C, challenge of Rag1−/− mice with E. coli mpk did not lead to translocation of E. coli mpk but did not protect the mice from translocation and dissemination of E. coli strain Nissle 1917 (Fig. 5C, panel II), as confirmed by analysis of E. coli strain Nissle 1917-specific pMUT1 and pMUT2 PCR (data not shown). Additionally, the mortality of mice was not reduced (Fig. 5D), but the increase in the level TNF-α in the serum was eliminated (Fig. 5E).
FIG. 5.
E. coli mpk cannot prevent translocation of E. coli strain Nissle 1917. Groups of at least four GF-raised Rag1−/− mice were challenged with 1 × 108 CFU of the commensal E. coli mpk (Ec mpk) 3 days before challenge with E. coli strain Nissle 1917 (EcN). Six days after E. coli strain Nissle 1917 challenge Rag1−/− mice were reconstituted with naïve T cells. (A) Numbers of CFU of E. coli mpk 3 days after challenge and total numbers of CFU (E. coli mpk and E. coli strain Nissle 1917) 7 days after E. coli strain Nissle 1917 challenge in feces. (B) Ratio of E. coli strain Nissle 1917 to E. coli mpk in feces 7 days after E. coli strain Nissle 1917 challenge. E. coli strain Nissle 1917 was identified by detection of the E. coli strain Nissle 1917-specific cryptic plasmids pMUT1 and pMUT2. (C) Numbers of CFU of E. coli strain Nissle 1917 in the MLN, liver, spleen, and lungs at 1 day after transfer. (D) Kaplan-Meyer survival curve for GF-raised Rag1−/− mice which were challenged with the commensal E. coli mpk 3 days before challenge with E. coli strain Nissle 1917 and T cell reconstituted 6 days after E. coli strain Nissle 1917 challenge. (E) TNF-α concentrations in sera after T-cell reconstitution. *, P < 0.05 (paired Student t test).
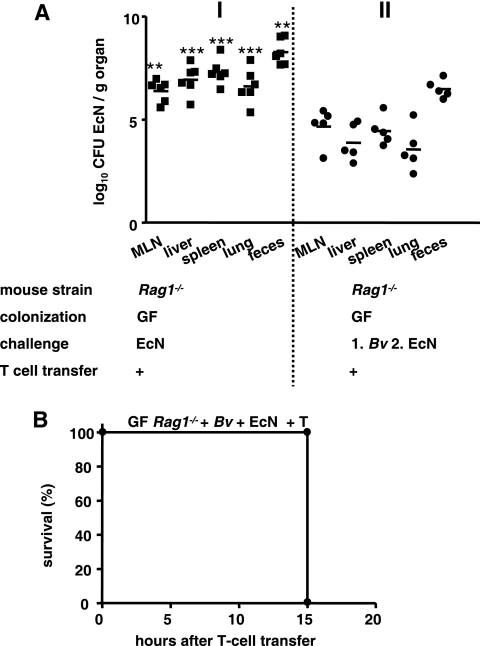
In contrast to the results for E. coli mpk, B. vulgatus mpk colonization resulted in inhibition of E. coli strain Nissle 1917 translocation, as indicated by the lower E. coli strain Nissle 1917 burden in the peripheral organs (Fig. 6A, panel II). However, the mortality of mice was unchanged (Fig. 6B).
FIG. 6.
B. vulgatus mpk reduces translocation of E. coli strain Nissle 1917, but not the mortality of mice. Groups of at least five GF-raised Rag1−/− mice were challenged with 1 × 108 CFU of the commensal B. vulgatus mpk (Bv) 3 days before challenge with E. coli strain Nissle 1917 (EcN). Six days after E. coli strain Nissle 1917 challenge Rag1−/− mice were reconstituted with naïve T cells. (A) Numbers of CFU of E. coli strain Nissle 1917 in peripheral organs and feces 7 days after challenge **, P < 0.01 for a comparison with E. coli strain Nissle 1917-challenged T-cell-reconstituted Rag1−/− mice; ***, P < 0.001 for a comparison with E. coli strain Nissle 1917-challenged T-cell-reconstituted Rag1−/− mice (one-way ANOVA, Bonferroni multiple-comparison post test). (B) Kaplan-Meier survival curve for GF-raised Rag1−/− mice which were challenged with the commensal B. vulgatus mpk 3 days before challenge with E. coli strain Nissle 1917 and T cell reconstituted 6 days after the E. coli strain Nissle 1917 challenge.
DISCUSSION
In order to examine the safety of probiotics, we used the probiotic E. coli strain Nissle 1917 in GF- and SPF-raised wild-type and Rag1−/− mouse models. We analyzed the contributions of the intestinal microbiota and the adaptive immune system to prevention of bacterial translocation across the intestinal epithelium, bacterial dissemination to various organs, and death. The clinical relevance of these investigations results (i) from the fact that probiotics have been administered to patients with therapy-related or disease-related immunosuppression and intestinal barrier dysfunction, in some cases resulting in severe side effects (3, 25), and (ii) from the fact that recent reports demonstrated the inflammatory potential of E. coli strain Nissle 1917 (6, 44).
Our results demonstrate that the intestinal microbiota prevents translocation of the probiotic E. coli strain Nissle 1917 even in the presence of a defective adaptive immune system. Conversely, in the absence of the intestinal microbiota, translocation of E. coli strain Nissle 1917 is prevented by a fully competent innate and adaptive immune system. However, when both the microbiota and adaptive immunity are defective, translocation of E. coli strain Nissle 1917 occurs. Once translocation of E. coli strain Nissle 1917 across the intestinal epithelium into internal organs has taken place, an attempt to rescue mice by adoptive transfer of immune cells (T cells) exacerbates the disease and accelerates mortality. Together, our results suggest that E. coli strain Nissle 1917 seems to overcome host innate immune defense mechanisms like lysis by the complement-protein complex or killing by phagocytes and that the defense against and clearance of translocated E. coli strain Nissle 1917 strongly depend on T-cell-mediated mechanisms.
Here we show that the intestinal microbiota is sufficient to prevent translocation of the probiotic E. coli strain Nissle 1917 to mouse organs. This finding suggests that the adaptive immunity is dispensable in this context, although lymphocytes have been shown to substantially impact the proliferation and differentiation of the intestinal epithelium (41). This suggestion is based on our findings that Rag1−/− mice raised under SPF conditions and exhibiting a physiologically mature microbiota did not have any bacteria in their MLN, liver, spleen, or lungs and that all of these mice survived oral challenge with high levels of E. coli strain Nissle 1917. Results of other investigators revealed that SPF-raised C3H/HeJZtm mice, which have a defective Toll-like receptor 4 (TLR4) allele and hence are defective for innate immunity, are also protected from translocation of E. coli strain Nissle 1917 (6).
Our findings also revealed that the microbiota is dispensable with regard to bacterial translocation in the presence of an intact adaptive immune system; GF-raised B6 mice were highly resistant to E. coli strain Nissle 1917 challenge, as were GF-raised Rag1−/− mice that were reconstituted with T cells prior to E. coli strain Nissle 1917 challenge. This finding was unexpected since GF-raised mice have various defects in their intestinal mucosa, including a smaller width, a sparse stroma in the lamina propria, wider microvillus brush borders, and small Peyer's patches (43).
An important finding of the present study is that a lack of both the intestinal microbiota and the adaptive immune system allows the probiotic E. coli strain Nissle 1917 to translocate and cause death in Rag1−/− mice. All GF-raised Rag1−/− mice succumbed the bacterial challenge within 7 days, and high numbers of CFU of E. coli strain Nissle 1917 were found in the MLN, liver, spleen, or lungs of the animals. Interestingly, GF-raised C3H/HeJZtm mice are also susceptible to translocation of E. coli strain Nissle 1917 (6). Thus, it seems that two of the three components (i.e., the microbiota and the innate and adaptive immune systems) need to be in a competent state to form an effective barrier against microbial invasion and to prevent disease. These findings may have important consequences for administration of probiotics in general and E. coli strain Nissle 1917 specifically.
This is in line with recent findings of Slack et al., who showed that adaptive immunity is critical for successful mutualism in TLR signaling-deficient mice and that TLR signaling is required for the normal elimination of low numbers of bacteria that are translocated from the intestinal lumen into the mucosa, but that commensal-specific serum IgG responses, induced in response to translocated intestinal bacteria, can restore effective bacterial clearance in TLR signaling-deficient mice (38). Slack et al. suggested that there is a flexible set point between innate immunity and adaptive immunity, which is determined by the functional performance of each system that protects the host (38). However, we eliminated the possibility that B cells have a major role in our animal model of Rag1−/− mice.
A major finding of the present study was that competition between E. coli strain Nissle 1917 and the intestinal mouse strain E. coli mpk did not prevent translocation and dissemination of E. coli strain Nissle 1917. This suggests that translocation of E. coli strain Nissle 1917 might be an active process that depends on E. coli strain Nissle 1917-specific fitness or virulence factors that enable E. coli strain Nissle 1917 to compete with E. coli mpk and to cross the intestinal barrier. However, our data provide a hint that anaerobic intestinal bacteria like, e.g., B. vulgatus might at least reduce translocation of E. coli mpk. Further studies are necessary to clarify the molecular mechanisms underlying these effects. The genome of E. coli strain Nissle 1917 has been described, and the data revealed a number of so-called pathogenicity islands and genes encoding adhesins (type 1 and F1C fimbriae, Iha, curli, AIDA-I/Sap-like), proteases (Sat and Tsh), and microcins, as well as multiple-gene clusters coding for proteins involved in iron acquisition (yersiniabactin, aerobactin, salmochelin, and Chu hemin receptor) that increase bacterial fitness (16). Whether the probiotic characteristics of E. coli strain Nissle 1917 are related to its increased fitness is an important question and challenges the concept of probiotic bacteria in general. In fact, it is crucial to elucidate whether there is a direct or indirect molecular relationship between factors that promote probiotic functions, fitness, and thus increased facultative pathogenicity in immunocompromised hosts (hosts with, e.g., a defective microbiota and T-cell deficiency).
Reconstitution of GF-raised Rag1−/− mice with naïve CD4+ T cells after E. coli strain Nissle 1917 challenge increased the mortality rate to 100% from the mortality rate of 72% observed for nonreconstituted mice. Compared to the results for nonreconstituted mice, T-cell reconstitution led to increased numbers of bacteria in various organs, more severe lung pathology, and significantly increased levels of TNF-α in the serum. Since transfer of Tnf-α−/− T cells did not reduce the high mortality rate in GF-raised Rag1−/− mice, other TNF-α-independent effects might account for the increased mortality. The concept suggested by Hotchkiss and Nicholson indicates that death from sepsis might be the result of a substantially impaired immune response that is due to extensive death of immune effector cells (21). Our results suggest that once translocation of bacteria across the intestinal epithelium into internal organs has taken place, the response of the adaptive immune system exacerbates the disease and accelerates mortality.
A previous in vitro study of E. coli strain Nissle 1917 revealed proinflammatory traits (44). Therefore, to determine the virulence of E. coli strain Nissle 1917 in GF-raised Rag1−/− mice on a molecular level, we tested isogenic flagellum and LPS mutants of E. coli wild-type strain Nissle 1917. Neither deletion of the flagella nor changes in the LPS structure of E. coli strain Nissle 1917 affected bacterial translocation and dissemination or mortality.
We hypothesized that the intestinal microbiota is essential for prevention of the translocation of E. coli strain Nissle 1917 across the intestinal barrier. Once translocated, E. coli strain Nissle 1917 seems to be able to evade host innate immune defenses like lysis by the complement protein complex or phagocytosis; a T-cell-mediated adaptive immune mechanism seems to be essential for control and clearance of translocated E. coli strain Nissle 1917. However, other studies have reported serum sensitivity of E. coli strain Nissle 1917 in humans (17). Recent studies provided evidence that translocation of certain E. coli strains might occur via novel transcellular pathways activated in enterocytes by inflammatory and metabolic stress (27). The data of Macutkiewicz et al. suggest that translocating E. coli strains associated with infections are not opportunistic extraintestinal pathogenic E. coli (ExPEC) strains but may comprise a separate group of E. coli strains (27).
Our results suggest that administration of probiotic E. coli strain Nissle 1917 preparations to immunocompromised patients that also have a defective intestinal microbiota after, e.g., antibiotic therapy may lead to severe adverse effects and therefore should not be recommended. A typical target population may be very-low-birth-weight preterm infants or patients after organ transplantation. Although a recent study (25) concluded that probiotic bacteria, such as Bifidobacterium and Lactobacillus, fed enterally to very-low-birth-weight preterm infants for 6 weeks reduced the incidence of death or necrotizing enterocolitis, the authors mentioned that “occurrences of sepsis even seemed more frequent in the study group” (25). Recently, Guenther et al. described severe sepsis in a preterm infant due to treatment with the probiotic E. coli strain Nissle 1917 (18). Although experiments with germfree, monocolonized, or gnotobiotic mice are not directly equivalent to the situation in patients, the data obtained with our model may explain why septic episodes have been observed in immunocompromised patients that had an immature or disrupted intestinal microbiota, were treated with probiotics, and exhibited T-cell-mediated pathogenesis that resulted in fatal sepsis.
Acknowledgments
We thank Sabine Schmidt, University of Ulm, for use of an expert gnotobiotic animal breeding facility.
This project was supported by the DFG (grants FR 2087/4-2 and SFB 766), by the ZEM, and by the IZKF/BMBF.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 26 April 2010.
REFERENCES
- 1.Backhed, F., S. Normark, E. K. Schweda, S. Oscarson, and A. Richter-Dahlfors. 2003. Structural requirements for TLR4-mediated LPS signalling: a biological role for LPS modifications. Microbes Infect. 5:1057-1063. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart, D. C., and S. R. Carding. 2007. Inflammatory bowel disease: cause and immunobiology. Lancet 369:1627-1640. [DOI] [PubMed] [Google Scholar]
- 3.Besselink, M. G., H. C. van Santvoort, E. Buskens, M. A. Boermeester, H. van Goor, H. M. Timmerman, V. B. Nieuwenhuijs, T. L. Bollen, B. van Ramshorst, B. J. Witteman, C. Rosman, R. J. Ploeg, M. A. Brink, A. F. Schaapherder, C. H. Dejong, P. J. Wahab, C. J. van Laarhoven, E. van der Harst, C. H. van Eijck, M. A. Cuesta, L. M. Akkermans, and H. G. Gooszen. 2008. Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet 371:651-659. [DOI] [PubMed] [Google Scholar]
- 4.Billips, B. K., A. J. Schaeffer, and D. J. Klumpp. 2008. Molecular basis of uropathogenic Escherichia coli evasion of the innate immune response in the bladder. Infect. Immun. 76:3891-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjarnason, A., S. N. Adler, and I. Bjarnason. 2008. Probiotic prophylaxis in predicted severe acute pancreatitis. Lancet 372:114-115. [DOI] [PubMed] [Google Scholar]
- 6.Bleich, A., J. P. Sundberg, A. Smoczek, R. von Wasielewski, M. F. de Buhr, L. M. Janus, G. Julga, S. N. Ukena, H. J. Hedrich, and F. Gunzer. 2008. Sensitivity to Escherichia coli Nissle 1917 in mice is dependent on environment and genetic background. Int. J. Exp. Pathol. 89:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blum, G., R. Marre, and J. Hacker. 1995. Properties of Escherichia coli strains of serotype O6. Infection 23:234-236. [DOI] [PubMed] [Google Scholar]
- 8.Blum-Oehler, G., S. Oswald, K. Eiteljorge, U. Sonnenborn, J. Schulze, W. Kruis, and J. Hacker. 2003. Development of strain-specific PCR reactions for the detection of the probiotic Escherichia coli strain Nissle 1917 in fecal samples. Res. Microbiol. 154:59-66. [DOI] [PubMed] [Google Scholar]
- 9.Boudeau, J., A. L. Glasser, S. Julien, J. F. Colombel, and A. Darfeuille-Michaud. 2003. Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn's disease. Aliment. Pharmacol. Ther. 18:45-56. [DOI] [PubMed] [Google Scholar]
- 10.Cannon, J. P., T. A. Lee, J. T. Bolanos, and L. H. Danziger. 2005. Pathogenic relevance of Lactobacillus: a retrospective review of over 200 cases. Eur. J. Clin. Microbiol. Infect. Dis. 24:31-40. [DOI] [PubMed] [Google Scholar]
- 11.Caviglia, R., I. Boskoski, and M. Cicala. 2008. Long-term treatment with infliximab in inflammatory bowel disease: safety and tolerability issues. Expert Opin. Drug Saf. 7:617-632. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Y., K. Chou, E. Fuchs, W. L. Havran, and R. Boismenu. 2002. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc. Natl. Acad. Sci. U. S. A. 99:14338-14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flint, H. J., E. A. Bayer, M. T. Rincon, R. Lamed, and B. A. White. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121-131. [DOI] [PubMed] [Google Scholar]
- 14.Frank, D. N., and N. R. Pace. 2008. Gastrointestinal microbiology enters the metagenomics era. Curr. Opin. Gastroenterol. 24:4-10. [DOI] [PubMed] [Google Scholar]
- 15.Frick, J. S., K. Fink, F. Kahl, M. J. Niemiec, M. Quitadamo, K. Schenk, and I. B. Autenrieth. 2007. Identification of commensal bacterial strains that modulate Yersinia enterocolitica and dextran sodium sulfate-induced inflammatory responses: implications for the development of probiotics. Infect. Immun 75:3490-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grozdanov, L., C. Raasch, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, and U. Dobrindt. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grozdanov, L., U. Zahringer, G. Blum-Oehler, L. Brade, A. Henne, Y. A. Knirel, U. Schombel, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, E. T. Rietschel, and U. Dobrindt. 2002. A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J. Bacteriol. 184:5912-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guenther, K., E. Straube, W. Pfister, A. Guenther, and A. Huebler. 2010. Severe sepsis after probiotic treatment with Escherichia coli Nissle 1917. Pediatr. Infect. Dis. J. 29:188-189. [DOI] [PubMed] [Google Scholar]
- 19.Hilbert, D. W., K. E. Pascal, E. K. Libby, E. Mordechai, M. E. Adelson, and J. P. Trama. 2008. Uropathogenic Escherichia coli dominantly suppress the innate immune response of bladder epithelial cells by a lipopolysaccharide- and Toll-like receptor 4-independent pathway. Microbes Infect. 10:114-121. [DOI] [PubMed] [Google Scholar]
- 20.Hooper, L. V. 2009. Do symbiotic bacteria subvert host immunity? Nat. Rev. Microbiol. 7:367-374. [DOI] [PubMed] [Google Scholar]
- 21.Hotchkiss, R. S., and D. W. Nicholson. 2006. Apoptosis and caspases regulate death and inflammation in sepsis. Nat. Rev. Immunol. 6:813-822. [DOI] [PubMed] [Google Scholar]
- 22.Kerneis, S., A. Bogdanova, J. P. Kraehenbuhl, and E. Pringault. 1997. Conversion by Peyer's patch lymphocytes of human enterocytes into M cells that transport bacteria. Science 277:949-952. [DOI] [PubMed] [Google Scholar]
- 23.Kruis, W., P. Fric, J. Pokrotnieks, M. Lukas, B. Fixa, M. Kascak, M. A. Kamm, J. Weismueller, C. Beglinger, M. Stolte, C. Wolff, and J. Schulze. 2004. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ledoux, D., V. J. Labombardi, and D. Karter. 2006. Lactobacillus acidophilus bacteraemia after use of a probiotic in a patient with AIDS and Hodgkin's disease. Int. J. STD AIDS 17:280-282. [DOI] [PubMed] [Google Scholar]
- 25.Lin, H. C., C. H. Hsu, H. L. Chen, M. Y. Chung, J. F. Hsu, R. I. Lien, L. Y. Tsao, C. H. Chen, and B. H. Su. 2008. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: a multicenter, randomized, controlled trial. Pediatrics 122:693-700. [DOI] [PubMed] [Google Scholar]
- 26.Lupp, C., M. L. Robertson, M. E. Wickham, I. Sekirov, O. L. Champion, E. C. Gaynor, and B. B. Finlay. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:204. [DOI] [PubMed] [Google Scholar]
- 27.Macutkiewicz, C., G. Carlson, E. Clark, U. Dobrindt, I. Roberts, and G. Warhurst. 2008. Characterisation of Escherichia coli strains involved in transcytosis across gut epithelial cells exposed to metabolic and inflammatory stress. Microbes Infect. 10:424-431. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Medina, M., X. Aldeguer, M. Lopez-Siles, F. Gonzalez-Huix, C. Lopez-Oliu, G. Dahbi, J. E. Blanco, J. Blanco, L. J. Garcia-Gil, and A. rfeuille-Michaud. 2009. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm. Bowel Dis. 15:872-882. [DOI] [PubMed] [Google Scholar]
- 29.Mazmanian, S. K., J. L. Round, and D. L. Kasper. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620-625. [DOI] [PubMed] [Google Scholar]
- 30.Mombaerts, P., J. Iacomini, R. S. Johnson, K. Herrup, S. Tonegawa, and V. E. Papaioannou. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68:869-877. [DOI] [PubMed] [Google Scholar]
- 31.Neurath, M. F., I. Fuss, M. Pasparakis, L. Alexopoulou, S. Haralambous, K. H. Meyer zum Buschenfelde, W. Strober, and G. Kollias. 1997. Predominant pathogenic role of tumor necrosis factor in experimental colitis in mice. Eur. J. Immunol. 27:1743-1750. [DOI] [PubMed] [Google Scholar]
- 32.Nikfar, S., R. Rahimi, F. Rahimi, S. Derakhshani, and M. Abdollahi. 2008. Efficacy of probiotics in irritable bowel syndrome: a meta-analysis of randomized, controlled trials. Dis. Colon Rectum 51:1775-1780. [DOI] [PubMed] [Google Scholar]
- 33.Quigley, E. M. 2006. New perspectives on the role of the intestinal flora in health and disease. J. Gastrointestin. Liver Dis. 15:109-110. [PubMed] [Google Scholar]
- 34.Rahimi, R., S. Nikfar, F. Rahimi, B. Elahi, S. Derakhshani, M. Vafaie, and M. Abdollahi. 2008. A meta-analysis on the efficacy of probiotics for maintenance of remission and prevention of clinical and endoscopic relapse in Crohn's disease. Dig. Dis. Sci. 53:2524-2531. [DOI] [PubMed] [Google Scholar]
- 35.Rembacken, B. J., A. M. Snelling, P. M. Hawkey, D. M. Chalmers, and A. T. Axon. 1999. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 354:635-639. [DOI] [PubMed] [Google Scholar]
- 36.Sartor, R. B. 2004. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology 126:1620-1633. [DOI] [PubMed] [Google Scholar]
- 37.Schlee, M., J. Wehkamp, A. Altenhoefer, T. A. Oelschlaeger, E. F. Stange, and K. Fellermann. 2007. Induction of human beta-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect. Immun. 75:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slack, E., S. Hapfelmeier, B. Stecher, Y. Velykoredko, M. Stoel, M. A. Lawson, M. B. Geuking, B. Beutler, T. F. Tedder, W. D. Hardt, P. Bercik, E. F. Verdu, K. D. McCoy, and A. J. Macpherson. 2009. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325:617-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soutourina, O. A., and P. N. Bertin. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 27:505-523. [DOI] [PubMed] [Google Scholar]
- 40.Stappenbeck, T. S., L. V. Hooper, and J. I. Gordon. 2002. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U. S. A. 99:15451-15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stromberg, P. E., C. A. Woolsey, A. T. Clark, J. A. Clark, I. R. Turnbull, K. W. McConnell, K. C. Chang, C. S. Chung, A. Ayala, T. G. Buchman, R. S. Hotchkiss, and C. M. Coopersmith. 2009. CD4+ lymphocytes control gut epithelial apoptosis and mediate survival in sepsis. FASEB J. 23:1817-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturm, A., K. Rilling, D. C. Baumgart, K. Gargas, T. Abou-Ghazale, B. Raupach, J. Eckert, R. R. Schumann, C. Enders, U. Sonnenborn, B. Wiedenmann, and A. U. Dignass. 2005. Escherichia coli Nissle 1917 distinctively modulates T-cell cycling and expansion via Toll-like receptor 2 signaling. Infect. Immun. 73:1452-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, G. R., and P. C. Trexler. 1971. Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut 12:230-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ukena, S. N., A. M. Westendorf, W. Hansen, M. Rohde, R. Geffers, S. Coldewey, S. Suerbaum, J. Buer, and F. Gunzer. 2005. The host response to the probiotic Escherichia coli strain Nissle 1917: specific up-regulation of the proinflammatory chemokine MCP-1. BMC Med. Genet. 6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waidmann, M., O. Bechtold, J. S. Frick, H. A. Lehr, S. Schubert, U. Dobrindt, J. Loeffler, E. Bohn, and I. B. Autenrieth. 2003. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 125:162-177. [DOI] [PubMed] [Google Scholar]
- 46.Zyrek, A. A., C. Cichon, S. Helms, C. Enders, U. Sonnenborn, and M. A. Schmidt. 2007. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell. Microbiol. 9:804-816. [DOI] [PubMed] [Google Scholar]