Abstract
Type IV pili (Tfp), which mediate multiple phenotypes ranging from adhesion to motility, are one of the most widespread virulence factors in bacteria. However, the molecular mechanisms of Tfp biogenesis and associated functions remain poorly understood. One of the underlying reasons is that the roles played by the numerous genes involved in Tfp biology are unclear because corresponding mutants have been studied on a case-by-case basis, in different species, and using different assays, often generating heterogeneous results. Therefore, we have recently started a systematic functional analysis of the genes involved in Tfp biology in a well-characterized clinical isolate of the human pathogen Neisseria meningitidis. After previously studying 16 genes involved in Tfp biogenesis, here we report the characterization of 7 genes that are dispensable for piliation and potentially involved in Tfp biology. Using a battery of assays, we assessed piliation and each of the Tfp-linked functions in single mutants, double mutants in which filament retraction is abolished by a concurrent mutation in pilT, and strains overexpressing the corresponding proteins. This showed that each of the seven genes actually fine-tunes a Tfp-linked function(s), which brings us one step closer to a global view of Tfp biology in the meningococcus.
Pili, also known as fimbriae, are hair-like filaments extending from the surface of many bacteria. No other pili are present in as many species as type IV pili (Tfp) (20, 32). These flexible, thin (5- to 8-nm), long (several-micrometer) filaments, which often interact laterally to form bundles, share sequence and structural characteristics. They are essentially polymers of one protein, generically named pilin, that is synthesized as a precursor with a conserved N-terminal leader peptide cleaved by a dedicated prepilin peptidase. Although the lengths of the leader peptide and mature protein define two distinct pilus subtypes named type IVa (Tfpa) and type IVb (Tfpb), pilins have similar three-dimensional (3D) structures (9). They consist of a long hydrophobic N-terminal α-helix and a globular head with a conserved α/β roll fold (29). Consequently, all atomic models for Tfp have in common a helical organization in which the N-terminal α-helices represent the major assembly interface between pilin subunits and are buried within the interior of the filament (9). Surprisingly, in spite of this rather simple composition and structure, Tfp can mediate numerous functions. In addition to their role in promoting attachment to a variety of surfaces (a property that they share with other pili), Tfp often mediate bacterial aggregation, uptake of DNA during transformation, and twitching motility (20, 32). This functional versatility is probably a consequence of the capacity of Tfp to be retracted, which generates remarkable mechanical force (19).
Tfp have been and continue to be intensively studied, mostly in Pseudomonas aeruginosa, Neisseria gonorrhoeae, and Neisseria meningitidis for Tfpa and in enteropathogenic Escherichia coli and Vibrio cholerae for Tfpb (20, 32). However, the molecular mechanisms of Tfp biogenesis and associated functions remain poorly understood. The first and probably the major reason is the complexity of this biological system. From 10 proteins (in V. cholerae) up to 18 proteins (in P. aeruginosa) are necessary for Tfp biogenesis (32). Furthermore, proteins dispensable for Tfp biogenesis (the exact number of which is unclear) are often key players in Tfp biology, since in the corresponding mutants Tfp-mediated functions are dramatically affected. The best known example is PilT (44), a hexameric ATPase that powers Tfp retraction by disassembling pilin subunits from the pilus base (39). In addition to directly promoting twitching motility (23, 41), PilT-mediated pilus retraction affects most aspects of Tfp biology. In Neisseria species, for example, a pilT mutant is noncompetent for DNA transformation (47) and incapable of progressing from the initial stage of localized adherence to the late stage of diffuse adherence (35). It also displays increased bacterial aggregation (47), which indicates that pilus retraction is sometimes a disruptive force. Another example is PilX, a protein processed by the prepilin peptidase that colocalizes with Tfp in N. meningitidis. It is a minor (low-abundance) pilin with a 3D structure typical of type IV pilins and most likely assembled within filaments as major pilins are (14). A pilX mutant displays a selective loss of Tfp-linked phenotypes (13), since it is unable to form aggregates and hence is unable to adhere to human cells because interbacterial interactions are essential for localized adherence. Aggregation is restored in a double pilXT mutant, in which filament retraction is abolished by a concurrent mutation in pilT, suggesting that PilX participates in the formation of aggregates by counterbalancing PilT-mediated pilus retraction (13). A structure-function analysis suggested that pilus retraction is hindered when PilX subunits in filaments from different bacteria brace against each other (14).
The second, somewhat paradoxical, reason for our fragmentary understanding of Tfp biology is the fact that these filaments have been studied in many bacterial species (20, 32). However, most of these species are phylogenetically distant, and very different assays are often used. Therefore, different and hardly compatible phenotypes have often been assigned to mutants with mutations in genes sharing high sequence conservation and identical genomic organizations (32). For example, while a pilU mutant of P. aeruginosa is hyperpiliated and does not twitch (45), an N. gonorrhoeae pilU mutant is normally piliated and exhibits twitching motility but has altered aggregation and adhesion to human cells (30). To add to the complexity, limited genetic changes can affect Tfp-mediated functions even in different strains in the same species. For example, expression of different pilE alleles in N. meningitidis modulates adhesion to human cells (28). Therefore, en bloc extrapolation of existing results to other species (or even different strains of the same species) is risky, at best.
While it is uncertain whether these diverse findings reflect actual differences in the studied bacteria or are merely due to different experimental procedures, it is clear that a global view of Tfp biology is yet to be achieved for any piliated strain. This prompted us to start a systematic functional analysis of Tfp biology in a clinical isolate of the human pathogen N. meningitidis. This strain, 8013, is a good model for several reasons. First, it is heavily piliated and displays all the phenotypes classically associated with Tfp. Second, we have resources and experimental procedures to test and/or quantify piliation and each Tfp-linked function. Third, we have designed in 8013 the NeMeSys platform for large-scale identification of gene function. It consists of a manual annotation of 8013's genome and a large archived library of defined mutants, fully integrated in a bioinformatic database (36). This platform has been instrumental for the identification and functional characterization of 16 genes important for piliation in N. meningitidis, which generated important results with general implications for understanding Tfp biogenesis (6, 7, 36). Here, we have extended our systematic phenotypic analysis to seven genes that are dispensable for Tfp biogenesis, including the previously characterized pilX, whose sequence and/or previous reports suggest that they play a role in Tfp biology (comP, pilT, pilT2, pilU, pilV, pilX, and pilZ). This led to many interesting observations reported here and, together with our previous work, brings us one step closer to a global view of Tfp biology in N. meningitidis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type (WT) strain of N. meningitidis used in this study is a sequenced variant of 8013 (an encapsulated serogroup C clinical isolate) expressing a highly adhesive pilin variant and no opacity proteins (28, 36). N. meningitidis was grown on GCB agar plates (Difco) containing Kellogg's supplements and, when required, 100 μg/ml kanamycin, 60 μg/ml spectinomycin, 5 μg/ml rifampin, and 3 μg/ml erythromycin. Plates were incubated at 37°C in a moist atmosphere containing 5% CO2. E. coli DH5α and TOP10 (Invitrogen) were used for cloning experiments, whereas BL21(DE3)pLysS (Stratagene) was used for protein expression and purification. Transformants were grown in liquid or solid Luria-Bertani medium (Difco) containing, when required, 100 μg/ml ampicillin, 100 μg/ml spectinomycin, and 50 μg/ml kanamycin.
pilT2, pilV, pilX, and pilZ mutans came from the NeMeSys archived library of defined transposition mutants (10, 36). The pilT mutant was described previously (35). The pilU mutant was engineered using an in vitro transposon mutagenesis method described elsewhere (33). A comP mutant and a second pilV mutant (a gift from P. Morand and X. Nassif) were generated by cloning a spectinomycin resistance cassette (15) into the corresponding genes. In order to minimize secondary variations, all the mutations were retransformed in the WT strain, and mutants (expressing the WT pilE allele as checked by sequencing) were stored at −80°C. All experiments were performed with bacteria from these frozen stocks grown on plates. The double mutants containing a concurrent mutation in the pilT gene were constructed by transforming the above-described mutants (except for comP/pilT, which we could not obtain) with chromosomal DNA extracted from a pilT mutant. To complement the mutants, genes placed under the transcriptional control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter, functional both in E. coli and N. meningitidis, were inserted in a specific chromosomal locus by allelic exchange (21). This was done as previously described for pilX (13). Genes were amplified using pairs of primers containing overhangs corresponding to PacI restriction sites (underlined). We used comP-IndF (CCTTAATTAAGGAGTAATTTTATGACTGATAATCGGGGGTTT) and comP-IndR (CCTTAATTAACTACTTAAATAACTTGCAGTCC) for comP, pilT-IndF (CCTTAATTAAGGAGTAATTTTATGCAGATTACCGACTTACTC) and pilT-IndR (CCTTAATTAATCAGAAACTCATACTTTCGCT) for pilT, pilT2-IndF (CCTTAATTAAGGAGTAATTTTATGACCGCAAAGGAAGAACTG) and pilT2-IndR (CCTTAATTAATCAGAGCAGTTCCAAATCGG) for pilT2, pilU-IndF (CCTTAATTAAGGAGTAATTTTATGAATACCGATAACCTGCAC) and pilU-IndR (CCTTAATTAATCAGGAAATGAGGTTGAGAC) for pilU, pilV-IndF (CCTTAATTAAGGAGTAATTTTATGAAAAACGTTCAAAAAGGC) and pilV-IndR q(CCTTAATTAATTAGTCGAAGCCGGGGCAGGA) for pilV, and pilZ-IndF (CCTTAATTAAGGAGTAATTTTATGTCAGACGGACAAAATATTC) and pilZ-IndR (CCTTAATTAATTACATGGTAAACGTAGGTCTGC) for pilZ. To create strains expressing His6-tagged PilZ proteins, we used the same strategy except that a His6 tag was added in the forward or reverse primers (indicated in lowercase). We therefore used pilZ-IndF and pilZHisC-IndR (CGCTTAATTAATTAgtggtggtggtggtggtgCATGGTAAACGTAGGTCTGC) to create a pilZ allele expressing a C-terminally tagged protein and pilZHisN-IndF (CGCTTAATTAAGGAGTAATTTTATGcaccaccaccaccaccacTCAGACGGACAAAATATTCCG) together with pilZ-IndR to create a pilZ allele expressing an N-terminally tagged protein. PCR fragments were first cloned into pCR8/GW/TOPO (Invitrogen), verified by sequencing, and subcloned into pGCC4 restricted with PacI (21). All the corresponding proteins were expressed in E. coli upon induction with 0.4 mM IPTG (Merck Chemicals) as assessed by SDS-PAGE and Coomassie blue staining. These vectors were then introduced into N. meningitidis, in the chromosome of which they integrated by homologous recombination. For comP and pilT mutants, which are noncompetent, the above vectors were first transformed in the WT strain, which was subsequently transformed with chromosomal DNA extracted from a comP or pilT mutant. Expression of the corresponding proteins in N. meningitidis was induced by growing the strains for 16 h on GCB agar plates containing 0.5 mM IPTG and assessed by immunoblotting (see below).
SDS-PAGE, antisera, and immunoblotting.
N. meningitidis whole-cell protein extracts were prepared as described previously (6). E. coli whole-cell protein extracts were prepared by resuspending pellets directly in Laemmli sample buffer (Bio-Rad). When necessary, proteins were quantified using the Bio-Rad protein assay as suggested by the manufacturer (Bio-Rad). Separation of the proteins by SDS-PAGE and subsequent blotting to Amersham Hybond ECL membranes (GE Healthcare) were done using standard molecular biology techniques (38). Blocking, incubation with primary/secondary antibodies, and detection using Amersham ECL Plus and/or Advance reagents (GE Healthcare) were done following the manufacturer's instructions. Alternatively, SDS-polyacrylamide gels were stained using Bio-Safe Coomassie blue stain (Bio-Rad).
Antisera against ComP, PilC1, PilE, PilT, PilU, PilV, PilX, and PilZ were used at dilutions of between 1/2,000 and 1/5,000. Amersham ECL horseradish peroxidase (HRP)-linked secondary antibodies (GE Healthcare) were used at a 1/10,000 dilution. It should be noted that detection of PilT2, the only protein for which we do not have a specific antibody, was performed with the anti-PilT antibody in a pilT mutant background owing to the high sequence identity between these two proteins. Antisera against PilC1, PilE, PilT, and PilX have been described previously (13, 25, 26). A rabbit antiserum specific for PilZ was produced against a purified recombinant protein. The entire pilZ gene was amplified using 981pETF (GGAATTCCATATGTCAGACGGACAAAATA) and 981pETR (CCGCTCGAGCATGGTAAACGTAGGTCTGC), which contained overhangs corresponding to NdeI and XhoI restriction sites. The fragment was first cloned into pCRII-TOPO, checked by sequencing, and then subcloned into pET-20 (Novagen) restricted with NdeI and XhoI. This introduced a His6 tag at the C terminus of the recombinant protein, which was expressed in E. coli BL21(DE3)pLysS and purified on an Ni-nitrilotriacetic acid (NTA) column. Antisera specific for ComP and PilV (a gift from P. Morand and X. Nassif) were produced in a similar way. We have purified antisera against PilV, PilX, and PilZ by immunoaffinity using the corresponding proteins. The PilU-specific antiserum (a gift from G. Duménil and X. Nassif) was generated against synthetic peptides.
Tfp detection, quantification, and purification.
As previously described (7), Tfp were visualized by immunofluorescence (IF) microscopy on a Nikon Eclipse E600 microscope after they were stained with the 20D9 monoclonal antibody (34). Piliation was quantified using this antibody and a previously described whole-cell enzyme-linked immunosorbent assay (ELISA) (13).
Pilus purification by ammonium sulfate precipitation was carried as previously described (14), with one minor modification. Bacteria from one heavily inoculated agar plate were first resuspended by gentle pipetting in phosphate-buffered saline (PBS) and adjusted to a given optical density at 600 nm (OD600) in order to perform accurate CFU counts (no pilus shearing occurs at this stage). Suspensions containing equivalent numbers of bacteria were pelleted by a 2-min centrifugation at 12,000 × g, pellets were resuspended in 1 ml of 0.15 M ethanolamine (pH 10.5), and pilus purification was carried as before (14). In addition, we designed a pilus immunopurification procedure using the 20D9 antibody. Bacteria from one heavily inoculated agar plate were first resuspended in 1 ml PBS, vortexed for 2 min to shear Tfp, and adjusted to a given OD600. Cells and large debris were pelleted by centrifugation at 16,000 × g for 7 min at 4°C. Eight microliters of 20D9 antibody was then added to 800 μl supernatant, which was incubated on a wheel for 2 h at room temperature. In the meantime, Dynabeads coated with protein A (Invitrogen) were washed twice in sodium phosphate buffer and then once in PBS following the manufacturer's instructions. Thirty microliters of beads was then added to each sample, which was further incubated for 1 h on a wheel at room temperature. Beads with bound 20D9-Tfp immunocomplexes were captured using a DynaMag magnet (Invitrogen), supernatant was removed, and beads were resuspended in PBS. After five such washes, beads were resuspended in 30 μl Laemmli sample buffer and incubated for 5 to 10 min at 95°C to release the bound immunocomplexes. Beads were captured again as described above, and the supernatant containing the 20D9-Tfp immunocomplex was analyzed by SDS-PAGE.
Transformation assay.
Natural competence for DNA transformation was quantified as follows. N. meningitidis grown on agar plates was resuspended in liquid GCB containing 5 mM MgCl2 (GCB transfo) at an OD550 of 1 (approximately 108 CFU/ml). The numbers of CFU/ml were systematically assessed by performing counts. Two hundred microliters of the previous suspension was mixed with 1 μg of Rifr chromosomal DNA in 24-well plates. This DNA was extracted from a mutant of strain 8013 spontaneously resistant to rifampin. After incubation for 30 min at 37°C on an orbital shaker, GCB transfo (1.8 ml) was then added to the wells and the plates were further incubated for 3 h at 37°C without shaking. Transformants were quantified by counting the number of Rifr CFU obtained by plating appropriate dilutions on plates containing rifampin.
Aggregation assay.
N. meningitidis grown on agar plates was resuspended at an OD600 of 0.1 or 0.02 in 500 μl RPMI containing 10% fetal bovine serum GOLD (PAA Laboratories) and incubated at 37°C in 24-well plates. Aggregates forming on the bottoms of the wells were visualized by phase-contrast microscopy using a Nikon TS100F microscope. Digital images were recorded using a Sony HDR-CX11 high-definition (HD) camcorder mounted onto the microscope.
Twitching motility assay.
An assay was designed to assess twitching motility by phase-contrast microscopy. We observed bacterial motility within aggregates that were attached either to the bottom of the wells in 24-well plates in aggregation assays (see above) or to human cells in adhesion assays (see below). Short HD videos (approximately 30 s) were recorded using the camcorder mounted onto the microscope.
Adherence assay.
Adhesion of meningococci to pooled human umbilical vein endothelial cells (HUVEC) (Lonza) was done as described previously (13). In brief, monolayers of 105 cells in 24-well plates were infected with 1 ml of N. meningitidis resuspended in RPMI-10% serum (RPMI-S) at an OD600 of 0.01 (approximately 2 × 107 CFU/ml). Numbers of added bacteria were systematically determined by counting CFU. In some experiments, the cells were fixed before infection with 2.5% paraformaldehyde in PBS (35). After 30 min, unbound bacteria were removed and new RPMI-S was added. The infection was continued for up to 4 h, with the medium being replaced every hour. At the endpoint, several washes were done, and adherent bacteria were recovered by scraping the wells and plated on agar plates to perform CFU counts. Digital images and HD videos were recorded using the above-described setup.
RESULTS
General features of ComP, PilT, PilT2, PilU, PilV, PilX, and PilZ proteins.
Earlier screens of the NeMeSys collection of mutants identified pilX and pilZ mutants, which, although piliated, were dramatically affected in aggregation and/or adhesion (7, 13). A mining of the complete genome of strain 8013 (36) revealed only five additional genes that either play a role in Tfp biology in N. meningitidis (pilT) (13, 35) or are likely to play a role based either on experimental evidence for other species (comP, pilU, and pilV) (30, 45, 46, 48) or solely on sequence homology (pilT2). Out of these seven genes, which are virtually identical in the closely related pathogen N. gonorrhoeae, three (pilT, pilU, and pilZ) are widely conserved in Tfpa-expressing species (32). Mutants with mutations in these seven genes are piliated (36) as shown by immunofluorescence (IF) microscopy using the 20D9 monoclonal antibody specific for fibers of strain 8013 (34). Interestingly, the proteins encoded by these seven genes can be cataloged in three classes based on their sequence features.
The pilZ mutant was originally identified as being unable to form aggregates, but it was not studied further (7). PilZ is predicted to be a cytoplasmic protein with very limited homology to the Pfam domain PF07238 found in proteins binding the secondary messenger cyclic di-GMP (c-di-GMP) (5, 37). We raised a specific serum against PilZ whose affinity is high as seen in immunoblots with the purified protein, but we could not detect PilZ in the WT strain or in a pilZind strain that contains a second chromosomal copy of pilZ under the transcriptional control of an IPTG-inducible promoter. Even the construction of strains expressing tagged PilZ-His6 or His6-PilZ proteins and the use of a commercial anti-His antibody did not allow detection of this protein. This suggests that PilZ either is expressed at very low levels or has an extremely short half-life.
ComP, PilV, and the previously characterized PilX each harbor a distinctive N-terminal leader sequence (see Fig. S1A in the supplemental material) found in type IV pilins (9, 13, 14). Similarity between the mature forms of PilE (the major pilus subunit), ComP, PilV, and PilX is limited to the 27 mainly hydrophobic N-terminal amino acids that are key for assembly into Tfp (9). In addition, each protein harbors C-terminal cysteines and is likely to contain a disulfide bond-delimited D region found in most pilins and minor pilins (9, 14). As previously done for PilX, we showed by immunoblotting that ComP and PilV are processed by the prepilin peptidase PilD, since only unprocessed precursors were detected in a pilD mutant (Fig. 1 A). Furthermore, like PilX, both ComP and PilV copurify with Tfp, as they could be detected in pilus preparations (Fig. 1B). Therefore, it is likely that ComP, PilV, and PilX are all minor pilins. Importantly, copurification of each minor pilin with Tfp was mainly independent of the others, except for ComP, which was much more abundant in the pili of a pilV mutant (Fig. 1B).
FIG. 1.
Immunoblot analysis of the minor pilins ComP, PilV, and PilX. (A) Immunoblot detection of ComP, PilV, and PilX (as a control) in whole-cell protein extracts in PilD+ and PilD− genetic backgrounds. Equal amounts of proteins were loaded in each lane. For ComP, which could not be detected in the WT strain, this was done in an overexpressing comPind strain that contains a second copy of comP under the transcriptional control of an IPTG-inducible promoter. (B) Immunoblot detection of ComP, PilV, and PilX in “classical” pilus preparations from the WT strain and various mutants. Equal amounts of pili were loaded in each lane, as assessed by PilE immunodetection (data not shown). There was one major cross-reacting species in each immunoblot (indicated by an asterisk). As suggested by its size and shape, it is possible that this band corresponds to PilE, which is the overwhelmingly dominant protein in pilus preparations.
PilT2 and PilU show a very high degree of amino acid conservation (60%) with the retraction motor PilT (see Fig. S1B in the supplemental material). These proteins, which differ from PilT mainly by the presence of terminal extensions, belong to the family of type II/IV secretion system ATPases (31). In contrast to the pilU gene, which is found downstream of pilT in most bacteria expressing Tfpa (30, 45), pilT2 is specific to Neisseria and has not been previously studied. This gene is part of a putative operon that contains the pilZ gene (see Fig. S1C in the supplemental material). While in most piliated species, this putative operon starts with holB (which encodes a subunit of the DNA polymerase III), in Neisseria it starts with pilT2, which is separated from holB by only 34 bp.
Assessing and quantifying piliation.
Although N. meningitidis mutants with mutations in these 7 genes express Tfp as assessed by IF microscopy (36), it was important to characterize their piliation further.
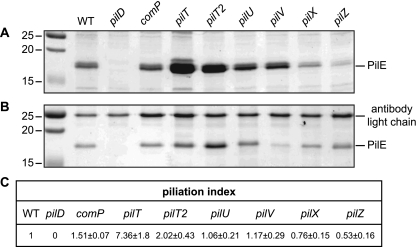
We first assessed whether pili could be purified using a classical procedure during which filaments sheared by vortexing are precipitated using ammonium sulfate (46). Pilus preparations obtained from equivalent numbers of CFU were fairly pure, since the 17-kDa pilin (PilE) was overwhelmingly the dominant species detected on Coomassie blue-stained gels after SDS-PAGE (Fig. 2 A). Compared to the yield of pili obtained from the WT strain, pilus yields in the seven mutants were either (i) lower (slightly in a pilX mutant and dramatically in a pilZ mutant, in which PilE was barely detectable), (ii) similar (in the comP, pilV, and pilU mutants), or (iii) higher (in the pilT and pilT2 mutants). Surprisingly, the very low Tfp yields in the pilZ mutant were not consistent with the high numbers of filaments seen by IF microscopy. To show that pili from the pilZ mutant could be purified, we designed a small-scale affinity purification procedure using the 20D9 antibody. In brief, pili sheared by vortexing as described above were incubated with this antibody, and filaments were immunoprecipitated using magnetic beads coated with protein A. After several washes to remove the unbound components in the sample, pilus immunopreparations (IPs) were separated by SDS-PAGE and gels were stained with Coomassie blue (Fig. 2B). Analysis of the pilus IPs in positive and negative controls confirmed that this new purification procedure is efficient. The only protein species that could be detected were the 17-kDa PilE and the light and heavy chains of the antibody, of which only the 25-kDa light chain is visible in Fig. 2B. Although the minute amounts of antibody that were used (this is a scarce resource) meant that only a small fraction of pili could be recovered, pili could be immunoprecipitated from each mutant (including the pilZ mutant), confirming that the pilZ mutant is piliated.
FIG. 2.
Qualitative (A and B) and quantitative (C) assays of piliation in N. meningitidis comP, pilT, pilT2, pilU, pilV, pilX, and pilZ mutants. The WT strain and a nonpiliated pilD mutant were included as positive and negative controls, respectively. (A and B) Tfp purified from the different strains using a “classical” shearing/ammonium sulfate precipitation method (A) or a shearing/immunoprecipitation method (B) were separated by SDS-PAGE and stained with Coomassie blue. Samples were prepared from equivalent numbers of CFU, and identical volumes were loaded in each lane. (C) Tfp were quantified by a whole-cell ELISA using a monoclonal antibody specific for strain 8013 fibers. Equivalent numbers of CFU were applied to the well of a microtiter plate, and Tfp were quantified by measuring the OD450. Results are expressed as CFU mutant/CFU WT strain giving an OD450 of 0.4 and are the means ± standard deviations from 4 to 12 independent experiments. Ratios for the WT strain (positive control) and the nonpiliated pilD mutant (negative control) have been set at 1 and 0, respectively. A ratio smaller than 1 indicates that the mutant is less piliated than the WT strain.
Next, we quantified piliation in the different mutants by a whole-cell ELISA procedure that uses the 20D9 antibody (13). In brief, after preparation of suspensions containing equivalent numbers of CFU for each strain, serial dilutions were applied to the wells of a microtiter plate and reacted with 20D9, which was itself reacted with a peroxidase-linked secondary antibody. The amount of bound secondary antibody was then quantified spectrophotometrically. While the WT strain reacted strongly, there was an almost complete absence of background with a nonpiliated pilD mutant. We determined, using exponential regression models, the piliation index for each mutant as the ratio of WT CFU to mutant CFU giving an OD450 of 0.4 (Fig. 2C). A ratio smaller than 1 indicates that the mutant is less piliated than the WT strain (i.e., more CFU are needed to obtain the same OD450) and vice versa. We found that the pilX and pilZ mutants displayed 1.4- and 2.1-fold decreases in piliation, respectively. Piliation in the comP, pilV, and pilU mutants was not affected, while the pilT and pilT2 mutants were hyperpiliated, with approximately 7.4- and 2-fold increases in piliation, respectively. Therefore, of the seven studied proteins, PilX and PilZ slightly affect Tfp assembly/stability, while PilT and PilT2 are antagonists of pilus formation since piliation is increased in their absence.
Quantifying competence for DNA transformation.
N. meningitidis is naturally competent for transformation throughout its growth cycle. Competence, which is a Tfp-linked phenotype, is a multisequential process during which DNA is taken up in the periplasm, translocated across the inner membrane, and integrated into the chromosome by homologous recombination (2). We quantified competence in the seven mutants by counting the number of rifampin-resistant colonies obtained after transformation of equivalent numbers of CFU with 1 μg of chromosomal DNA purified from a mutant of strain 8013 spontaneously resistant to rifampin (13). Similar results were obtained with chromosomal DNA purified from an erythromycin-resistant mutant (data not shown). The results were entirely consistent with those for N. gonorrhoeae, in which several of these mutants have been analyzed (1, 2, 47, 48). While almost 0.02% of the recipient cells were transformed for the WT strain, transformability of a nonpiliated pilD mutant was reduced more than 300-fold (Fig. 3). comP and pilT mutants were noncompetent. We confirmed that this was due to the absence of ComP or PilT by showing that competence was restored in complemented comP/comPind and pilT/pilTind strains, with transformabilities 5.1- and 2.5-fold higher than that in the WT strain, respectively. The pilX mutant, as reported previously (13), had a 13.2-fold reduced transformability, which was statistically significant (P = 0.0039) as assessed by a two-tailed Student t test. Competence in the complemented pilX/pilXind strain was restored and was 6.3-fold higher than that in the WT strain. The pilT2, pilU, and pilZ mutants were as competent as the WT strain. Finally, as in N. gonorrhoeae (1), the pilV mutant displayed a 2.4-fold-higher transformability than the WT strain (P = 0.011). In conclusion, four of the seven proteins studied modulate competence for natural DNA transformation. ComP and PilT are essential for this phenotype, PilX is necessary for full competence, and PilV has a negative impact, since in its absence transformation is more efficient.
FIG. 3.
Quantification of the competence for DNA transformation in N. meningitidis comP, pilT, pilT2, pilU, pilV, pilX, and pilZ mutants. The WT strain and a nonpiliated pilD mutant were included as positive and negative controls, respectively. Equivalent numbers of recipient cells were transformed using 1 μg of chromosomal DNA purified from a Rifr strain, and Rifr transformants were counted. Results are expressed as percentages of recipient cells transformed and are the means ± standard deviations from four to six independent experiments.
Assessing bacterial aggregation.
The ability to form bacterial aggregates, a Tfp-linked phenotype key for meningococcal virulence, was assessed in liquid medium. The mutants were incubated for 2 h at 37°C under static conditions, and aggregates were observed by phase-contrast microscopy (7, 13). While no aggregates were seen for the pilD mutant, we found that the comP, pilT2, pilU, and pilV mutants produced regular and round aggregates similar to those produced by the WT strain (Fig. 4 A). In contrast, as previously described (13), the pilT mutant displayed highly irregular aggregates. Round aggregates were restored in the complemented pilT/pilTind strain (data not shown), indicating that aggregate morphology is influenced by pilus dynamics. Finally, no aggregates were seen for the pilX and pilZ mutants, even after prolonged incubation. Because the pilZ gene is likely to be part of an operon (see Fig. S1C in the supplemental material), we showed that the nonaggregative phenotype is not due to a polar effect by demonstrating that aggregation is restored in the complemented pilZ/pilZind strain. Interestingly, aggregation was also restored in the pilZ/his6-pilZind and pilZ/pilZind-his6 strains described above (data not shown). This shows that the corresponding PilZ-His6 and His6-PilZ proteins are produced and functional, despite the fact that we were unable to detect them by immunoblotting.
FIG. 4.
Aggregation as assessed by phase-contrast microscopy (scale bars, 10 μm). (A) Aggregates observed after 2 h in N. meningitidis comP, pilT, pilT2, pilU, pilV, pilX, and pilZ mutants. The WT strain and a nonpiliated pilD mutant were included as positive and negative controls, respectively. Aggregates restored in double pilXT and pilZT mutants are also shown. (B) Aggregation kinetics in pilT2 and pilV mutants. The WT strain was included as a control. Pictures were taken at 0, 10, and 45 min.
Subsequently, since PilT-powered filament retraction has a disruptive role in the formation of bacterial aggregates, we introduced a concurrent pilT mutation into the mutants and tested them for aggregation. All the double mutants displayed highly irregular aggregates similar to those seen in the pilT mutant (data not shown). Importantly, as previously shown for the pilXT mutant (13), the pilZT mutant formed aggregates despite the absence of PilZ (Fig. 4A), which indicates that the mutation in pilT is epistatic to that in pilZ with respect to aggregation.
We also monitored aggregation kinetics for the mutants that formed aggregates (Fig. 4B). While the comP and pilU mutants displayed aggregative kinetics similar to those of the WT strain, we found that the pilT2 and pilV mutants formed aggregates significantly faster. While it took approximately 45 min for very small aggregates to become visible in the WT strain, aggregates in the pilT2 and pilV mutants became visible in as little as 10 min and were already large after 45 min. Normal aggregation kinetics were restored in both complemented strains pilT2/pilT2ind and pilV/pilVind (data not shown), confirming that the observed phenotypes were indeed due to the lack of expression of PilT2 or PilV. In the pilT mutant, kinetics were impossible to assess because its irregular aggregates are resistant to dissociation and were present from the beginning of the assay. In conclusion, five of the seven proteins studied modulate Tfp-mediated formation of bacterial aggregates. PilX and PilZ are essential for this phenotype (unless filament retraction is abolished), while PilT, PilT2, and PilV antagonize it to various extents.
Assessing twitching motility.
Twitching motility, which occurs under humid conditions on both organic and inorganic surfaces, is powered by Tfp retraction (20). Unlike in P. aeruginosa (40), observation of twitching motility zones at the interstitial surface between agar and plastic after subsurface stab inoculation is impractical in N. meningitidis. Furthermore, other methods (e.g., observation of bacteria at the edges of colonies growing on the surface of agar plates) are poorly discriminative. We therefore designed a robust assay to readily assess this phenotype in the meningococcus. By coupling a high-definition (HD) digital camcorder to the microscope, we documented twitching motility by recording in real time bacterial movement within aggregates attached to the plastic in 24-well plates. When WT aggregates were observed, bacteria exhibited continuous and vigorous jerky movements (see Video S1 in the supplemental material). This movement is clearly distinct in amplitude and speed from the Brownian motion exhibited by isolated diplococci that can be seen in the same video. This movement indeed results from twitching motility as confirmed by observing a pilT mutant. While isolated pilT diplococci display Brownian motion, the aggregates formed by this mutant are completely still (see Video S2 in the supplemental material). When the other four mutants that formed aggregates (the comP, pilT2, pilU, and pilV mutants) were analyzed in this way, we found that they all exhibit jerky movements (see Videos S3, S4, S5, and S6 in the supplemental material), indicating that the corresponding proteins play no detectable role in twitching motility.
Assessing and quantifying adhesion to human cells.
The Tfp-linked phenotype possibly most important for meningococcal virulence is the ability to adhere strongly to human cells (43). As in our previous studies (7, 13), we used human umbilical vein endothelial cells (HUVEC) to quantify the adhesion abilities of the seven mutants. When output results were normalized to the initial inocula, there was more than a 1,500-fold decrease in adhesion for a nonpiliated pilD mutant compared to the WT strain (Fig. 5 A). We found that most of the mutants, but not the pilX and pilZ mutants, adhered to HUVEC as well as the WT strain. As previously demonstrated (13), the pilX mutant was as dramatically affected for adhesion as a nonpiliated mutant. Unexpectedly, the pilZ mutant, although it does not form aggregates, which has so far always been associated with abolished adhesion, adhered 10-fold better than a nonpiliated mutant (P = 1.28 × 10−8). Moreover, the pilZ mutant displayed a morphologically unique adhesion (Fig. 6). Unlike the WT strain, for which round and dense aggregates of bacteria were adhering to the cells, which is typical of Tfp-mediated adherence (35), the pilZ mutant adhered in fragmented clusters in which there was little contact between the diplococci. Adhesion in the complemented pilZ/pilZind strain was normal (data not shown), confirming that the impaired adherence in the pilZ mutant was due to the lack of expression of PilZ.
FIG. 5.
Quantification of adhesion to HUVEC. After a 30-min contact during which standard numbers of bacteria adhered to standard numbers of cells, nonadherent bacteria were removed by replacing the medium. After further incubation (with the medium being regularly replaced every hour), cells were washed and recovered by scraping, and adherent bacteria were counted. Results, expressed as CFU of adhering bacteria, were normalized for an inoculum of 107 CFU. (A) Adhesion or N. meningitidis comP, pilT, pilT2, pilU, pilV, pilX, and pilZ mutants to HUVEC after 270 min of infection. The WT strain and a nonpiliated pilD mutant were included as positive and negative controls, respectively. Restored adhesion in pilXT and pilZT double mutants is also shown. Results are the means ± standard deviations from 3 to 10 independent experiments. (B) Adhesion of the pilU mutant to HUVEC after 30 and 90 min of infection. The WT strain was included as a control. Results are the means ± standard deviations from five independent experiments.
FIG. 6.
Adhesion of a pilZ mutant to HUVEC as observed by phase-contrast microscopy (scale bars, 10 μm). Images were taken after 150 min of infection. The WT strain and a nonpiliated pilD mutant were included as positive and negative controls, respectively.
Next, since PilT-powered filament retraction has an important role in adhesion, we quantified the adhesive abilities of all the double mutants containing a concurrent mutation in the pilT gene. Interestingly, as previously reported for the pilX/T mutant (13), we found that the pilZT double mutant adhered to HUVEC as well as the pilT mutant (Fig. 5A). In other words, the mutation in pilT is epistatic to that in pilZ with respect to adhesion, because N. meningitidis can adhere to human cells in the absence of PilZ when PilT is also absent. We therefore tested by immunoblotting whether the copurification with Tfp of the Pil proteins known to modulate adhesion to human cells, i.e., PilC1, PilX, and PilV (13, 27, 46), was affected in the absence of PilZ. The answer was negative (Fig. 7), which rules out the possibility that impaired adhesion in the pilZ mutant is due to a reduced association of PilC1, PilX, and PilV with the filaments.
FIG. 7.
Immunoblot detection of PilV, PilX, and PilC1 in “classical” pilus preparations from a pilZ mutant. The WT strain and a pilE mutant were included as positive and negative controls, respectively. Equal amounts of pili were loaded in each lane, as assessed by PilE immunodetection (data not shown). As in Fig. 1, there was a major cross-reacting species (indicated by an asterisk) in the immunoblots in which minor pilins were detected, which might correspond to PilE.
To further delineate adhesion in the adhering mutants, we monitored it by phase-contrast microscopy over time. This revealed important differences. Compared to the WT strain, more bacteria were seen on cells for the pilU mutant at early time points. We therefore quantified the adhesive abilities of the pilU mutant and the WT strain after 30 and 90 min of infection (Fig. 5B), which showed, respectively, 3.8- and 2.7-fold increases in adherent CFU in the pilU mutant (P = 0.053 and 0.014 at 30 and 90 min, respectively). This suggests that PilU has a negative impact on initial adhesion, since in its absence more bacteria adhere at early time points. Similarly, it became readily apparent on closer inspection that adhering pilV aggregates exhibit an increase in twitching motility, which was documented by producing HD movies. While a few bacteria exhibited limited movement within adhering aggregates in the WT strain (see Video S7 in the supplemental material), in the pilV mutant virtually all the meningococci exhibited frenetic, jerky movements (see Video S8 in the supplemental material). This movement clearly corresponded to twitching motility, since adhering aggregates of the pilT mutant were still (see Video S9 in the supplemental material). This increase in movement was not actively induced by the human cells, since it was also observed on cells fixed with paraformaldehyde prior to infection (data not shown). On inspection of frames in these movies, another difference was also apparent: pilV aggregates are looser than those formed by the WT strain (compare Videos S7 and S8 in the supplemental material).
In conclusion, five of the seven proteins studied modulate Tfp-mediated adherence to human cells. PilX and PilZ are essential for high numbers of bacteria to adhere (unless filament retraction is abolished by a concurrent pilT mutation), while PilU, PilV, and PilT (35) affect this phenotype in a qualitative fashion.
Phenotypic alterations resulting from gene overexpression.
The above findings came from the study of phenotypic alterations resulting from gene mutation. However, although this is the most popular and efficient path to identifying gene function, significant functional information can also be extracted from the study of phenotypic effects resulting from gene overexpression. In Tfp biology, this has been shown for the fimT and fimU genes in P. aeruginosa (4). Given the fact that some of the proteins studied could be grouped into functional classes, it was interesting to determine what phenotypic effects, if any, would result from overexpression of some proteins in heterologous mutant backgrounds. Since PilT, PilT2, and PilU are paralogs, we wondered whether overexpression of PilT2 or PilU could cross-complement the defects in the pilT mutant. We therefore constructed pilT/pilT2ind and pilT/pilUind strains (which have a second chromosomal copy of the pilT2 or pilU gene under the transcriptional control of an IPTG-inducible promoter) and tested their competence and twitching motility. While competence and twitching motility were restored in the complemented pilT/pilTind strain, both the pilT/pilT2ind and pilT/pilUind strains behaved as the pilT mutant did (data not shown). This indicates that overexpression of PilT2 and PilU is not able to cross-complement the phenotypic defects resulting from the absence of PilT, suggesting that there is no major functional overlap between these three proteins.
Similarly, because all the corresponding proteins are likely to be minor pilins, we tested whether overexpression of PilV or PilX could cross-complement the competence defect in the comP mutant and whether ComP and PilV could cross-complement the defects in the pilX mutant. The answer to the first question was negative; the comP/pilVind and comP/pilXind mutants remained noncompetent like the comP mutant (data not shown). In contrast, while the pilX/pilVind mutant remained nonaggregative, the pilX/comPind mutant formed aggregates (Fig. 8 A). These aggregates, however, were morphologically unique, since they were flat, loose, and fragile, tending to disappear upon very moderate agitation. The possibility to restore aggregates in a pilX mutant without the concurrent introduction of a pilT mutation offered us the opportunity to assess twitching motility in the absence of PilX. HD movies of the pilX/comPind aggregates showed that they displayed jerky movements (data not shown), confirming that PilX is dispensable for twitching motility. Consistent with the close link between aggregation and adhesion to human cells, we found that there was a 4.6-fold increase (P = 0.043) in adherent CFU in the pilX/comPind strain compared to the pilX mutant (Fig. 8B). Interestingly, in the pilX/comPind strain there was also an increase (P = 0.022) in the number of transformants to WT levels (Fig. 8C). Finally, when we tested pilX/pilVind for transformability, we found a reduction in transformability compared to the pilX mutant. Although it was of low statistical significance, this was consistent with the negative impact of PilV on transformation. Therefore, although there appears to be no major functional redundancy between the minor pilins, they can modulate each other's functions, both positively and negatively.
FIG. 8.
Aggregation, competence, and/or adhesion in cross-complemented pilX/comPind and pilX/pilVind strains. (A) Aggregation in the pilX/comPind and pilX/pilVind mutants as observed by phase-contrast microscopy after 2 h of growth (scale bars, 10 μm). The pilX/pilXind strain was included as a control. (B) Adhesion of the pilX/comPind strain to HUVEC after 270 min of infection. The WT strain and the pilD and pilX mutants were included as controls. The results, expressed as CFU of adhering bacteria normalized for an inoculum of 107 CFU, are the means ± standard deviations from four to six independent experiments. (C) Quantification of the competence for DNA transformation in pilX/comPind and pilX/pilVind strains. The WT strain, the pilX/pilXind strain, and the pilD and pilX mutants were included as controls. Results are expressed as percentages of recipient cells transformed and are the means ± standard deviations from three to six independent experiments.
DISCUSSION
Tfp biology is a complex system involving a large number of genes, many of which are conserved in both sequence and genomic organization in piliated species (32). An integrated molecular view of Tfp biology remains an elusive goal because of important differences between the various model species in which these genes have been functionally characterized. We have therefore started a systematic functional analysis in a sequenced clinical isolate of N. meningitidis. After recently reporting the analysis of 16 genes essential for Tfp biogenesis (6, 7), here we have extended our effort. We have systematically studied the phenotypic alterations resulting from mutation or overexpression of seven genes dispensable for piliation, showing that they all modulate Tfp-mediated functions (Table 1).
TABLE 1.
Summary of the phenotypic alterations in N. meningitidis comP, pilT, pilT2, pilU, pilV, pilX, and pilZ mutants
| Tfp biology feature | Alterationa in mutant: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| pilT | pilT2 | pilU | comP | pilV | pilX | pilXT | pilZ | pilZT | |
| Piliation | ↑ | ↑ | ↓ | ↑ | ↓ | ↑ | |||
| Competence | Nil | Nil | ↑ | ↓ | Nil | Nil | |||
| Aggregation | ↑ | ✓ | ✓ | Nil | ↑ | Nil | ↑ | ||
| Twitching motility | Nil | ↑ | Nil | ND | Nil | ||||
| Adhesion to cells | ✓ | ✓ | ✓ | Nil | ✓ | ↓ | ✓ | ||
When there is no indication, the corresponding feature is unaffected compared to that in the WT strain. ↑ and ↓, the feature is positively or negatively affected, respectively. Nil, the feature is abolished. ✓, the feature is altered in a qualitative fashion (see text for details). ND, not determined.
We previously reported that an N. meningitidis pilZ mutant was piliated, since we could visualize its filaments by IF microscopy (7). Here, we unambiguously show that PilZ is dispensable for piliation, although the filaments in this mutant could not be readily purified using a classical precipitation method. This is similar to what has been reported for a P. aeruginosa mutant unable to glycosylate the major pilin (42), in which case the poor pilus yields were attributed to altered physical properties of the filaments. In addition, this offers a plausible explanation for the apparent discrepancy between our findings and those of Alm et al., who reported that a P. aeruginosa pilZ mutant is nonpiliated based on the fact that no pilin could be detected in pilus preparations (3). Nevertheless, PilZ plays an important role in Tfp biology in N. meningitidis, since the pilZ mutant displays dramatic defects in aggregation and adhesion (Table 1). Interestingly, the findings that aggregation and adhesion are restored in a pilZT double mutant suggest that PilZ participates in these phenotypes by somehow counterbalancing PilT-mediated filament retraction. These findings are similar, but not identical, to those with the pilX mutant (Table 1). PilZ does not interfere with PilX presentation within the filaments, and the intrinsically different properties of PilZ and PilX suggest that PilZ (a predicted cytoplasmic protein) does not affect aggregation directly as the minor pilin PilX does (14). What the molecular basis of PilZ's role in Tfp biology could be is thus still a matter of speculation. However, despite limited homology to a motif found in proteins binding the important secondary messenger c-di-GMP (5), several observations argue against a regulatory function for PilZ through binding of c-di-GMP. First, the N. meningitidis 8013 genome encodes no proteins containing known motifs for c-di-GMP synthesis and degradation. Second, the crystal structure of PilZ in Xanthomonas campestris (56% amino acid conservation with N. meningitidis PilZ) showed that it lacks an N-terminal motif essential for c-di-GMP binding (16). Third, PilZ from P. aeruginosa (64% amino acid conservation) does not bind c-di-GMP (22). Two alternative possibilities could be envisioned to explain the dramatic phenotypic defects in the pilZ mutant. One could speculate that PilZ slows down filament retraction by interfering directly or indirectly with PilT and that in its absence bacterial aggregates could not resist the disruptive forces of pilus retraction. Alternatively, the phenotypic defects of the pilZ mutant could be due to its slightly decreased piliation. This is, however, less likely, because it has been shown in N. gonorrhoeae by modulating piliation that a moderate decrease in piliation leads to merely a 2-fold decrease in adherence (17).
We have previously shown that of the eight genes in the N. meningitidis genome that encode proteins harboring the distinctive prepilin N-terminal leader sequence, five (pilE, pilH, pilI, pilJ, and pilK) are essential for Tfp biogenesis (7), while pilX is dispensable but key for Tfp-mediated functions (13, 14). We demonstrate here that the last two genes (comP and pilV) are dispensable for piliation but play important roles in Tfp biology, similar to those in N. gonorrhoeae (46, 48). Our results support the notion that, like PilX (14), ComP and PilV are minor pilins that assemble within the filaments in a way similar to that for PilE and exert their role(s) from that location. Importantly, ComP, PilV, and PilX all have different functions (Table 1). As previously reported (13), PilX is essential for aggregation and hence adhesion to human cells. Interestingly, it is now clear that PilX also plays a role in competence, because its reduced competence cannot simply be a consequence of its abolished aggregation or slightly reduced piliation, since the pilZ mutant, which exhibits similar defects, is normally competent. The second minor pilin, ComP, is essential for competence for DNA transformation, which is consistent with results for N. gonorrhoeae (2, 48). Finally, PilV presents multiple and rather subtle phenotypic alterations. PilV is an intrinsic antagonist of both competence, which is consistent with the case for N. gonorrhoeae (1), and aggregation. While PilV might impinge on competence by antagonizing accumulation of ComP within the filaments, as suggested for the gonococcus (1), such a scenario cannot explain its negative role in aggregation, since PilX levels in filaments are independent of PilV. In addition, PilV is important for normal adherence to human cells in both the meningococcus and the gonococcus, but in very different ways. While in N. gonorrhoeae a pilV mutant displays a dramatic reduction in adherence (46), an N. meningitidis pilV mutant adheres as well as the WT strain but the adhering aggregates are looser and exhibit a striking increase in twitching motility. This phenotype, never previously observed, is not actively induced by the cells, because it was also observed on fixed cells. A unifying molecular mechanism explaining such a vast array of unrelated phenotypes is difficult to imagine at this stage. The simple possibility that aggregation is faster in a pilV mutant because of a slowing down of pilus retraction seems incompatible with the “hypertwitching” phenotype seen on human cells. However, there is one attractive possibility, i.e., that of PilV being a factor contributing directly or indirectly to the cohesion of aggregates. In the absence of PilV, aggregates might be less compact, and looser aggregates would in turn lead to an apparent increase in twitching motility since there would be less resistance to PilT-mediated retraction of the filaments. Importantly, the adhesion of looser and “hypertwitching” pilV aggregates is expected to be less efficient and hence more fragile, which is supported by their low resistance to shear stress as recently observed in adhesion experiments performed in a laminar flow chamber (24). This fragility could, at least partly, account for the impaired adhesion observed in N. gonorrhoeae (46). Such a scenario could be tested, since it is expected that adhering aggregates of a pilVT mutant, which are very compact due to the absence of disruptive forces generated by pilus retraction, should display restored resistance to shear stress. Nevertheless, it remains to be seen how this could be reconciled with the faster aggregation seen in liquid medium.
The last three genes that we have characterized (pilT, pilT2, and pilU) form the most homogeneous class, since they encode paralogous proteins belonging to the family of type II/IV secretion system ATPases (31). Because this class includes one of the most intensively studied genes in Tfp biology, which encodes the retraction motor PilT, it was tempting to speculate that pilU and pilT2 encode proteins modulating Tfp dynamics. Quantification of the filaments showed that PilT2 is indeed an antagonist of filament formation/stability, but to a lesser extent than PilT. The simplest explanation is that PilT2 participates in pilus retraction synergistically to PilT. Therefore, in the absence of PilT2, pilus retraction would be slowed down, which is entirely consistent with the phenotype of the pilT2 mutant. This scenario remains to be tested, possibly by using optical tweezers (18) to measure the kinetics and force generated by Tfp retraction in a pilT2 mutant. On the other hand, how a modulatory role of PilU in pilus retraction may account for the faster adhesion of the pilU mutant is unclear. However, due to the close dependence of adhesion on bacterial aggregation, it would not be surprising that this faster adhesion results from altered aggregative properties. Since the pilU mutant does not aggregate faster than the WT strain, its faster adhesion might be due to the formation of larger aggregates, which cannot be readily tested using the currently available assays. Intriguingly, these results further widen the incredibly vast array of roles attributed to PilU in different species. Our results are most similar to those for N. gonorrhoeae, although there is a major difference since the gonococcal pilU mutant, although hyperadherent, is apparently nonaggregative, which is an unusual phenotypic combination (30). They are also similar to what has been described for a pilU mutant of Pseudomonas stutzeri (11), except for the reduced transformability in latter species. In contrast, our findings are totally different from those for P. aeruginosa, in which a pilU mutant is hyperpiliated and does not twitch (45), and for Dichelobacter nodosus, in which the mutant does not twitch but is normally piliated (12). While the reason(s) for such disparate findings is unclear, these observations strengthen our original assumption that an integrated view of Tfp biology in any species is achievable only through a systematic functional analysis of each pil gene in the species. Interestingly, the finding that even when overexpressed PilT2 or PilU cannot substitute for PilT suggests that these proteins are unlikely to form separate retraction motors, as has been recently suggested for Myxococcus xanthus, where several PilT paralogs exist (8). Rather, if PilT2 or PilU indeed modulates pilus retraction, this is more likely to occur through interaction with PilT hexamers (39); this subject awaits further experiments.
In conclusion, together with our previous analysis of genes required for Tfp biogenesis in N. meningitidis, the findings reported here show that at least 23 proteins (ComP, PilC1, PilC2, PilD, PilE, PilF, PilG, PilH, PilI, PilJ, PilK, PilM, PilN, PilO, PilP, PilQ, PilT, PilT2, PilU, PilV, PilW, PilX, and PilZ) are directly involved in Tfp biology in this human pathogen. Interestingly, since the wide range of phenotypic defects associated with the seven mutants analyzed in this study are not obscured by the absence of pili, this will promote further characterization of the corresponding proteins. This is expected to shed light on the poorly understood molecular mechanisms of the important functions mediated by Tfp that make these filaments one of the most widespread virulence factors in the bacterial world.
Supplementary Material
Acknowledgments
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BBSRC) and the Agence Nationale pour la Recherche (ANR). Daniel R. Brown was supported by a Doctoral Training Grant from the BBSRC.
We thank G. Duménil, P. Morand, and X. Nassif (U570 INSERM, France) for the kind gift of reagents. We are grateful to A. M. Misic and K. T. Forest (University of Wisconsin-Madison) and to C. Recchi (Imperial College London) for critical reading of the manuscript.
This paper is dedicated to the memory of Jean-Marc Reyrat, our longtime friend and colleague, who died on 28 October 2009 at the age of 42.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 3 May 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Aas, F. E., C. Lovold, and M. Koomey. 2002. An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to type IV pilus expression. Mol. Microbiol. 46:1441-1450. [DOI] [PubMed] [Google Scholar]
- 2.Aas, F. E., M. Wolfgang, S. Frye, S. Dunham, C. Lovold, and M. Koomey. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., A. J. Bodero, P. D. Free, and J. S. Mattick. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:46-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm, R. A., and J. S. Mattick. 1996. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:3809-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3-6. [DOI] [PubMed] [Google Scholar]
- 6.Carbonnelle, E., S. Helaine, X. Nassif, and V. Pelicic. 2006. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol. Microbiol. 61:1510-1522. [DOI] [PubMed] [Google Scholar]
- 7.Carbonnelle, E., S. Helaine, L. Prouvensier, X. Nassif, and V. Pelicic. 2005. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occuring after pilus assembly, essential for fiber stability and function. Mol. Microbiol. 55:54-64. [DOI] [PubMed] [Google Scholar]
- 8.Clausen, M., V. Jakovljevic, L. Sogaard-Andersen, and B. Maier. 2009. High-force generation is a conserved property of type IV pilus systems. J. Bacteriol. 191:4633-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig, L., M. E. Pique, and J. A. Tainer. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363-378. [DOI] [PubMed] [Google Scholar]
- 10.Geoffroy, M., S. Floquet, A. Métais, X. Nassif, and V. Pelicic. 2003. Large-scale analysis of the meningococcus genome by gene disruption: resistance to complement-mediated lysis. Genome Res. 13:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graupner, S., N. Weger, M. Sohni, and W. Wackernagel. 2001. Requirement of novel competence genes pilT and pilU of Pseudomonas stutzeri for natural transformation and suppression of pilT deficiency by a hexahistidine tag on the type IV pilus protein PilAI. J. Bacteriol. 183:4694-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han, X., R. M. Kennan, J. K. Davies, L. A. Reddacliff, O. P. Dhungyel, R. J. Whittington, L. Turnbull, C. B. Whitchurch, and J. I. Rood. 2008. Twitching motility is essential for virulence in Dichelobacter nodosus. J. Bacteriol. 190:3323-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helaine, S., E. Carbonnelle, L. Prouvensier, J.-L. Beretti, X. Nassif, and V. Pelicic. 2005. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol. Microbiol. 55:65-77. [DOI] [PubMed] [Google Scholar]
- 14.Helaine, S., D. H. Dyer, X. Nassif, V. Pelicic, and K. T. Forest. 2007. 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 104:15888-15893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klee, S. R., X. Nassif, B. Kusecek, P. Merker, J. L. Beretti, M. Achtman, and C. R. Tinsley. 2000. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect. Immun. 68:2082-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, T. N., K. H. Chin, J. H. Liu, A. H. Wang, and S. H. Chou. 2009. XC1028 from Xanthomonas campestris adopts a PilZ domain-like structure without a c-di-GMP switch. Proteins 75:282-288. [DOI] [PubMed] [Google Scholar]
- 17.Long, C. D., S. F. Hayes, J. P. M. van Putten, H. A. Harvey, M. A. Apicella, and H. S. Seifert. 2001. Modulation of gonococcal piliation by regulatable transcription of pilE. J. Bacteriol. 183:1600-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maier, B. 2005. Using laser tweezers to measure twitching motility in Neisseria. Curr. Opin. Microbiol. 8:344-349. [DOI] [PubMed] [Google Scholar]
- 19.Maier, B., L. Potter, M. So, C. D. Long, H. S. Seifert, and M. P. Sheetz. 2002. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. U. S. A. 99:16012-16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 21.Mehr, I. J., C. D. Long, C. D. Serkin, and H. S. Seifert. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merighi, M., V. T. Lee, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65:876-895. [DOI] [PubMed] [Google Scholar]
- 23.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 24.Mikaty, G., M. Soyer, E. Mairey, N. Henry, D. Dyer, K. T. Forest, P. Morand, S. Guadagnini, M. C. Prévost, X. Nassif, and G. Duménil. 2009. Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog. 5:e1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morand, P. C., E. Bille, S. Morelle, E. Eugène, J. L. Beretti, M. Wolfgang, T. F. Meyer, M. Koomey, and X. Nassif. 2004. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 23:2009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morand, P. C., P. Tattevin, E. Eugène, J.-L. Beretti, and X. Nassif. 2001. The adhesive property of the type IV pilus-associated component PilC1 of pathogenic Neisseria is supported by the conformational structure of the N-terminal part of the molecule. Mol. Microbiol. 40:846-856. [DOI] [PubMed] [Google Scholar]
- 27.Nassif, X., J.-L. Beretti, J. Lowy, P. Stenberg, P. O'Gaora, J. Pfeifer, S. Normark, and M. So. 1994. Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 91:3769-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nassif, X., J. Lowy, P. Stenberg, P. O'Gaora, A. Ganji, and M. So. 1993. Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol. Microbiol. 8:719-725. [DOI] [PubMed] [Google Scholar]
- 29.Parge, H. E., K. T. Forest, M. J. Hickey, D. A. Christensen, E. D. Getzoff, and J. A. Tainer. 1995. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378:32-38. [DOI] [PubMed] [Google Scholar]
- 30.Park, H. S., M. Wolfgang, and M. Koomey. 2002. Modification of type IV pilus-associated epithelial cell adherence and multicellular behavior by the PilU protein of Neisseria gonorrhoeae. Infect. Immun. 70:3891-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051-3072. [DOI] [PubMed] [Google Scholar]
- 32.Pelicic, V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827-837. [DOI] [PubMed] [Google Scholar]
- 33.Pelicic, V., S. Morelle, D. Lampe, and X. Nassif. 2000. Mutagenesis of Neisseria meningitidis by in vitro transposition of Himar1 mariner. J. Bacteriol. 182:5391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pujol, C., E. Eugène, L. de Saint Martin, and X. Nassif. 1997. Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect. Immun. 65:4836-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pujol, C., E. Eugène, M. Marceau, and X. Nassif. 1999. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. U. S. A. 96:4017-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rusniok, C., D. Vallenet, S. Floquet, H. Ewles, C. Mouzé-Soulama, D. Brown, A. Lajus, C. Buchrieser, C. Médigue, P. Glaser, and V. Pelicic. 2009. NeMeSys: a resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol. 10:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryjenkov, D. A., R. Simm, U. Romling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310-30314. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Satyshur, K. A., G. A. Worzalla, L. S. Meyer, E. K. Heiniger, K. G. Aukema, A. M. Misic, and K. T. Forest. 2007. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure 15:363-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semmler, A., C. Whitchurch, and J. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145:2863-2873. [DOI] [PubMed] [Google Scholar]
- 41.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smedley, J. G., E. Jewell, J. Roguskie, J. Horzempa, A. Syboldt, D. B. Stolz, and P. Castric. 2005. Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect. Immun. 73:7922-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virji, M. 2009. Pathogenic Neisseriae: surface modulation, pathogenesis and infection control. Nat. Rev. Microbiol. 7:274-286. [DOI] [PubMed] [Google Scholar]
- 44.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 45.Whitchurch, C. B., and J. S. Mattick. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13:1079-1091. [DOI] [PubMed] [Google Scholar]
- 46.Winther-Larsen, H. C., F. T. Hegge, M. Wolfgang, S. F. Hayes, J. P. M. van Putten, and M. Koomey. 2001. Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc. Natl. Acad. Sci. U. S. A. 98:15276-15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hébert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321-330. [DOI] [PubMed] [Google Scholar]
- 48.Wolfgang, M., J. P. van Putten, S. F. Hayes, and M. Koomey. 1999. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol. Microbiol. 31:1345-1357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.