Abstract
Pantoea agglomerans is an ecologically diverse taxon that includes commercially important plant-beneficial strains and opportunistic clinical isolates. Standard biochemical identification methods in diagnostic laboratories were repeatedly shown to run into false-positive identifications of P. agglomerans, a fact which is also reflected by the high number of 16S rRNA gene sequences in public databases that are incorrectly assigned to this species. More reliable methods for rapid identification are required to ascertain the prevalence of this species in clinical samples and to evaluate the biosafety of beneficial isolates. Whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) methods and reference spectra (SuperSpectrum) were developed for accurate identification of P. agglomerans and related bacteria and used to detect differences in the protein profile within variants of the same strain, including a ribosomal point mutation conferring streptomycin resistance. MALDI-TOF MS-based clustering was shown to generally agree with classification based on gyrB sequencing, allowing rapid and reliable identification at the species level.
Pantoea agglomerans (20) is a ubiquitous plant-epiphytic bacterium that belongs to the family Enterobacteriaceae. While several strains are commercialized for biological control of plant diseases (23), the species also includes two phytopathogenic pathovars that carry distinctive virulence plasmids (32). P. agglomerans has a Jekyll-Hyde nature, being described also as an opportunistic human pathogen (30), which raises biosafety regulatory issues for the utilization of beneficial isolates (45). Clinical reports predominantly involve septicemia following penetrating trauma (16, 56) or nosocomial infections (14, 55). Clinical pathogenicity of this species has not been confidently confirmed (unfulfilled Koch's postulates). Infections attributed to P. agglomerans are typically of a polymicrobial nature involving patients affected by other diseases (14) and may represent secondary contamination of wounds. Standard clinical diagnostics and identification rely mainly on biochemical profiling analysis or alternatively on 16S rRNA gene sequencing, despite the inadequacy of these techniques for precise discrimination within the Enterobacter and Pantoea genera (5, 20, 39). Problems with correct identification have been observed for automated systems such as the API 20E (24, 39) and Vitek-2/GNI+ (39, 40) (both from bioMerieux) or the Phoenix (11, 38) and Crystal identification systems (40, 48) (both from BD Diagnostic Systems).
P. agglomerans is a composite taxon conglomerating former Enterobacter agglomerans, Erwinia milletiae, and Erwinia herbicola strains. Accurate identification is complicated by the unsettled taxonomy of the “P. agglomerans-E. herbicola-E. agglomerans” complex (45). Recent analyses based on gyrB sequencing, multilocus sequence analysis (MLSA) (4), and fluorescent amplified fragment length polymorphisms (fAFLP) (45) indicate that strains belonging to Enterobacter or Erwinia archived in culture collections are often erroneously assigned to P. agglomerans and are likely also misidentified in clinical diagnostics. False classifications of environmental P. agglomerans strains as related Pantoea species, including human- or plant-pathogenic P. ananatis, are also common (45). Inadequate biochemical identification methods and uncertainty regarding current taxonomy are revealed also by the excessive number of 16S rRNA gene sequences incorrectly assigned to P. agglomerans that can be retrieved from GenBank (Fig. 1). Sequencing of housekeeping genes, MLSA, and fAFLP are labor-intensive, time-consuming, and impractical approaches as routine diagnostic tools.
FIG. 1.
Taxonomy of putative P. agglomerans isolates based on 16S rRNA gene sequences retrieved from GenBank under the currently accepted species name or under the old basonyms Enterobacter agglomerans and Erwinia herbicola. Out of a total of 331 complete or partial sequences found, 263 could be aligned over their 1,240-bp central region resulting in a minimum evolution tree. For the analysis, gaps and missing data were eliminated only in pairwise sequence comparisons, resulting in a total of 1,114 positions. Nodal supports were assessed by 1,000 bootstrap replicates. Only bootstrap values greater than 50% are shown. The scale bar represents the number of base substitutions per site. The number of “P. agglomerans” sequences clustering with a given reference strain in shown in parentheses. Reference strains and clades containing reference strains are marked in bold, and the corresponding accession numbers are indicated between brackets. For the genus Erwinia the following reference strains were used: E. persicina HK204 [NR_026049.1], E. rhapontici 2OP2 [FJ595873], E. billingiae Eb661 [AM055711], E. tasmaniensis Et2/99 [AM292080], and E. amylovora FAW 23482 [AY456711].
Whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (31) is an emerging technology for identification of bacteria (26, 46), fungi (17, 33), viruses (29, 51), insects (41), and helminths (42). MALDI-TOF MS-based identification can accurately resolve bacterial identity at the genus, species, and in some taxa subspecies levels (e.g., Salmonella enterica serovars, Listeria genotypes) (1, 18). Identity is based on unique mass/charge ratio (m/z) fingerprints of proteins, which are ionized using short laser pulses directed to bacterial cells obtained from a single colony embedded in a matrix. After desorption, ions are accelerated in vacuum by a high electric potential and separated on the basis of the time taken to reach a detector, which is directly proportional to the mass-to-charge ratio of an ion. This technique has been shown to deliver reproducible protein mass fingerprints starting from an aliquot of a single bacterial colony within minutes and without any prior separation, purification, or concentration of samples. Whole-cell MALDI-TOF MS is a reliable technique across broad conditions (e.g., different growth media, cell growth states), with limited variability in mass-peak signatures within a selected mass range (2,000 < m/z < 20,000) that does not affect reliability of identification (28, 31). MALDI-TOF MS profiles primarily represent ribosomal proteins, which are the most abundant cellular proteins and are synthesized under all growth conditions (47). MALDI-TOF MS identification profiles derived from several characterized strains for a given species are used to develop reference spectra (e.g., SuperSpectrum; AnagnosTec GmbH, Potsdam, Germany), and they include a subset of characteristic and reproducible markers. MALDI-TOF MS identification databases are currently available for a relatively wide range of clinical bacteria, and this method has become an accepted tool for routine clinical diagnostics due to enhanced simplicity, rapidity, and reliability. However, environmental bacteria, such as Pantoea, have not been widely evaluated using MALDI-TOF MS and are largely absent from identification databases, limiting the practical reach of this new technology.
Our objectives were to develop a robust method for rapid identification of P. agglomerans and related bacteria based on MALDI-TOF MS and to compare MALDI-TOF MS results against those obtained from a phylogenetic analysis based on gyrB sequencing as well as against biochemical identification methods.
MATERIALS AND METHODS
Screening and analysis of total available Pantoea 16S rRNA gene sequences.
The NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/nuccore) was scrutinized for 16S rRNA gene sequences of putative P. agglomerans isolates by searching for entries beneath the currently accepted species name and under the old basonyms Enterobacter agglomerans and Erwinia herbicola. Out of a total of 394 complete or partial sequences found, 263 were at least 1,240 bp long and were retained for the analysis together with 21 reference sequences of relevant species of Enterobacteriaceae. The resulting ClustalW (52) alignment was employed to construct a minimum evolution tree using the Molecular Evolutionary Genetics Analysis (MEGA) program, version 4.0 (50).
Bacterial strains.
A collection of 53 strains received as P. agglomerans, a Pantoea sp., or E. agglomerans from research and culture collections and 20 reference strains belonging to closely related Pantoea species and other Enterobacteriaceae were compared (Table 1 ). The streptomycin-resistant, commercial biocontrol strain Pantoea vagans C9-1S (formerly P. agglomerans [45]) and a variant strain, P. vagans C9-1W, lacking the 530-kb pPag3 plasmid (49), were included to evaluate the sensitivity and robustness of MALDI-TOF MS. For standardized spectral acquisition and generation of “SuperSpectra,” as determined by SARAMIS (spectral archive and microbial identification system) (AnagnosTec GmbH), bacteria were grown on LB agar at 28°C for 24 to 48 h. Tryptic soy agar (TSA) and Mueller-Hinton agar (MHA) were used in parallel to assess the influence of different media on species recognition. Alongside MALDI-TOF MS analysis, biochemical profiling and conventional Sanger sequencing of the gyrB gene were performed for all strains.
TABLE 1.
Strains used in this studyb
| Strain designationa | Received as | Origin | Host | Properties | Other designations or notes | Phoenixc | Reference |
|---|---|---|---|---|---|---|---|
| Brenneria | |||||||
| LMG 2694T | B. nigrifluens | United States | Juglans regia | Phytopathogen | No | 57 | |
| LMG 2709T | B. rubrifaciens | United States | Juglans regia | Phytopathogen | No | 58 | |
| Enterobacter | |||||||
| LMG 5336 | E. agglomerans | United States | N/A | Clinical | P. agglomerans ATCC 29904 | Yes | |
| LMG 5337 | E. agglomerans | United States | Human | Clinical | P. agglomerans ATCC 29915 | No | |
| LMG 5339 | E. agglomerans | United States | Chicken | Veterinarial | P. agglomerans ATCC 29917 | Yes | |
| LMG 5341 | E. agglomerans | United States | Human | Clinical | P. agglomerans ATCC 29919 | Yes | |
| LMG 5345 | E. agglomerans | United States | Human | Clinical | P. agglomerans ATCC 29923 | Yes | |
| Erwinia | |||||||
| CFBP 1232T | E. amylovora | United Kingdom | Pyrus communis | Phytopathogen | (Yes) | 2 | |
| CFBP 4342 | E. herbicola pv. gypsophilae | Netherlands | Gypsophila paniculata | Phytopathogen | No | ||
| CFBP 3618T | E. rhapontici | United Kingdom | Rheum rhabarbarum | Environmental | No | 27 | |
| CFBP 3622T | E. persicina | Japan | Lycopersicon esculentum | Environmental | Yes | 21 | |
| LMG 2613T | E. billingiae | United Kingdom | Pyrus communis | Environmental | Yes | 35 | |
| Pantoea | |||||||
| ATCC 27981 | P. agglomerans | United States | Human | Clinical | CDC 865-74 | Yes | |
| ATCC 27985 | P. agglomerans | United States | Human | Clinical | CDC 811-75 | Yes | |
| ATCC 27987 | P. agglomerans | United States | Human | Clinical | CDC 1400-74 | Yes | |
| ATCC 27988 | P. agglomerans | United States | Human | Clinical | CDC 1377-71 | Yes | |
| ATCC 27989 | P. agglomerans | United States | Human | Clinical | CDC 1300-74 | Yes | |
| ATCC 27990 | P. agglomerans | United States | Human | Clinical | CDC 1134-72 | Yes | |
| ATCC 27991 | P. agglomerans | United States | Human | Clinical | CDC 358-74 | Yes | |
| ATCC 27992 | P. agglomerans | United States | Human | Clinical | CDC 1269-74 | Yes | |
| ATCC 27993 | P. agglomerans | United States | Human | Clinical | CDC 4-74 | Yes | |
| ATCC 27994 | P. agglomerans | United States | Human | Clinical | CDC 3072-74 | Yes | |
| ATCC 27995 | P. agglomerans | United States | Human | Clinical | CDC 4854-73 | Yes | |
| ATCC 27996 | P. agglomerans | United States | Insect | Environmental | CDC 5474-71 | No | |
| ATCC 27998 | P. agglomerans | United States | Human | Clinical | CDC 1741-71 | Yes | |
| ATCC 29001 | P. agglomerans | United States | Human | Clinical | CDC 164-75 | Yes | |
| ATCC 29918 | P. agglomerans | United States | Human | Clinical | CDC 4388-71 | Yes | |
| C9-1 | P. agglomerans | United States | Apple tree | Biocontrol | Wild-type | Yes | 22 |
| C9-1S | P. agglomerans | United States | Apple tree | Biocontrol | BlightBan C9-1 (Smr) | No | 22 |
| C9-1W | P. agglomerans | United States | Apple tree | Biocontrol | ΔpPag3 variant of C9-1 | No | 49 |
| CECT 4842 | P. agglomerans | Sweden | Industrial | Environmental | Yes | ||
| CIP 102343 | P. agglomerans | Algeria | Human | Clinical | Yes | ||
| CIP 102701 | P. agglomerans | France | Human | Clinical | Yes | ||
| CIP 82.100 | P. agglomerans | Canada | Cereal plant | Environmental | ATCC 33243, type strain of former Erwinia herbicola | No | 27 |
| CIP A181 | P. agglomerans | France | Human | Clinical | Yes | ||
| CPA-2 | P. agglomerans | Spain | Apple tree | Biocontrol | No | 37 | |
| E325 | P. agglomerans | United States | Apple tree | Biocontrol | BloomTime | No | 44 |
| Eh1087 | P. agglomerans | New Zealand | Apple tree | Biocontrol | Produces phenazine | No | 25 |
| Eh239 | P. agglomerans | United States | Barley plant | Biocontrol | No | 9 | |
| Eh252 | P. agglomerans | United States | Malus pumila | Biocontrol | Yes | 54 | |
| Eh318 | P. agglomerans | United States | Apple tree | Biocontrol | No | 59 | |
| Eh454 | P. agglomerans | United States | Barley plant | Biocontrol | No | 9 | |
| Eh460 | P. agglomerans | United States | Barley plant | Biocontrol | No | 9 | |
| EM13cb | P. agglomerans | Spain | Human | Clinical | Yes | ||
| EM15cb | P. agglomerans | Spain | Human | Clinical | Yes | ||
| EM17cb | P. agglomerans | Spain | Human | Clinical | Yes | ||
| EM21cb | P. agglomerans | Spain | Human | Clinical | Yes | ||
| EM22cb | P. agglomerans | Spain | Human | Clinical | Yes | ||
| EM2cb | P. agglomerans | Spain | Human | Clinical | No | ||
| EPS486 | P. agglomerans | Spain | Apple tree | Environmental | EM486 | Yes | |
| EPS595 | P. agglomerans | Spain | Pyrus communis | Environmental | EM595 | No | |
| EPS125 | P. agglomerans | Spain | Pyrus communis | Biocontrol | No | 3 | |
| LMG 1286T | P. agglomerans | Zimbabwe | Human | Clinical | Type strain: ATCC 27155T, CECT 850T | Yes | 20 |
| LMG 2557 | P. agglomerans | United Kingdom | Pyrus communis | Environmental | NCPPB 1269 | No | |
| LMG 2595 | P. agglomerans | South Africa | Allium cepa | Environmental | No | ||
| LMG 2941 | P. agglomerans | N/A | Malus sylvestris | Environmental | No | ||
| P10c | P. agglomerans | New Zealand | Apple tree | Biocontrol | BlossomBless | No | 53 |
| SC-1 | P. agglomerans | United States | Cotton plant | Phytopathogen | No | 34 | |
| VA21971 | P. agglomerans | Switzerland | Human | Clinical | Yes | ||
| ATCC 43348 | P. agglomerans pv. gypsophilae | United States | Gypsophila paniculata | Phytopathogen | Crown gall disease | No | 13 |
| LMG 20103 | P. ananatis | South Africa | Eucalyptus tree | Phytopathogen | No | ||
| LMG 2665T | P. ananatis | Brazil | Ananas comosus | Phytopathogen | Yes | 36 | |
| LMG 2676 | P. ananatis | United States | Puccinia graminis f. sp. tritici | Phytopathogen | No | 43 | |
| LMG 5342 | P. ananatis | United States | Human | Clinical | P. ananatis ATCC 29920 | Yes | 36 |
| LMG 2603T | P. dispersa | Japan | Soil | Environmental | No | 20 | |
| LMG 2605 | P. dispersa | Tanzania | Vigna unguiculata | Environmental | Yes | ||
| LMG 2770 | P. dispersa | United States | Human | Clinical | No | ||
| CFBP 3614T | P. stewartii subsp. indologenes | India | Setaria italica | Phytopathogen | LMG 2632 | Yes | 36 |
| CFBP 3517T | P. stewartii subsp. stewartii | United States | Zea mays var. rugosa | Phytopathogen | LMG 2715 | Yes | 36 |
| LMG 24195 | P. vagans | Uruguay | Eucalyptus tree | Phytopathogen | BCC 013 | 7 | |
| LMG 24196 | P. vagans | Argentina | Eucalyptus tree | Phytopathogen | BCC 015 | Yes | 7 |
| LMG 24199T | P. vagans | Uganda | Eucalyptus tree | Phytopathogen | BCC 105T | 7 | |
| LMG 5343 | Pantoea sp. | United States | Human | Clinical | P. agglomerans ATCC 29921 | Yes | |
| Tatumella | |||||||
| LMG 23359 | T. citrea | Philippines | Pineapple plant | Environmental | Yes | 8 | |
| LMG 22097 | T. punctata | Japan | Orange tree | Environmental | Yes | 8 |
T, type strain.
N/A, information not available.
Indicates whether or not Phoenix analysis has been performed for this strain. Parentheses indicate failure to grow using the standard Phoenix protocol.
gyrB sequence analysis.
Amplification and sequencing of the gyrB gene were performed as described previously (45) by means of the HotStarTaq master mix kit (Qiagen, Basel, Switzerland) and the ABI PRISM BigDye Terminators version 1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA), respectively. Two degenerate primers were used to amplify and sequence a 970-bp region of the gyrB gene, gyr-320 (5′-TAARTTYGAYGAYAACTCYTAYAAAGT-3′) and rgyr-1260 (5′-CMCCYTCCACCARGTAMAGTTC-3′) (15). The phylogenetic tree was generated on the basis of a 740-bp fragment of the gyrB amplicon. DNA sequences were aligned with ClustalW (52). Sites presenting alignment gaps were excluded from analysis. The Molecular Evolutionary Genetics Analysis (MEGA) program, version 4.0 (50), was used to calculate evolutionary distances and to infer a tree based on the neighbor-joining (NJ) method using the maximum composite likelihood (MCL) model. Nodal robustness of the inferred tree was assessed by 1,000 bootstrap replicates. GenBank accession numbers for sequences used in this work were GU225728 and GU225729, FJ617346 to FJ617453, FJ617355 to FJ617459, FJ617361 to FJ617483, FJ617385 to FJ617486, FJ617389 to FJ617491, FJ617393 to FJ617496, FJ617398 to FJ617402, FJ617404 to FJ617405, FJ617404 to FJ617405, FJ617408 to FJ617413, FJ617416 to FJ617419, FJ617422, FJ617424, FJ617425 to FJ617427, EF988757 to EF988758, EF988768, and EU145275.
Biochemical identification.
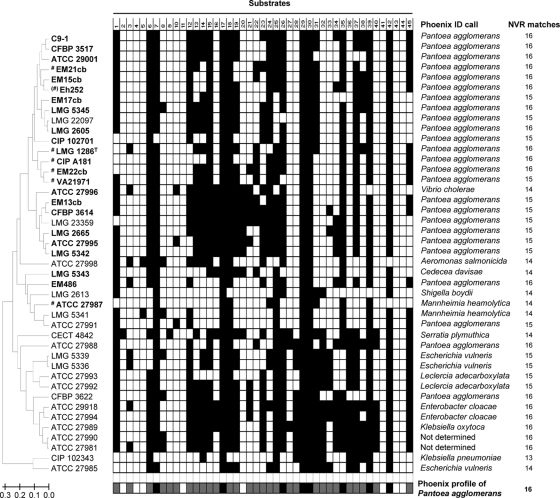
Automated biochemical identification of strains was performed using the Phoenix 100 ID/AST system (V5.66A) with NMC/ID-51 panels and the EpiCenter (V5.66A/V4.61A) microbiology data management system (BD Biosciences, Sparks, MD), following manufacturers' protocols. Each panel contained 45 substrates (Fig. 2) and included two fluorescent positive-control wells. Identification was determined using the Phoenix software by comparing the patterns of positive and negative reactions of individual samples with those of species contained in the commercial database. The current Phoenix database contains 90 genera, 324 species, and five CDC enteric groups.
FIG. 2.
Phoenix 100 ID/AST system (BD Diagnostic Systems) results for a subset of 43 strains chosen among those considered in the study. A UPGMA dendrogram (on the left) was generated based on the binary biochemical data (shown in the middle) of the selected strains. Positive reactions are indicated by a black square, whereas an empty square means that for that strain the reaction was negative after 48 h incubation. Names of strains belonging to the Pantoea genus according to gyrB sequencing are in boldface, while those belonging to P. agglomerans are preceded by a number sign (#). The projected Phoenix profile for P. agglomerans is shown below; a grey square indicates that either a positive or a negative result can be expected for that reaction. Identification provided by the Phoenix 100 ID/AST system is shown on the right side, followed by the number of nonvariable reactions (NVR, Nmax=16) matching the expected biochemical profile of P. agglomerans. Substrates evaluated were as follows: 1, arginine-arginine-AMC (AMC is 7-amido-4-methylcoumarin); 2, glycine-proline-AMC; 3, glycine-AMC; 4, glutaryl-glycine-arginine-AMC; 5, l-arginine; 6, l-glutamic acid-AMC; 7, l-leucine-AMC; 8, l-phenylalanine-AMC; 9, l-proline-AMC; 10, l-pyroglutamic acid-AMC; 11, l-tryptophan-AMC; 12, lysine-alanine-AMC; 13, acetate; 14, adonitol; 15, citrate; 16, colistin; 17, d-mannitol; 18, α-ketoglutaric acid; 19, malonate; 20, polymyxin B; 21, tiglic acid; 22, Mu-N-acetyl-β-d-glucosaminide; 23, gamma-l-glutamyl-Na; 24, l-proline-Na; 25, p-nitrophenol-β-d-glucoside; 26, bis(p-nitrophenol) phosphate; 27, β-allose; 28, β-gentiobiose; 29, dextrose; 30, d-fructose; 31, d-galactose; 32, d-gluconic acid; 33, d-mellibiose; 34, sorbitol; 35, sucrose; 36, galacturonic acid; 37, l-arabinose; 38, l-rhamnose; 39, methyl-β-glucoside; 40, maltulose; 41, N-acetylgalactosamine; 42, N-acetylglucosamine; 43, ornithine; 44, urea; and 45, esculin.
MALDI-TOF MS spectrum acquisition.
Cells from a single bacterial colony grown on LB agar for 24 h were transferred to a target spot of a steel target plate using a disposable loop, overlaid with 0.5 μl of a 2,5-dihydroxybenzoic acid (DHB) matrix (AnagnosTec GmbH, Potsdam, Germany), and air dehydrated within 1 to 2 min at 24 to 27°C. Protein mass fingerprints were obtained using a MALDI-TOF mass spectrometry Axima confidence machine (Shimadzu-Biotech Corp., Kyoto, Japan), with detection in the linear, positive mode at a laser frequency of 50 Hz and within a mass range of 2,000 to 20,000 Da. Acceleration voltage was 20 kV, and the extraction delay time was 200 ns. A minimum of 20 laser shots per sample was used to generate each ion spectrum. For each bacterial sample, a total of 50 protein mass fingerprints were averaged and processed using the Launchpad version 2.8 software (Shimadzu-Biotech Corp.). For peak acquisition, the average smoothing method was chosen, with a smoothing filtering width of 50 channels. Peak detection was performed with the threshold-apex peak detection method using the adaptive voltage threshold which roughly follows the signal noise level, and subtraction of the baseline was set with a baseline subtraction filter width of 500 channels. For each sample, a list of the significant spectrum peaks was generated that included the m/z values for each peak, mass deviations, and signal intensity. Calibration was conducted for each target plate using spectra of the reference strain Escherichia coli K-12 (GM48 genotype). E. coli K-12 was deposited on each plate in two fixed positions, and the calibration was performed at the beginning of each plate acquisition. At the end, a second measurement of K-12 as a control was performed.
MALDI-TOF MS spectrum analysis and SuperSpectrum generation.
Generated protein mass fingerprints were first imported in SARAMIS and analyzed using the following presetting parameters: mass range, from 2,000 to 20,000 Da; allowed mass deviation, 800 ppm. The spectra were related through cluster analysis by applying the single-link agglomerative algorithm of SARAMIS. Distance trees were compared to the neighbor-joining phylogenetic gyrB tree. As reference spectra for a rapid identification, SARAMIS uses so-called “SuperSpectra” consisting of taxon-specific biomarkers. SuperSpectrum generation was based on recovered mass signal markers with an absolute intensity of at least 200 mV included in the 2,000- to 20,000-Da mass range. To create the SuperSpectrum of a species, only the protein mass fingerprints of the strains clustering with the respective type strain in the gyrB tree were used (with species clustering indicated by the associated bracket). Using the SuperSpectrum tool, a subset of protein masses found in at least 90% of the strains of one species were selected and tested for their discriminatory power by comparing them to all of the database entries. Dependent on the amount of remaining species-identifying marker masses, each was given a numeric value in order to get a maximum total number of points not higher than 1,250. A set of 20 to 40 marker masses is normally sufficient to obtain a specific identification to the species level. The identification results obtained using MALDI-TOF MS were compared to those obtained using DNA sequencing combined with a BLAST similarity search and the outcome of the biochemical analysis using the Phoenix 100 ID/AST system.
Nucleotide sequence accession numbers.
Sequences newly obtained as a result of this study were deposited in GenBank under accession numbers GU225728 and GU225729.
RESULTS AND DISCUSSION
Only 151 of 263 (i.e., 57%) 16S rRNA sequences retrieved from NCBI listed as belonging to P. agglomerans isolates clustered indeed with type strain P. agglomerans LMG 1286T. The remaining ones could be assigned either to other Pantoea spp. (29 sequences), to the genus Erwinia (20 sequences), or to other taxa of the Enterobacteriaceae (62 sequences) or did not cluster with any of the chosen reference species or genera and did not produce significant matches with reliable 16S rRNA sequences at NCBI (i.e., blastn ≤ 97%) (Fig. 1). These results underscore the inadequacy of current biochemical and molecular identification methods employed in clinical diagnostics for P. agglomerans and the common use of obsolete taxonomy. Two ways incorrect sequences may appear in GenBank are blanket relocation to P. agglomerans of some species within the “P. agglomerans-E. herbicola-E. agglomerans” complex or a posteriori sequencing of isolates biochemically misidentified as P. agglomerans. The presence of such rogue data in the GenBank 16S rRNA gene database constitutes a potential pitfall for anyone trying to identify P. agglomerans only on the basis of 16S rRNA gene sequences.
Automated biochemical identification using the Phoenix 100 ID/AST system was less accurate than gyrB sequencing or MALDI-TOF MS and returned uncertain strain identification within P. agglomerans and related species. Of the 23 Pantoea strains analyzed, 19 were identified biochemically as P. agglomerans (Fig. 2), although only six of them could accurately be assigned to P. agglomerans using gyrB sequence analysis (Fig. 3). The Phoenix 100 ID/AST system was unable to separate the different species within the genus, as none of the other recognized Pantoea species is currently present in its database. Conversely, five strains belonging to other genera were incorrectly assigned to P. agglomerans using biochemical analysis: Erwinia persicina LMG 3622, Enterobacter sp. ATCC 27988 and ATCC 27991, Tatumella punctata LMG 22097, and Tatumella citrea LMG 23359, the latter two previously belonging to the former “Japanese species” of Pantoea (4, 8). For the first three strains this outcome is the consequence of imprecise biochemical profiling, as 29 out of 45 reactions within the NMC/ID-51 panel are allowed to deliver a variable result while still retaining the identification as P. agglomerans (Fig. 2). This is most evident in Enterobacter sp. ATCC 27988, where all 16 nonvariable reactions correspond to the Phoenix profile of P. agglomerans but where 14 out of the 29 reactions which allow a variable result are different between this strain and P. agglomerans LMG 1286T. On the other hand, Tatumella strains LMG 22097 and LMG 23359 have biochemical profiles which are closely related to those of Pantoea and only recently have DNA-DNA hybridization and phenotypic tests allowed the transfer of these species to the genus Tatumella (8). One strain belonging to P. agglomerans (ATCC 27987) was incorrectly assigned biochemically to Mannheimia haemolytica, while P. ananatis ATCC 27996 and Pantoea sp. LMG 5343 were misidentified as Vibrio cholerae and Cedecea davisae, respectively. For all three strains the number of nonvariable reactions matching the Phoenix profile of P. agglomerans fell short of 15, which is apparently among the minimum prerequisites for a positive identification (Fig. 2). Taken together, these results suggest that the taxonomical confusion within the former “P. agglomerans-E. herbicola-E. agglomerans” complex (19) contributed at least in part to generate imprecise biochemical profiles for the identification of P. agglomerans.
FIG. 3.
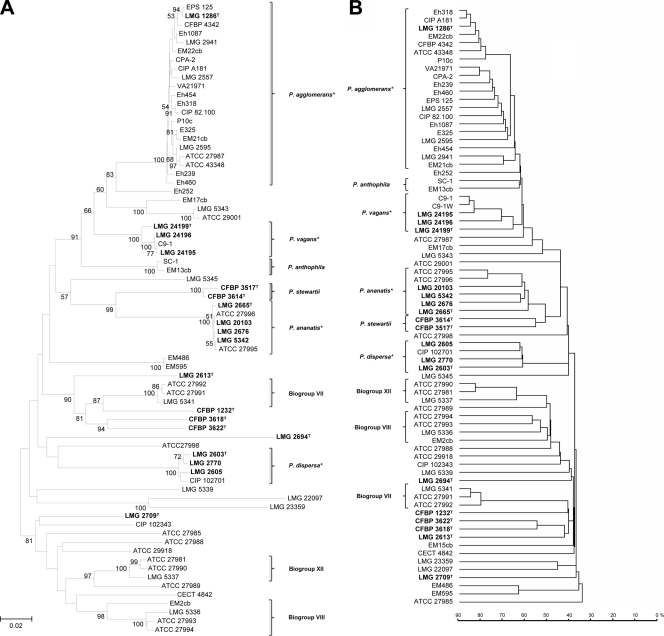
Comparison between dendrograms derived from gyrB sequencing (A) and MALDI-TOF MS protein mass fingerprints (B). With the exception of the reference strains marked in bold, all other isolates were received as P. agglomerans, a Pantoea sp., or E. agglomerans from culture collections. The gyrB phylogenetic tree was generated on the basis of a 740-bp fragment of the gyrB amplicon using 1,000 bootstrap replicates of the neighbor-joining (NJ) method with the maximum composite likelihood (MCL) model without choosing any outgroup. Sites presenting alignment gaps were excluded from analysis. The MALDI-TOF MS dendrogram was constructed based on the protein mass fingerprint patterns of all analyzed strains using the single-link clustering algorithm implemented in SARAMIS. This algorithm is proprietary to the database provider and does not provide the option to perform bootstrap analysis. Correct clustering of each strain was confirmed by running at least four replicates per isolate, but, for clarity and conciseness, graphics were reduced to show a single representative spectrum per strain. The main clusters obtained in both phylogenetic trees were highly similar; thus, the two methods were comparable in discriminating P. agglomerans from other species and in recognizing previously misidentified strains. The diversity within the single species is apparently exaggerated by MALDI-TOF (deeper strain branches) and is not reflecting the actual phylogenetic distances. *, for P. agglomerans, P. vagans, P. ananatis, and P. dispersa, brackets indicate the isolates clustering with the respective type strain, which were used to create the SuperSpectrum of the corresponding species.
As expected, phylogenetic analysis based on gyrB sequences (Fig. 3) provided greater discriminatory power than either biochemical or 16S rRNA gene sequencing to describe the Pantoea group (45). Confirming the uncertainty of P. agglomerans identification, only 20 of the 53 strains received from culture collections as P. agglomerans, Pantoea spp., or E. agglomerans clustered with type strain LMG 1286T according to gyrB sequencing. Seven strains previously assigned to P. agglomerans were found to fit in the MLSA groups of Pantoea recently described as new species (i.e., C9-1 as P. vagans; EM13cb and SC-1 as P. anthophila; LMG 5343 and ATCC 29001 as P. brenneri; and EM17cb as P. conspicua), or to belong to a novel subspecies of P. agglomerans (Eh252) (4, 6, 7, 45). The remaining strains were reassigned to other Pantoea species or Enterobacteriaceae, although precise identification was not possible in all cases.
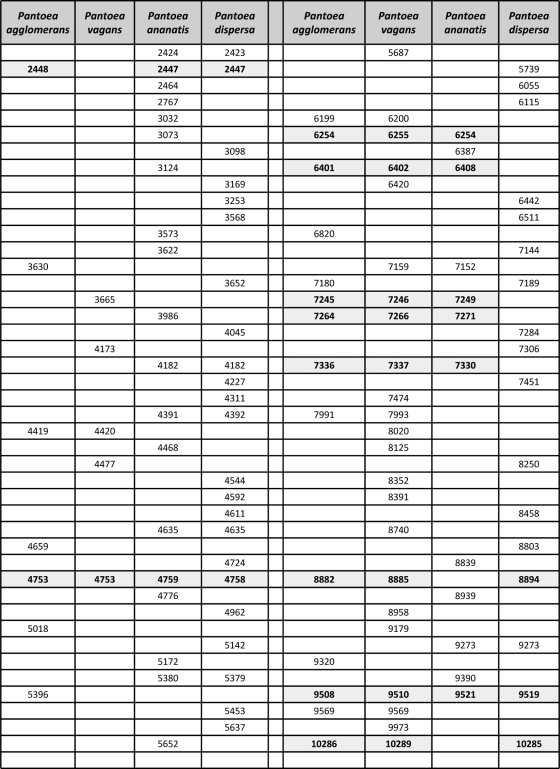
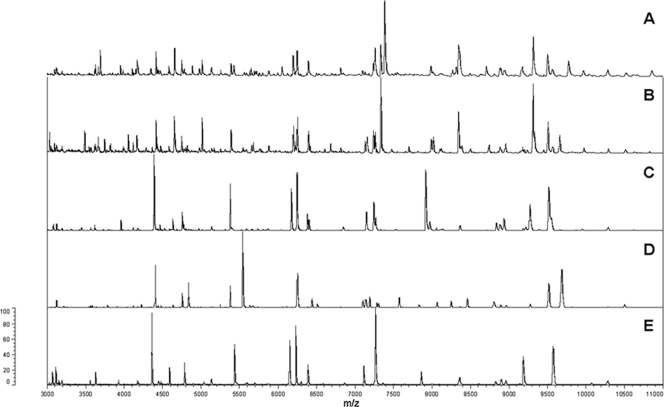
MALDI-TOF MS delivered results which were almost equivalent to those of gyrB sequencing in terms of species grouping (Fig. 3). Only a single strain (ATCC 27987) was found to be intermediate, being offset from the P. agglomerans (sensu stricto) group when a mass range of 2 to 20 kDa was used (Fig. 3) but not when the lower limit of the mass range was raised to 3 kDa. ATCC 27987 was confirmed as P. agglomerans (sensu stricto) using fAFLP, although it must be noted that it was the only isolate genetically assigned to P. agglomerans for which the biochemical signature obtained with the Phoenix 100 ID/AST system was noticeably dissimilar from those of the other strains of the species, leading not only to a wrong automated identification based on the number of correct nonvariable reaction but also to an apparently incorrect clustering based on the overall reaction pattern (Fig. 2). The protein mass fingerprints of the 21 P. agglomerans (sensu stricto) strains and strains of P. ananatis, P. dispersa, and P. vagans were used to generate a SuperSpectrum with identifying mass peaks for each species (Fig. 4). A typical MALDI-TOF MS spectrum of P. agglomerans contained about 150 ion peaks between 2,000 and 20,000 Da, with the highest intensity peaks found between 4,000 and 11,000 Da. Comparison of these protein mass fingerprints defined a set of 21 markers present in at least 90% of all protein mass fingerprints for confident identification of P. agglomerans. While a subset of masses were shared by different Pantoea species, a combination of discriminatory signals provided a unique species-level signature (Fig. 4). MALDI-TOF MS spectra of strains having an identity that could not be genetically confirmed as Pantoea (e.g., LMG 5339) showed widely divergent profiles compared to strains confidently assigned to this genus (Fig. 5). These divergent signal patterns were reflected in both the MALDI-TOF MS and gyrB trees, with the related strains clustering well outside the genus Pantoea (Fig. 3). On the basis of 16S rRNA gene sequencing, strain LMG 5339 showed 99.5% identity to Buttiauxella agrestis DSM 4586 (45).
FIG. 4.
Characteristic masses (in daltons) recognized as markers for the identification of P. agglomerans, P. vagans, P. ananatis, and P. dispersa. Two or more masses are considered equivalent and are shown on the same line if their values deviate by less than 800 ppm from the average. The 800-ppm mass accuracy setting defining the spectral bin size was evaluated/validated by AnagnosTec GmbH. Even if the error margin in a data set analyzed on a single MALDI-TOF MS instrument is usually smaller (around 400 ppm), a larger bin size is necessary to allow, within the SARAMIS database, the comparison of data collected with different instruments in different laboratories. An 800-ppm mass accuracy still allows one to distinguish mass differences of 14 Da for proteins of 10 kDa, which is normally sufficient to detect single-amino-acid exchanges in proteins of that size. Masses shared by three or more Pantoea species are marked in bold.
FIG. 5.
MALDI-TOF MS protein mass fingerprints of representative strains belonging to different Pantoea species: (A) P. agglomerans LMG 1286T, (B) P. vagans LMG 24199T, (C) P. ananatis LMG 2676, and (D) P. dispersa LMG 2603T. Similar and diverging marker masses for the identification of these Pantoea species are listed in Fig. 4. For comparison, the spectrum of strain LMG 5339 received from the BCCM/LMG collection under the incorrect name of P. agglomerans is shown at the bottom (E). Both gyrB sequencing and MALDI-TOF MS clearly demonstrate the misnaming of this strain. Relative intensities of ions (percentages) are shown on the y axis and the masses (in daltons) of the ions are shown on the x axis. The m/z values represent mass-to-charge ratios.
MALDI-TOF MS analysis was able to discriminate strains within Pantoea and to segregate related strains into separate species/clades with the same level of accuracy as gyrB sequencing and more sensitively than either biochemical or 16S rRNA gene sequencing approaches. Clustering between strains within a group was not identical in the two methods, but these fluctuations at the subgroup level are inconsequential as long as the aim is to ensure that strains remain within a species. Misidentified strains previously grouped into P. agglomerans were accurately assigned to P. ananatis, P. dispersa, or the recently defined species P. vagans or to discrete clades following Brenner's biogroups (10). For example, strains could be assigned to biogroup VII (LMG 5336, ATCC 27993, ATCC 27994, and EM2cb), biogroup VIII (LMG 5341, ATCC 27991, and ATCC 27992), and biogroup XII (LMG 5337, ATCC 27981, and ATCC 27990) using both MALDI-TOF MS and gyrB sequencing (Fig. 3). Furthermore, MALDI-TOF MS was in agreement with gyrB sequencing regarding strains clustering together as P. stewartii (CFBP 3517 and CFPB3614), P. anthophila (SC-1 and EM13cb) or a probable novel Pantoea species (EM486 and EM595) (45).
Our standardized MALDI-TOF MS protocol analysis using strains grown on LB agar proved to deliver highly repeatable results, with only a few replicates that do not immediately cluster with the other measurements performed on the same isolate. Even so, plotting of all the replicates easily allows one to discard substandard measurements which, in this case, still cluster within the same species and hence would retain all prerequisites for successful identification at this taxonomical level (see Fig. S1 in the supplemental material). The use of alternative media such as TSA or MHA shows that the alteration of the growth parameters does have a certain influence on the composition of the mass spectra obtained. This is reflected in the merging of nearby strain measurements in the dendrogram and leads to a loss of resolution that does not allow the unambiguous recognition of each single strain anymore. However, all measurements within the same species are still kept in a tight cluster, thereby preserving the conditions for unequivocal identification at species level (see Fig. S2 in the supplemental material).
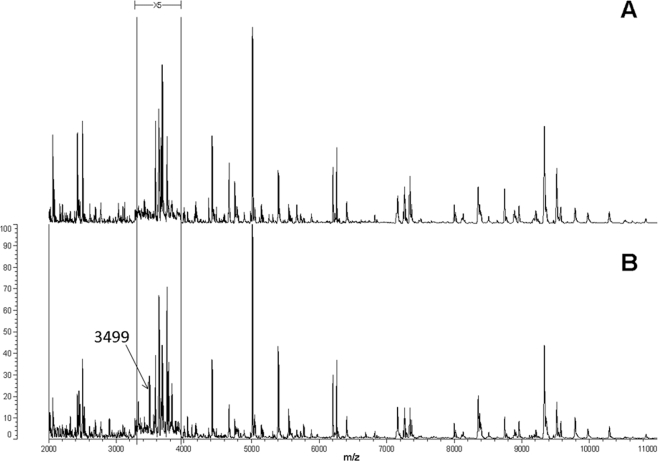
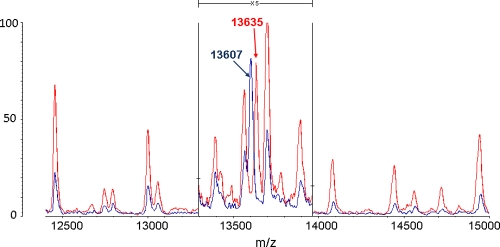
Identification using MALDI-TOF MS not only provided robust recognition but was also able to detect protein profile differences within P. vagans C9-1 (4.88-Mb genome) resulting from a large genomic alteration such as curing of the 530-kb megaplasmid pPag3 (pigmentless variant C9-1W, 4.35-Mb genome) (49), as shown for instance for the loss of the 3,499-Da mass signal (Fig. 6). Other masses missing from the plasmid-cured variant C9-1W were at 2,133, 2,140, 2,309, 2,396, 3,197, 3,552, 3,570, 4,295, and 4,850 Da, while no peak was found exclusively in the wild type. Since distinctive marker signals for P. vagans (Fig. 4) were unaffected by the loss of the plasmid and the number of missing masses was relatively low, neither the species-level assignment of this variant nor its position in the MALDI-TOF MS dendrogram was altered with respect to the wild type (Fig. 3). This can be explained by the fact that the main signals in MALDI-TOF MS were reported to be ribosomal proteins (47), and thus only a fraction of masses disappears from the profile following the loss of pPag3. Indeed, a number of characteristic masses recognized as markers for the identification of the considered Pantoea species are compatible with the predicted masses of Pantoea sp. At-9b ribosomal proteins deposited in the UniProt database (http://www.uniprot.org/) within the allowed mass deviation of 800 ppm (Table 2). Acquisition of the 135-kb virulence plasmid pPATH (32) also did not confound accurate identification of the phytopathogenic strain ATCC 43348 as P. agglomerans using MALDI-TOF MS. Moreover, replacement of the DHB matrix with sinapinic acid allowed us to identify the K43R (lysine to arginine) point mutation in 30S ribosomal protein S12 (12) that confers streptomycin resistance in the commercial biocontrol strain P. vagans C9-1S (Fig. 7).
FIG. 6.
Comparison of MALDI-TOF MS protein mass fingerprints of pigmentless P. vagans variant C9-1W (A) lacking the 530-kb plasmid pPag3 and the related wild-type strain P. vagans C9-1 (B). The masses (in daltons) of the ions are shown on the x axis. The m/z values represent mass-to-charge ratios. Relative intensities of ions (percentages) on the y axis are 5-fold exaggerated between 3,300 and 3,950 Da to highlight the disappearance of the 3,499-Da mass signal in C9-1W. Other masses missing from the plasmidless variant C9-1W were at 2,133, 2,140, 2,309, 2,396, 3,197, 3,552, 3,570, 4,295, and 4,850 Da, but most of the signals have low intensities and cannot easily be visualized in the protein profile. Marker masses for the identification of P. vagans are listed in Fig. 4.
TABLE 2.
Compatibility of Pantoea spp. markers with the predicted masses of ribosomal Pantoea sp. At-9b proteinsa
| Measured masses (Da) |
Pantoea sp. At-9b |
||||||
|---|---|---|---|---|---|---|---|
| P. agglomerans | P. vagans | P. ananatis | P. dispersa | Ribosomal protein | Accession no.b | Predicted massc | Modificationd |
| 4,391 | 4,392 | 50S L36 | C8QHD9 | 4,392 | |||
| 4,419 | 4,420 | 50S L36 | C8QHD9 | 4,392 | +acetyl | ||
| 5,396 | 50S L34 | C8Q9X2 | 5,396 | ||||
| 5,453 | 50S L36 | C8Q3U8 | 5,585 | −met | |||
| 6,387 | 50S L33 | C8QC26 | 6,372 | +CH3 | |||
| 6,387 | L32 | C8Q6B9 | 6,362 | +acetyl | |||
| 6,442 | L30 | C8QHE2 | 6,574 | −met | |||
| 7,245 | 7,246 | 7,249 | L29 | C8QHF2 | 7,219 | +acetyl | |
| 7,264 | 7,266 | 7,271 | 50S L35 | C8Q6T8 | 7,269 | ||
| 7,284 | 50S L35 | C8Q6T8 | 7,269 | +CH3 | |||
| 7,991 | 7,993 | 50S L31 | C8QBY0 | 7,957 | +acetyl | ||
| 8,882 | 8,885 | 8,894 | L28 | C8QC27 | 9,006 | −met | |
| 8,958 | L27 | C8QDH1 | 9,097 | −met | |||
| 9,179 | S16 | C8QFQ0 | 9,174 | ||||
| 10,286 | 10,289 | 10,285 | S19 | C8QHF6 | 10,416 | −met | |
Compatibility of the markers for the identification of Pantoea spp. (see Fig. 4) with the predicted masses of ribosomal proteins of Pantoea sp. At-9b deposited in the UniProt database (http://www.uniprot.org/) within the allowed mass deviation of 800 pm. Three single, common posttranslational modifications (N-terminal methionine loss, methylation, and acetylation) were considered to estimate the possible mass variations starting from the predicted mass of the protein in Pantoea sp. At-9b.
UniProt accession code.
Predicted mass without posttranslational modifications.
Modifications: −met = N-terminal methionine removal, −131 Da; +CH3= methylation, +14 Da; +acetyl = acetylation, +30 Da.
FIG. 7.
MALDI-TOF MS identification of the spontaneous lysine-to-arginine mutation in 30S ribosomal protein S12 (encoded by the rpsL gene) leading to streptomycin resistance in commercial biocontrol strain P. vagans C9-1S (BlightBan C9-1). The 13,607-Da mass signal found in wild-type strain C9-1 (in blue) corresponds to the predicted mass of RpsL in Pantoea sp. At-9b (molecular mass = 13,737 Da; UniProt accession code, C8QDZ1), assuming the removal of the N-terminal methionine (−131 Da) and matrix-assisted protonation (+1 Da) following the laser pulse. In streptomycin-resistant strain C9-1S (in red) the peak is shifted at 13,635 Da, whereas the positions of the other peaks remain constant. This variation (+28 Da) corresponds precisely to the difference in molecular mass between arginine (molecular mass = 174.2 Da) and lysine (molecular mass = 146.2 Da). The masses (in daltons) of the ions are shown on the x axis. The m/z values represent mass-to-charge ratios. For improved clarity, the relative intensities of ions (percentages) on the y axis are 5-fold exaggerated between 13,300 and 13,970 Da.
The primary drawback of MALDI-TOF MS for bacterial identification and diagnostics is the dearth of reference species in databases, which are limited primarily to clinical species while environmental bacteria such as Pantoea are largely absent. Unfortunately, there are at present no public repositories for diagnostic protein patterns, which are currently archived only within commercial databases such as that contained in SARAMIS. A further potential problem is the need of isolated colonies for MALDI-TOF MS analysis, a fact that may still represent a limitation for fastidious or slow-growing species (e.g., Bordetella sp., Borrelia sp., Neisseria gonorrhoeae, or Mycoplasma spp. in diagnostic laboratories). In such instances direct molecular or serological methods may still retain the advantage, although these methods often require some a priori assumption about the nature of the organism to be identified.
This study demonstrates both the accuracy and the simplicity of whole-cell MALDI-TOF MS for identification of the complex P. agglomerans group and related taxa. Strain identification using the unique protein profiles generated was achieved with minimal labor and materials and within a few minutes for MALDI-TOF MS sample preparation and analysis. This offers an attractive alternative to the relatively high investment required for single-locus validation, PCR amplification, and sequencing. Investment in a MALDI-TOF mass spectrometer is comparable to that needed for a 16-capillary DNA-sequencing machine, but it requires a fraction of the operating costs and consumables. We also demonstrated the application of MALDI-TOF MS for clustering analysis of Pantoea, almost equivalent to gyrB phylogenetic analysis. The unique ICMS fingerprints for Pantoea species developed in this study will facilitate more accurate and rapid identification of isolates from environmental and clinical samples using this technology.
Supplementary Material
Acknowledgments
We thank Viviana Rossi for support with biochemical and molecular identifications, Cinzia Benagli for MALDI-TOF MS technical support (ICM Bellinzona), and Theo Smits (ACW Wädenswil) for helpful discussion and critical review of the manuscript. We are grateful to Teresa Coutinho (FABI, University of Pretoria, South Africa) for the kind gift of the P. vagans LMG strains.
This work was supported by the Swiss Federal Office for Agriculture (BLW Fire Blight Biocontrol Project) and the Swiss Secretariat for Education and Research (SBF C06.0069). It was conducted within the framework of the European Science Foundation funded research network COST Action 873.
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barbuddhe, S. B., T. Maier, G. Schwarz, M. Kostrzewa, H. Hof, E. Domann, T. Chakraborty, and T. Hain. 2008. Rapid identification and typing of Listeria species using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5402-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billing, E., L. A. E. Baker, J. E. Crosse, and C. M. E. Garrett. 1961. Characteristics of English isolates of Erwinia amylovora (Burrill) Winsow et al. J. Appl. Microbiol. 24:195-211. [Google Scholar]
- 3.Bonaterra, A., M. Mari, L. Casalini, and E. Montesinos. 2003. Biological control of Monilinia laxa and Rhizopus stolonifer in postharvest of stone fruit by Pantoea agglomerans EPS125 and putative mechanisms of antagonism. Int. J. Food Microbiol. 84:93-104. [DOI] [PubMed] [Google Scholar]
- 4.Brady, C., I. Cleenwerck, S. N. Venter, M. Vancanneyt, J. Swings, and T. A. Coutinho. 2008. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 31:447-460. [DOI] [PubMed] [Google Scholar]
- 5.Brady, C., S. Venter, I. Cleenwerck, M. Vancanneyt, J. Swings, and T. Coutinho. 2007. A FALFP system for the improved identification of plant-pathogenic and plant-associated species of the genus Pantoea. Syst. Appl. Microbiol. 30:413-417. [DOI] [PubMed] [Google Scholar]
- 6.Brady, C. L., I. Cleenwerck, S. N. Venter, K. Engelbeen, P. de Vos, and T. A. Coutinho. 2009. Emended description of the genus Pantoea and description of four novel species from human clinical samples, Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov. and Pantoea conspicua sp. nov., and transfer of Pectobacterium cypripedii (Hori 1911) Brenner et al. 1973 emend. Hauben et al. 1998 to the genus Pantoea emend. as Pantoea cypripedii comb. nov. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] doi: 10.1099/ijs.0.017301-0. [DOI] [PubMed]
- 7.Brady, C. L., S. N. Venter, I. Cleenwerck, K. Engelbeen, M. Vancanneyt, J. Swings, and T. A. Coutinho. 2009. Pantoea vagans sp. nov., Pantoea eucalypti sp. nov., Pantoea deleyi sp. nov. and Pantoea anthophila sp. nov. Int. J. Syst. Evol. Microbiol. 59:2339-2345. [DOI] [PubMed] [Google Scholar]
- 8.Brady, C. L., S. N. Venter, I. Cleenwerck, K. Vandemeulebroecke, P. de Vos, and T. A. Coutinho. 2010. Transfer of Pantoea citrea, Pantoea punctata and Pantoea terrea to the genus Tatumella emend. as Tatumella citrea comb. nov., Tatumella punctata comb. nov. and Tatumella terrea and description of Tatumella morbirosei sp. nov. Int. J. Syst. Evol. Microbiol. 60:484-494. [DOI] [PubMed] [Google Scholar]
- 9.Braun-Kiewnick, A., B. J. Jacobsen, and D. C. Sands. 2000. Biological control of Pseudomonas syringae pv. syringae, the causative agent of basal kernel blight of barley, by antagonistic Pantoea agglomerans. Phytopathology 90:368-375. [DOI] [PubMed] [Google Scholar]
- 10.Brenner, D. J., G. R. Fanning, J. K. Leete Knutson, A. G. Steigerwalt, and M. I. Krichevsky. 1984. Attempts to classify Herbicola group-Enterobacter agglomerans strains by deoxyribonucleic acid hybridization and phenotypic tests. Int. J. Syst. Bacteriol. 34:45-55. [Google Scholar]
- 11.Carroll, K. C., B. D. Glanz, A. P. Borek, C. Burger, H. S. Bhally, S. Henciak, and D. Flayhart. 2006. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterobacteriaceae. J. Clin. Microbiol. 44:3506-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiou, C.-S., and A. L. Jones. 1995. Molecular analysis of high-level streptomycin resistance in Erwinia amylovora. Phytopathology 95:324-328. [Google Scholar]
- 13.Cooksey, D. A. 1986. Galls of Gypsophila paniculata caused by Erwinia herbicola. Plant Dis. 70:464-468. [Google Scholar]
- 14.Cruz, A. T., A. C. Cazacu, and C. H. Allen. 2007. Pantoea agglomerans, a plant pathogen causing human disease. J. Clin. Microbiol. 45:1989-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dauga, C. 2002. Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: a model molecule for molecular systematic studies. Int. J. Syst. Evol. Microbiol. 52:531-547. [DOI] [PubMed] [Google Scholar]
- 16.De Champs, C., S. Le Seaux, J. J. Dubost, S. Boisgard, B. Sauvezie, and J. Sirot. 2000. Isolation of Pantoea agglomerans in two cases of septic monoarthritis after plant thorn and wood sliver injuries. J. Clin. Microbiol. 38:460-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Respinis, S., G. Vogel, C. Benagli, M. Tonolla, O. Petrini, and G. J. Samuels. 2010. MALDI-TOF MS of Trichoderma: a model system for the identification of microfungi. Mycol. Prog. 9:79-100. [Google Scholar]
- 18.Dieckmann, R., R. Helmuth, M. Erhard, and B. Malorny. 2008. Rapid classification and identification of salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:7767-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewing, W. H., and M. A. Fife. 1972. Enterobacter agglomerans (Beijerinck) comb. nov. (the herbicola-lathyri bacteria). Int. J. Syst. Bacteriol. 22:4-11. [Google Scholar]
- 20.Gavini, F., J. Mergaert, A. Beji, C. Mielcarek, D. Izard, K. Kersters, and J. De Ley. 1989. Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int. J. Syst. Bacteriol. 39:337-345. [Google Scholar]
- 21.Hao, M. V., D. J. Brenner, A. G. Steigerwalt, Y. Kosako, and K. Kornagata. 1990. Erwinia persicinus, a new species isolated from plants. Int. J. Syst. Bacteriol. 40:379-383. [DOI] [PubMed] [Google Scholar]
- 22.Ishimaru, C. A., E. J. Klos, and R. R. Brubaker. 1988. Multiple antibiotic production by Erwinia herbicola. Phytopathology 78:746-750. [Google Scholar]
- 23.Johnson, K. B., and V. O. Stockwell. 1998. Management of fire blight: a case study in microbial ecology. Annu. Rev. Phytopathol. 36:227-248. [DOI] [PubMed] [Google Scholar]
- 24.Kämpfer, P., A. Nienhüser, G. Packroff, F. Wernicke, A. Mehling, K. Nixdorf, S. Fiedler, C. Kolauch, and M. Esser. 2008. Molecular identification of coliform bacteria isolated from drinking water reservoirs with traditional methods and the Colilert-18 system. Int. J. Hyg. Environ. Health 211:374-384. [DOI] [PubMed] [Google Scholar]
- 25.Kearns, L. P., and H. K. Mahanty. 1998. Antibiotic production by Erwinia herbicola Eh1087: its role in inhibition of Erwinia amylovora and partial characterization of antibiotic biosynthesis genes. Appl. Environ. Microbiol. 64:1837-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krader, P., and D. Emerson. 2004. Identification of archaea and some extremophilic bacteria using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Extremophiles 8:259-268. [DOI] [PubMed] [Google Scholar]
- 27.Kwon, S.-W., S.-J. Go, H.-W. Kang, J.-C. Ryu, and J.-K. Jo. 1997. Phylogenetic analysis of Erwinia species based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 47:1061-1067. [DOI] [PubMed] [Google Scholar]
- 28.Lay, J. O. 2001. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20:172-194. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, J. K., M. Bendahmane, T. J. Smith, R. N. Beachy, and G. Siuzdak. 1998. Identification of viral mutants by mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 95:8596-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lind, E., and J. Ursing. 1986. Clinical strains of Enterobacter agglomerans (synonyms: Erwinia herbicola, Erwinia milletiae) identified by DNA-DNA-hybridization. Acta Pathol. Microbiol. Immunol. Scand. B 94:205-213. [DOI] [PubMed] [Google Scholar]
- 31.Maier, T., S. Klepel, U. Renner, and M. Kostrzewa. 2006. Fast and reliable MALDI-TOF MS-based microorganism identification. Nat. Methods 3. doi: 10.1038/nmeth870. [DOI]
- 32.Manulis, S., and I. Barash. 2003. Pantoea agglomerans pvs. gypsophilae and betae, recently evolved pathogens? Mol. Plant Pathol. 4:307-314. [DOI] [PubMed] [Google Scholar]
- 33.Marklein, G., M. Josten, U. Klanke, E. Müller, R. Horré, T. Maier, T. Wenzel, M. Kostrzewa, G. Bierbaum, A. Hoerauf, and H.-G. Sahl. 2009. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medrano, E. G., and A. A. Bell. 2007. Role of Pantoea agglomerans in opportunistic bacterial seed and boll rot of cotton (Gossypium hirsutum) grown in the field. J. Appl. Microbiol. 102:134-143. [DOI] [PubMed] [Google Scholar]
- 35.Mergaert, J., L. Hauben, M. C. Cnockaert, and J. Swings. 1999. Reclassification of non-pigmented Erwinia herbicola strains from trees as Erwinia billingiae sp. nov. Int. J. Syst. Bacteriol. 49:377-383. [DOI] [PubMed] [Google Scholar]
- 36.Mergaert, J., L. Verdonck, and K. Kersters. 1993. Transfer of Erwinia ananas (synonym, Erwinia uredovora) and Erwinia stewartii to the genus Pantoea emend. as Pantoea ananas (Serrano 1928) comb. nov. and Pantoea stewartii (Smith 1898) comb. nov., respectively, and description of Pantoea stewartii subsp. indologenes subsp. nov. Int. J. Syst. Bacteriol. 43:162-173. [Google Scholar]
- 37.Nunes, C., J. Usall, N. Teixidó, E. Fons, and I. Viñas. 2002. Post-harvest biological control by Pantoea agglomerans (CPA-2) on Golden Delicious apples. J. Appl. Microbiol. 92:247-255. [DOI] [PubMed] [Google Scholar]
- 38.O'Hara, C. M. 2006. Evaluation of the Phoenix 100 ID/AST system and NID panel for identification of Enterobacteriaceae, Vibrionaceae, and commonly isolated nonenteric gram-negative bacilli. J. Clin. Microbiol. 44:928-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Hara, C. M., F. C. Tenover, and J. M. Miller. 1993. Parallel comparison of accuracy of API 20E, Vitek GNI, MicroScan Walk/Away Rapid ID, and Becton Dickinson Cobas Micro ID-E/NF for identification of members of the family Enterobacteriaceae and common gram-negative, non-glucose-fermenting bacilli. J. Clin. Microbiol. 31:3165-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Hara, C. M., G. L. Westbrook, and J. M. Miller. 1997. Evaluation of Vitek GNI+ and Becton Dickinson Microbiology systems Crystal E/NF identification systems for identification of members of the family Enterobacteriaceae and other gram-negative, glucose-fermenting and non-glucose-fermenting bacilli. J. Clin. Microbiol. 35:3269-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perera, M. R., R. D. Flores Vargas, and M. G. K. Jones. 2005. Identification of aphid species using protein profiling and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Entomol. Exp. Appl. 117:243-247. [Google Scholar]
- 42.Perera, M. R., V. A. Vanstone, and M. G. K. Jones. 2005. A novel approach to identify plant parasitic nematodes using MALDI-TOF mass spectrometry. Rapid Commun. Mass Spectrom. 19:1454-1460. [DOI] [PubMed] [Google Scholar]
- 43.Pon, D. S., C. E. Townsend, G. E. Wessman, C. G. Schmitt, and C. H. Kingsolver. 1954. A Xanthomonas parasitic on uredia of cereal rusts. Phytopathology 44:707-710. [Google Scholar]
- 44.Pusey, P. L., V. O. Stockwell, and D. R. Rudell. 2008. Antibiosis and acidification by Pantoea agglomerans strain E325 may contribute to suppression of Erwinia amylovora. Phytopathology 98:1136-1143. [DOI] [PubMed] [Google Scholar]
- 45.Rezzonico, F., T. H. M. Smits, E. Montesinos, J. E. Frey, and B. Duffy. 2009. Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruelle, V., B. El Moualij, W. Zorzi, P. Ledent, and E. De Pauw. 2004. Rapid identification of environmental bacterial strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 18:2013-2019. [DOI] [PubMed] [Google Scholar]
- 47.Ryzhov, V., and C. Fenselau. 2001. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 73:746-750. [DOI] [PubMed] [Google Scholar]
- 48.Sanjaq, S. 2007. Enterobacter sakazakii—Risikoprofil und Untersuchungen zum Nachweis in Säuglingsnahrungen. Ph.D. thesis. Justus-Liebig-Universitaet Giessen, Giessen, Germany.
- 49.Smits, T. H. M., F. Rezzonico, C. Pelludat, A. Goesmann, J. E. Frey, and B. Duffy. 2010. Genomic and phenotypic characterization of a non-pigmented variant of Pantoea vagans biocontrol strain C9-1 lacking the 530 kb megaplasmid pPag3. FEMS Microbiol. Lett. [Epub ahead of print.] doi: 10.1111/j.1574-6968.2010.01994.x. [DOI] [PubMed]
- 50.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 51.Tan, S. W., S. M. Wong, and R. M. Kini. 2000. Rapid simultaneous detection of two orchid viruses using LC- and/or MALDI-mass spectrometry. J. Virol. Methods 85:93-99. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanneste, J. L., D. C. Cornish, J. Yu, and M. D. Voyle. 2002. P10c: a new biological control agent for control of fire blight which can be sprayed or distributed using honey bees. Acta Hortic. 590:231-235. [Google Scholar]
- 54.Vanneste, J. L., J. Yu, and S. V. Beer. 1992. Role of antibiotic production by Erwinia herbicola Eh252 in biological control of Erwinia amylovora. J. Bacteriol. 174:2785-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Rostenberghe, H., R. Noraida, W. I. Wan Pauzi, H. Habsah, M. Zeehaida, A. R. Rosliza, I. Fatimah, N. Y. Nik Sharimah, and H. Maimunah. 2006. The clinical picture of neonatal infection with Pantoea species. Jpn. J. Infect. Dis. 59:120-121. [PubMed] [Google Scholar]
- 56.Vincent, K., and R. M. Szabo. 1988. Enterobacter agglomerans osteomyelitis of the hand from a rose thorn. A case report. Orthopedics 11:465-467. [DOI] [PubMed] [Google Scholar]
- 57.Wilson, E. E., M. P. Starr, and J. A. Berger. 1957. Bark canker, a bacterial disease of the Persian walnut tree. Phytopathology 47:669-673. [Google Scholar]
- 58.Wilson, E. E., F. M. Zeitoun, and D. L. Fredrickson. 1967. Bacterial phloem canker, a new disease of Persian walnut trees. Phytopathology 57:618-621. [Google Scholar]
- 59.Wright, S. A. I., C. H. Zumoff, L. Schneider, and S. V. Beer. 2001. Pantoea agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl. Environ. Microbiol. 67:284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.