Abstract
Histoplasma capsulatum is very prevalent in the environment and is one of the most common causes of mycoses in humans and diverse animals in Brazil. Multiple typing methods have been developed to study H. capsulatum epidemiology; however, there is limited information concerning comparisons of results obtained with different methods using the same set of isolates. To explore the diversity of H. capsulatum in Brazil and to determine correlations between the results of three different molecular typing techniques, we examined 51 environmental, animal, and human isolates by M13 PCR fingerprinting, PCR-restriction fragment length polymorphism (RFLP) analysis of the internal transcribed region 1 (ITS1)-5.8S-ITS2 region of the rDNA locus, and DNA sequencing and phylogenetic analysis of parts of four protein-encoding genes, the Arf (ADP ribosylation factor), H-anti (H antigen precursor), Ole (delta-9 fatty acid desaturase), and Tub1 (alpha-tubulin) genes. Each method identified three major genetic clusters, and there was a high level of concordance between the results of the typing techniques. The M13 PCR fingerprinting and PCR-RFLP analyses produced very similar results and separated the H. capsulatum isolates included in this study into three major groups. An additional approach used was comparison of our Brazilian ITS1-5.8S-ITS2 sequences with the sequences deposited previously in NCBI data banks. Our analyses suggest that H. capsulatum can be divided into different molecular types that are dispersed around the world. Our results indicate that the three methods used in this study are reliable and reproducible and that they have similar sensitivities. However, M13 PCR fingerprinting has some advantages over the other two methods as it is faster, cheaper, and more user friendly, which especially increases its utility for molecular typing of Histoplasma in situations where laboratory facilities are relatively limited.
Histoplasmosis is a serious community-acquired infection in the United States (28) and in certain countries of Latin America, where it is an especially significant problem in patients with AIDS (14). This disease is one of the most common systemic mycoses in Brazil, where epidemiological surveys carried out using the histoplasmin skin test have indicated that it is endemic in all areas surveyed (15). Data suggest that the numbers of cases of histoplasmosis in Brazil may be underestimated and that the areas where it is endemic are more widespread than previously thought.
Histoplasma capsulatum is a dimorphic fungus that grows as a mold and produces aerial hyphae at 25 to 30°C, but it undergoes morphogenesis to a yeast phase at 37°C. The filamentous phase of this organism is usually found in soil enriched with several compounds, such as nitrogen and phosphate compounds. When conidial or hyphal fragments are inhaled by humans or animals, H. capsulatum changes to the yeast form and continues to replicate as a yeast. Although H. capsulatum has been recognized as an important fungal pathogen in immunocompromised hosts, particularly AIDS patients (27), there are significant gaps in our knowledge of this species' epidemiology and pathogenesis. For instance, systemic histoplasmosis has been found in patients with AIDS who do not reside in regions where it is endemic (29), leading to the suggestion that the disease can result from reactivation of a previously acquired H. capsulatum infection. The clinical manifestations of histoplasmosis range from asymptomatic infections, mild flu-like symptoms, or pneumonia to a systemic disease involving the skin, brain, intestine, adrenal glands, and/or bone marrow (6). Importantly, diverse strains of H. capsulatum have been identified worldwide, and the strains vary in virulence. In addition to classical biochemical assays, distinctions between strains may be based on colony morphology or polymorphism of the genome (19).
Multiple typing methods have been developed to study the epidemiology of H. capsulatum. These methods are based on phenotypic characteristics, such as antigenic profiles (13) and multilocus enzyme electrophoresis results (2), or on DNA-based analysis. Most recently, typing has been accomplished by analysis of fatty acid profiles of H. capsulatum (34). Molecular typing methods are generally considered to have advantages over phenotypic methods in terms of the stability of genomic markers and greater levels of typeability. Several genotype-based methods, such as hybridization of target genes (probes), chromosomal DNA typing, restriction fragment length polymorphism (RFLP) analysis, random amplified polymorphic DNA (RAPD) analysis, and sequencing, have been described for H. capsulatum (4, 5, 7, 8, 11, 17, 19). Despite the abundance of previously developed molecular techniques, there is limited information concerning comparisons of the results obtained with different methods using the same set of isolates. In H. capsulatum, no single approach based on DNA assays has been the dominant method.
The current study was done to explore the diversity of H. capsulatum in Brazil and to determine the correlation between the results of three different molecular typing techniques. For this analysis, we used M13 PCR fingerprinting, PCR-RFLP analysis of the internal transcribed spacer 1 (ITS1)-5.8S-ITS2 region of the rDNA gene, and analysis of the nucleotide sequence polymorphism of four partial genes. M13 PCR fingerprinting (25) is based on generation of multiple PCR products with different electrophoretic mobilities. PCR fingerprinting primers are typically designed using repetitive DNA sequences (31), and the products facilitate detection of two types of genetic variations: (i) differences in the length of DNA and (ii) alterations in the sequence of the priming regions. PCR-RFLP analysis of the ITS1-5.8S-ITS2 region of the rDNA gene (9) involves use of a gene-specific PCR in combination with restriction digestion in order to generate highly stable and reproducible markers. Analysis of the nucleotide sequence polymorphism is based on the sequences of four partial protein-encoding genes (Arf, the H-anti gene, Ole, Tub1) (4). Additionally, to assess the utility of an assay to study the global epidemiology of the fungus, we performed a DNA sequencing analysis of the ITS1-5.8S-ITS2 region to compare the Brazilian H. capsulatum ITS1-5.8S-ITS2 sequences with sequences obtained for H. capsulatum strains isolated in other countries.
MATERIALS AND METHODS
Cultures.
Fifty-one H. capsulatum strains were obtained from the culture collection of the Laboratório de Micologia of the Instituto de Pesquisa Clínica Evandro Chagas, Fundação Oswaldo Cruz, Rio de Janeiro, Brazil. The sources and geographical origins of these strains are shown in Table 1. Fungi were identified by using a conventional mycological method, including assessment of morphology on potato dextrose agar (PDA) slants and microscopic observation of fungal fragments after 15 days of incubation at room temperature. All isolates were tested for conversion to the yeast phase on brain heart infusion (BHI) medium and/or Ham's F-12 medium at 37°C for at least 7 days. Culture supernatants from each isolate were subjected to exoantigen testing (20).
TABLE 1.
Genotypes of 51 H. capsulatum isolates from Brazil determined by three molecular methods
| Isolate | State | Year isolated | Source | M13 group | PCR-RFLP molecular type | Clade based on combined sequencing data |
|
|---|---|---|---|---|---|---|---|
| NJ method | MP method | ||||||
| AC02 | Rio de Janeiro | 1985 | Soil | 1 | I | BrHc1 | BrHc1 |
| AC05 | Rio de Janeiro | 1985 | Soil | 1 | I | BrHc1 | BrHc1 |
| B670 | Rio de Janeiro | 1990 | Human | 1 | I | BrHc1 | BrHc1 |
| CADAM | Rio de Janeiro | 1984 | Dog | 1 | I | BrHc1 | BrHc1 |
| CAO4 | Rio de Janeiro | 1984 | Dog | 1 | I | BrHc1 | BrHc1 |
| EP02 | Rio de Janeiro | 1981 | Soil | 1 | I | BrHc1 | BrHc1 |
| IGS4/5 | Rio de Janeiro | 1981 | Soil | 1 | I | BrHc1 | BrHc1 |
| IGS19 | Rio de Janeiro | 1981 | Soil | 1 | I | BrHc1 | BrHc1 |
| IT04 | Rio de Janeiro | 1983 | Soil | 1 | I | BrHc1 | BrHc1 |
| RPS45 | Rio de Janeiro | 1984 | Soil | 1 | I | BrHc1 | BrHc1 |
| RPS51 | Rio de Janeiro | 1984 | Soil | 1 | I | BrHc1 | BrHc1 |
| RPS86 | Rio de Janeiro | 1984 | Soil | 1 | I | BrHc1 | BrHc1 |
| RS01 | Rio de Janeiro | 1981 | Rat | 1 | I | BrHc1 | BrHc1 |
| RS09 | Rio de Janeiro | 1981 | Rat | 1 | I | BrHc1 | BrHc1 |
| RS36 | Rio de Janeiro | 1981 | Rat | 1 | I | BrHc1 | BrHc1 |
| TI01 | Rio de Janeiro | 1984 | Soil | 1 | I | BrHc1 | BrHc1 |
| TI05 | Rio de Janeiro | 1984 | Soil | 1 | I | BrHc1 | BrHc1 |
| TI14 | Rio de Janeiro | 1984 | Soil | 1 | I | BrHc1 | BrHc1 |
| 3237 | Rio de Janeiro | 1988 | Human | 1 | I | BrHc1 | BrHc1 |
| 3356 | Rio de Janeiro | 1988 | Human | 1 | I | BrHc1 | BrHc1 |
| 3416 | Rio de Janeiro | 1989 | Human | 1 | I | BrHc1 | BrHc1 |
| 3612 | Rio de Janeiro | 1989 | Human | 1 | I | BrHc1 | BrHc1 |
| 3688 | Rio de Janeiro | 1989 | Human | 1 | I | BrHc1 | BrHc1 |
| 4334 | Rio de Janeiro | 1990 | Human | 1 | I | BrHc1 | BrHc1 |
| 4631 | Rio de Janeiro | 1990 | Human | 1 | I | BrHc1 | BrHc1 |
| 6406 | Rio de Janeiro | 1991 | Human | 1 | I | BrHc1 | BrHc1 |
| 6503 | Rio de Janeiro | 1991 | Human | 1 | I | BrHc1 | BrHc1 |
| 9291 | Rio de Janeiro | 1993 | Human | 1 | I | BrHc1 | BrHc1 |
| 78642 | Rio de Janeiro | 1995 | Human | 1 | I | BrHc1 | BrHc1 |
| 84476 | Rio de Janeiro | 1998 | Human | 1 | I | BrHc1 | BrHc1 |
| 84502 | Rio de Janeiro | 1998 | Human | 1 | I | BrHc1 | BrHc1 |
| 84564 | Rio de Janeiro | 1998 | Human | 1 | I | BrHc1 | BrHc1 |
| ES62 | Espírito Santo | Unknown | Human | 1 | III | BrHc2 | BrHc2 |
| JIEF | Ceará | 2000 | Human | 1 | III | BrHc3 | BrHc1 |
| RE5646 | Pernambuco | 2000 | Human | 1 | I | BrHc3 | BrHc1 |
| RE9463 | Pernambuco | 2000 | Human | 1 | I | BrHc3 | BrHc1 |
| 385BG | Mato Grosso do Sul | 2000 | Human | 2 | I | BrHc1 | BrHc1 |
| SP2414 | São Paulo | 2004 | Human | 2 | I | BrHc1 | BrHc1 |
| SP49 | São Paulo | 2000 | Human | 2 | II | BrHc4 | BrHc3 |
| MS53 | Mato Grosso do Sul | 2000 | Human | 3 | I | BrHc2 | BrHc2 |
| 9414 | Rio de Janeiro | 1993 | Human | 3 | I | BrHc1 | BrHc1 |
| ES55 | Espírito Santo | Unknown | Human | 3 | III | BrHc2 | BrHc2 |
| ES56 | Espírito Santo | Unknown | Human | 3 | III | BrHc2 | BrHc2 |
| ES60 | Espírito Santo | Unknown | Human | 3 | III | BrHc2 | BrHc2 |
| GO764 | Goiás | 1999 | Human | 3 | III | BrHc2 | BrHc2 |
| GO1820 | Goiás | 1999 | Human | 3 | III | BrHc2 | BrHc2 |
| 157CS | Rio Grande do Sul | 2002 | Human | 3 | III | BrHc2 | BrHc2 |
| 187LCT | Rio Grande do Sul | 2002 | Human | 3 | III | BrHc2 | BrHc2 |
| 190CLC | Rio Grande do Sul | 2002 | Human | 3 | III | BrHc2 | BrHc2 |
| 184PRS | Rio Grande do Sul | 2002 | Human | 3 | III | BrHc2 | BrHc2 |
| 177CS | Rio Grande do Sul | 2002 | Human | 3 | III | BrHc2 | BrHc2 |
aBold type indicates the 15.7% of the H. capsulatum isolates without total pairwise concordance but with similar topology as determined by all methods used in this study.
DNA extraction.
Yeast cells were recovered from Ham's F-12 medium and used for DNA extraction (30), which involved spheroplasting, enzymatic cell lysis, phenol-chloroform-isoamyl alcohol protein precipitation, ethanol precipitation, and enzymatic treatments. After RNase and proteinase K treatment, DNA was purified by phenol-chloroform-isoamyl alcohol (25:24:1) extraction and ethanol precipitation. The integrity of DNA was verified by electrophoresis on a 1% agarose gel in TBE buffer (0.89 M Tris, 0.89 M boric acid, 0.02 M disodium EDTA; pH 8.4) and staining with ethidium bromide (Roche Molecular Biochemicals) at a final concentration of 0.5 μg/ml. DNA was quantified spectrophotometrically.
M13 PCR fingerprinting.
The minisatellite-specific core sequence of wild-type phage M13 (5′-GAGGGTGGCGGTTCT-3′) was used as a single primer in the PCR. According to Vassart and colleagues (25), this method using a phage M13 sequence detects hypervariable minisatellites in human and animal DNA (25). The amplification reactions were performed with a 50-μl mixture containing 25 ng of genomic DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM (each) dATP, dCTP, dGTP, and dTTP (Roche Diagnostics GmbH, Mannheim, Mannheim, Germany), 3 mM magnesium acetate, 30 ng of primer, and 2.5 U Amplitaq DNA polymerase (Applied Biosystems, Foster City, CA). PCR was performed in a Perkin-Elmer thermal cycler (model 480), and the program consisted of 35 cycles of 20 s of denaturation at 94°C, 1 min of annealing at 50°C, and 20 s of extension at 72°C, followed by a final extension for 6 min at 72°C. Amplification products were removed, concentrated to approximately 20 μl, separated by electrophoresis on 1.4% agarose gels (stained with ethidium bromide [final concentration, 0.5 μg/ml]) in 1× TBE buffer at 60 V for 14 cm, and visualized under UV light. All visualized bands on the gels were counted independent of the intensity, and the presence or absence of the amplified DNA bands was recorded (10).
PCR RFLP analysis (rDNA gene).
PCR analysis of the ITS1-5.8S-ITS2 region of the rDNA gene was performed using 50-μl (final volume) reaction mixtures. Each reaction mixture contained 50 ng of DNA, 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2; Applied Biosystems, Foster City, CA), 0.2 mM (each) dATP, dCTP, dGTP, and dTTP (Roche Diagnostics GmbH), 3 mM magnesium acetate, 1.5 U AmpliTaq DNA polymerase (Applied Biosystems), 50 ng of primer SR6R (5′-AAGTARAAGTCGTAACAAGG-3′), and 50 ng of primer LR1 (5′-GGTTGGTTTCTTTTCCT-3′). The PCR was performed in a Perkin-Elmer thermal cycler (model 480) with the following program: initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation for 45 s at 94°C, annealing for 1 min at 61°C, and extension for 2 min at 72°C and then a final extension at 72°C for 10 min. The PCR products (20 μl) were double digested with Sau96I (10 U/μl) and HhaI (20 U/μl) for 3 h or overnight at 37°C, and the amplification products were mixed with 0.2 volume of loading buffer and separated by 3% agarose gel electrophoresis at 100 V for 5 h. RFLP patterns were assigned visually based on the fragments obtained after electrophoresis.
Statistical analysis.
BioloMICS version 7.5.80 (BioAware, Hannut, Belgium) was used to determine the genetic relationship of the strains. The DNA bands of each fingerprinting pattern were defined manually with a band position tolerance of 0.9%, a setting shown in a previous study (data not shown) to optimally define molecular size marker bands as 100% identical. Similarity coefficients were calculated by using the Dice coefficient, and cluster analyses were performed using the neighbor-joining (NJ) algorithms by using the “Fuzzy Logic” and “Area Sensitive” options of the BioloMICS software. The same statistical program was used to determine the cophenetic correlation coefficient.
Sequencing.
Partial DNA sequences of four nuclear genes and of a region of the rDNA locus (ITS1-58.S-ITS2) (4) were used for phylogenetic analyses. Table 2 shows the primer sequences and the predicted sizes of the amplified products. PCRs were performed with 100 ng of a genomic DNA template using 50-μl reaction mixtures. The reaction mixtures consisted of each primer at a concentration of 0.45 mM, 1.0 U of AmpliTaq DNA polymerase (Perkin-Elmer), 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, and 0.2 mM deoxynucleotide triphosphates, and the following temperature profile was used: 32 cycles of DNA denaturation for 15 s at 94°C, annealing for 30 s, and extension for 1 min at 72°C, followed by a final extension for 5 min at 72°C. The annealing temperature was 65°C in the first cycle and was reduced at a rate of 0.7°C/cycle for the next 12 cycles; thereafter, the PCR was continued using an annealing temperature of 56°C for the remaining 20 cycles (touchdown PCR) (1). Automated sequencing was done using the Sequencing Platform at Fundação Oswaldo Cruz-PDTIS/FIOCRUZ in Brazil and the Sequencing Facility of the Albert Einstein College of Medicine in New York. Sequences of both strands were generated, edited with the Sequencher version 4.6 software package (Genes Codes Corporation, United States), and aligned by using the Mega version 4.0.2 software. The following settings were used for Clustal W: slow/accurate; gap opening penalty, 15.00; gap extension penalty, 6.66; delay divergent sequences, 30%; DNA transition weight, 0.50; DNA weight matrix, IUB; and negative matrix, OFF. The same set of H. capsulatum isolates was compared with sequences available from the NCBI/GenBank database in order to define the levels of relatedness among the Brazilian isolates and isolates from other countries.
TABLE 2.
PCR primers used for the Arf, H-anti, Ole, and Tub1 genes and the ITS region
| Locus | Encoded protein or region | Primer | Nucleotide sequence (5′-3′) | Length of product (bp) | GenBank accession no. |
|---|---|---|---|---|---|
| Arf | ADP ribosylation factor | arf1 | AGAATATGGGGCAAAAAGGA | 459 | L25117 |
| arf2 | CGCAATTCATCTTCGTTGAG | ||||
| H-anti gene | H antigen precursor | H-anti3 | CGCAGTCACCTCCATACTATC | 408 | U20346 |
| H-anti4 | GCGCCGACATTAACCC | ||||
| Ole | Delta-9 fatty acid desaturase | ole3 | TTTAAACGAAGCCCCCACGG | 424 | X85962 |
| ole4 | CACCACCTCCAACAGCAGCA | ||||
| Tub1 | Alpha-tubulin | tub1 | GGTGGCCAAATCGCAAACTC | 278 | M28358 |
| tub2 | GGCAGCTTTCCGTTCCTCAGT | ||||
| ITS | Internal transcribed spacers plus rRNA genes | ITS4 | TCCTCCGCTTATTGATATGC | 600 | U18363 |
| ITS5 | GGAAGTAAAAGTCGTAACAAGG |
Final data analysis for partial sequences of four protein-encoding genes.
The nucleotide diversity (π) and divergence for the four gene regions were estimated for the H. capsulatum isolates studied as described by Nei and Li (12). The evolutionary history was inferred using the neighbor-joining method (16). The percentages of replicate trees in which associated taxa clustered as determined by bootstrap analysis (500 replicates) are indicated below (22). The evolutionary distances were computed using the maximum composite likelihood method (21) and were expressed as numbers of base substitutions per site. All positions containing gaps and for which there were missing data were eliminated from the data set (complete deletion option). There were a total of 1,432 positions in the final data set for the combined analysis of the four genes. Phylogenetic analyses were conducted using MEGA 4.0.2.
Nucleotide sequence accession numbers.
All of the sequences generated in this study have deposited in the GenBank database under accession numbers GU320834 to GU321088.
RESULTS
A total of 51 H. capsulatum isolates, including 34 clinical, 5 veterinary, and 12 environmental isolates, were subjected to molecular typing. Data lists of the characteristics of the isolates studied included the state, the laboratory code, the clinical, veterinary, or environmental origin, and the isolation history (Table 1).
M13 PCR fingerprinting.
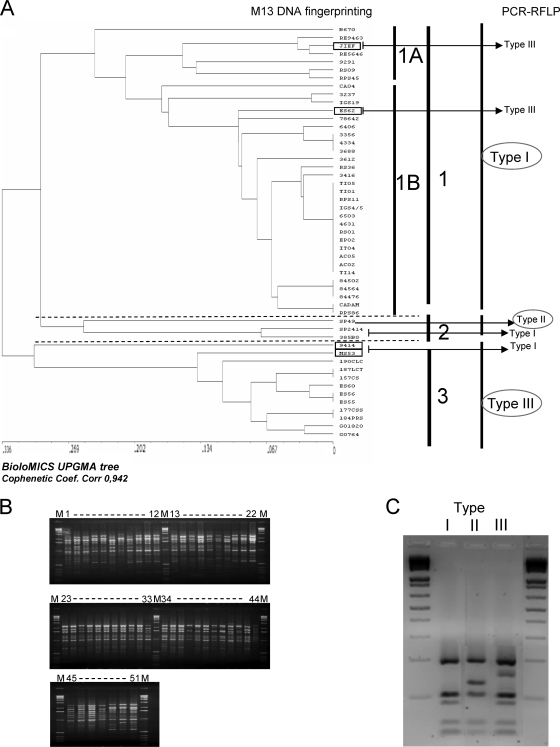
A total of 32 reproducible PCR fingerprinting profiles were obtained with primer M13 for the 51 H. capsulatum samples, indicating that there was a high level of genetic diversity among the H. capsulatum strains isolated from different geographic regions of Brazil. The genetic relationships determined with the unweighted-pair group method using average linkages (UPGMA) were represented by a dendrogram (cophenetic correlation coefficient, 0.942), and the discriminatory index calculated by using Simpson's index of diversity was 0.915 (3) (Fig. 1). Clustering of the 32 fingerprinting profiles resulted in three major groups (designated groups 1 to 3). The major group, group 1, was composed of 36 H. capsulatum isolates from different regions that exhibited higher levels of polymorphism. This group was divided into two subgroups, subgroups 1A and 1B. Subgroup 1A was composed of four H. capsulatum isolates from Rio de Janeiro and three isolates (JIEF, RE5646, and RE9463) from the northeast region of Brazil. Subgroup 1B was composed of H. capsulatum isolates from southeastern Brazil, including 28 isolates from Rio de Janeiro State obtained during several histoplasmosis outbreaks and epidemiological studies (18, 32, 33) and one isolate from Espírito Santo (ES62). Three isolates from southeastern Brazil comprised group 2. Strains SP 2414 from São Paulo and 385BG from Mato Grosso do Sul were closely related. However, H. capsulatum SP49, also from São Paulo State, had a completely different PCR fingerprinting profile than the other H. capsulatum strains included in this group. Group 3 included 12 strains, including 4 isolates (9414, ES60, ES56, and ES55) from the southeastern region of Brazil, 5 isolates (157CS, 187LCT, 190CLC, 184PRS, and 177CS) from the southern region, and 3 isolates (MS53, GO764, and GO1820) from the central region (Fig. 1).
FIG. 1.
(A) M13 fingerprints of 51 H. capsulatum isolates generated by BioloMICS version 7.5.80 (BioAware, Hannut, Belgium). (B) Electrophoretic profiles of 51 H. capsulatum isolates subjected to M13 PCR fingerprinting. (C) DNA banding patterns obtained for H. capsulatum isolates by PCR-RFLP analysis.
PCR-RFLP analysis.
PCR-RFLP amplification of the ITS region of the rDNA gene cluster with primers SR6R and LR1 generated an approximately 600-bp PCR product for all H. capsulatum isolates (26). Sau96I and HhaI were selected for RFLP analysis based on the presence of at least one cleavage site for H. capsulatum and on generation of several fragments that should be easily distinguished on a conventional agarose gel. Examination of the 600-bp PCR product containing the ITS1-5.8S-ITS2 sequence revealed species-specific variations in restriction enzyme (RE) cut sites. Double digestion with Sau96I and HhaI cut the ITS1-5.8S-ITS2 PCR product of H. capsulatum, and based on visual analysis of the results the isolates were divided into three molecular types, which were designated molecular types I, II, and III (Fig. 1B). Digestion produced 200-, 120-, 93-, 53-, and 30-bp fragments for molecular type I isolates, 200-, 150-, 120-, 53, and 30-bp fragments for molecular type II isolates, and 200-, 170-, 120-, 93-, 5-, and 30-bp fragments for molecular type III isolates. The majority of the patterns obtained in the ITS region analysis corresponded to the results for the three genetic groups generated by M13 PCR fingerprinting. Of the 38 H. capsulatum RFLP molecular type I isolates, 36 were classified as group 1 isolates by M13 PCR fingerprinting. The exceptions, H. capsulatum SP2414 and 385BG, clustered in group 2. H. capsulatum JIEF and ES62 (group 1 as determined by M13 PCR fingerprinting) were classified as molecular type III isolates. This cluster also included H. capsulatum SP49, which was in group 2 as determined by M13 PCR fingerprinting. Group 3 corresponded to molecular type III, except for isolates 9414 and MS53, which were in group 3 as determined by M13 PCR fingerprinting but were molecular type I isolates as determined by PCR-RFLP analysis. Of the 51 isolates, 74.5% were molecular type I isolates, 5.9% were molecular type II isolates, and 19.6% were molecular type III isolates (Table 1).
DNA polymorphism and phylogenetic analyses of protein-encoding genes.
The DNA of 51 Brazilian H. capsulatum isolates was amplified using previously described standardized conditions (11). As determined by the BLAST 2 program (24), all of the partial sequences of the Arf, H-anti, Ole, and Tub1 genes of Brazilian H. capsulatum isolates showed high levels of similarity with sequences previously deposited in the NCBI databank, which ranged from 97% to 100%, and the insertions and/or deletions observed in the sequences demonstrated that there were some genome changes in the partial sequences of the four genes studied in our isolates. Eighty-eight informative sites were distributed among the protein-encoding genes as follows: 20 positions in the Arf gene, 28 positions in the H-anti gene, 19 positions in the Ole gene, and 21 positions in the Tub1 gene.
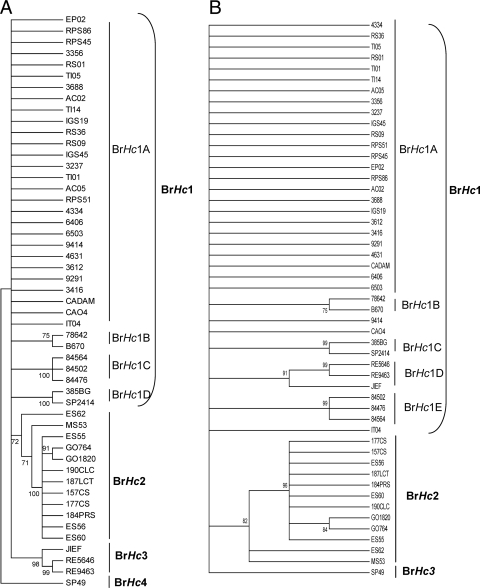
The phylogenetic tree for the four genes analyzed by either the neighbor joining (NJ) or maximum parsimony (MP) method (Fig. 2) contained the following three distinct clades: BrHc1 (with subclades BrHc1A, BrHc1B, BrHc1C, and BrHc1D), BrHc2, and BrHc3 (Fig. 2). Bootstrap values less than 70% are not shown in Fig. 2. The majority of the H. capsulatum soil and animal isolates from Rio de Janeiro were grouped in subclade BrHc1A. The majority of the clinical H. capsulatum isolates from the same locality also were in this clade or subclade. Subclade BrHc1B was comprised of two clinical isolates from Rio de Janeiro (78642 and B670). Three clinical isolates, also from Rio de Janeiro (84476, 84502, and 84564) formed subclade BrHc1C, which was the same topology found by M13 PCR fingerprinting. Subclade BrHc1D was represented by two isolates recovered from Mato Grosso do Sul (385BG) and São Paulo (SP 2414). Clade BrHc2 included 12 strains, including 4 isolates (ES55, ES56, ES60, and ES62) from the Espírito Santo states (southeast), 5 isolates (157CS, 187LCT, 190CLC, 184PRS, and 177CS) from Rio Grande do Sul (south), and 3 isolates (MS53, GO764, and GO1820) from central Brazil. Interestingly, the grouping of all of these isolates was similar to the grouping in PCR-RFLP molecular type III, except for MS53 from Mato Grosso do Sul (molecular type I). Three isolates from the northeast region, JIEF from Ceará and RE5646 and RE9463 from Pernambuco, were closely related genetically to clade BrHc1 only in the NJ analysis and formed clade BrHc3. All isolates were grouped in the three clades described above, except for H. capsulatum SP49 from São Paulo, which was considered a unique representative of another clade, clade BrHc4 (Fig. 2).
FIG. 2.
Evolutionary relationships of 51 taxa. (A) NJ tree based on analysis of combined data for the four loci. The branch lengths are proportional to distance. The evolutionary distances were computed using the maximum composite likelihood method. (B) Strict consensus for 176 MP trees derived from analysis the same data for the four loci. The MP tree was obtained using the close-neighbor interchange algorithm. The numbers at the nodes are percentages and indicate the levels of support based on 500 bootstrap replications of the parsimony procedure; only values greater than 70% are shown. There were a total of 1,432 positions in the final data set. Phylogenetic analyses were conducted using MEGA4.
Relatedness among Brazilian ITS1-5.8S-ITS2 sequences and other sequences from a public data bank.
We subjected the ITS1-5.8S-ITS2 sequences that we generated to BLAST analysis with data sets for H. capsulatum sequences in the NCBI public databank in order to assess the relationship between our Brazilian isolates and H. capsulatum isolates from other countries. We selected sequences that provided 100% query coverage and exhibited maximum similarity. The analysis of the evolutionary relationship of the 70 taxa (Brazilian isolates plus isolates from other countries) was conducted using Mega 4. The gene tree contained three distinct major clades designated clades A, B, and C. Analysis of the data sets identified demonstrated that the sequences of isolates from other countries comprised clade A (Fig. 3). Twenty-one of our Brazilian isolates were also included in this clade. Twenty-four of our isolates from Rio de Janeiro State were in clade B, and they shared 100% similarity with a previously described Brazilian isolate (accession no. AB055234) (5, 23) (Fig. 3). Six additional Rio de Janeiro State isolates were distinctly categorized as members of clade C.
FIG. 3.
Evolutionary relationships of 70 taxa (linearized) based on analysis of ITS1-5.8S-ITS2 of the rDNA gene. The evolutionary history was inferred using the neighbor-joining method. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are indicated at the nodes. The phylogenetic tree was linearized by assuming that the evolutionary rates in all lineages were equal. The tree is drawn to scale, and the branch lengths are in the same units as the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method and were in expressed as the number of base substitutions per site. There were a total of 418 positions in the final data set. Abbreviations: RJ, Rio de Janeiro; SP, São Paulo; PE, Pernambuco; ES, Espírito Santos; CE, Ceará; RS, Rio Grande do Sul. All Brazilian H. capsulatum sequences are indicated by normal type, and sequences of isolates from other countries are indicated by bold type. Phylogenetic analyses were conducted using MEGA4 (4).
DISCUSSION
This study of H. capsulatum isolates from different Brazilian regions was undertaken in an attempt to set up a network of mycology laboratories to study the distribution of this fungus and to determine if specific molecular genotypes predominate in the participating states of Brazil, as previously described for Cryptococcus neoformans isolates (9). We used three different molecular typing techniques with a set of 51 Brazilian environmental, animal, and human H. capsulatum isolates. Three major clusters were formed when M13 PCR fingerprinting, PCR-RFLP analysis of the ITS1-5.8S-ITS2 region of the rDNA locus, and phylogenetic analysis of four protein-encoding genes were used.
We reported previously that RAPD analysis was a useful tool for identifying molecular types of H. capsulatum in different geographic regions of Brazil (32). However, we show here that a different PCR-based typing method, M13 PCR fingerprinting, revealed high levels of polymorphism for isolates in a specific region. Furthermore, the relationship among the H. capsulatum isolates was not uniform in terms of geographical region. Although the DNA fingerprinting analysis also divided the set of Brazilian H. capsulatum isolates into three major groups (groups 1, 2, and 3), the groups exhibited a high level of polymorphism, especially groups 2 and 3, which were composed of H. capsulatum isolates from different Brazilian regions. Interestingly, H. capsulatum strain SP 49 from São Paulo formed a separate group. These data suggest that more than one genotype can be present in a geographic region. Meanwhile, we cannot eliminate the possibility that patients who initially acquired the fungus in one geographically distinct location migrated to another geographic location. The M13 PCR fingerprinting analysis revealed distinct polymorphic DNA profiles with high levels of discrimination. An exception was the H. capsulatum soil and animal isolates from Rio de Janeiro, since in both M13 PCR fingerprinting and PCR-RFLP analyses these isolates clustered in the same clade or group. These data suggested that these isolates could be autochthonous isolates and could be used as molecular markers for autochthonous strains from a region.
Similar results were obtained when a PCR-RFLP analysis was performed with this set of isolates. Three molecular types were generated. Of the 51 isolates examined by using PCR-RFLP, 6 (11.7%) grouped differently than they grouped when the M13 PCR fingerprinting method was used. The SP2414 isolate from São Paulo and the 365BG isolate from Mato Grosso do Sul were molecular type I isolates (based on PCR-RFLP profiles), but they clustered with group 2 when M13 PCR fingerprinting was used. The MS53 (Mato Grosso do Sul) and 9414 (Rio de Janeiro) isolates were molecular type I isolates (based on PCR-RFLP profiles); however, they were classified in group 3 when M13 PCR fingerprinting was used. Despite this disagreement between the M13 and PCR-RFLP typing results, these isolates were distinctly different from the other isolates when either method was used. A similar situation was observed with the ES62 and JIEF isolates, as they were categorized as molecular type III isolates by the PCR-RFLP method but were in group 1 when M13 PCR fingerprinting was used. These findings suggested that H. capsulatum molecular types are dispersed around the country. Thus, the previous hypothesis that a specific genotype is present exclusively in each Brazilian region (32) should be abandoned.
We previously reported that the population structure of H. capsulatum as determined by analysis of DNA sequences of partial protein-encoding genes (Arf, the H-anti gene, Ole, and Tub1) (5) was phylogenetically diverse in Brazil. The Brazilian isolates were placed in clade Lam A. It was also suggested that H. capsulatum comprises at least seven phylogenetic species, including two Latin American species (4). In this study, the phylogeny of 51 geographically diverse H. capsulatum isolates from different Brazilian states was evaluated using nucleotide sequences of the Arf, H-anti, Ole, and Tub1 genes. Phylogenetic analyses did not reveal differences in NJ and MP tree topologies and even showed that there was clustering of isolates RE5646, RE9463, and JIEF, which were closely related to clade BrHc1 in the NJ analysis. This finding was obtained in an MP analysis as well, although the topology was slightly different. In fact, this clustering corroborates the similar results obtained with all methods used in this work. This study allowed robust analysis of a set of Brazilian H. capsulatum isolates from different geographic regions, and there were three major clusters (clades BrHc1, BrHc2, and BrHc3), as well as clade BrHc4, in each of the four protein-encoding gene trees examined. These data corroborated our initial findings that demonstrated that the phylogenetic population in Brazil is varied (32). Although the genetic separation among the three major clades is considerable, certain isolates exhibited geographic clonality, suggesting that there is partial clonal spread of H. capsulatum within Brazil. However, migration, trips, and tourism could be responsible for some of the diversity identified. Interestingly, all of the H. capsulatum isolated in Rio de Janeiro State were classified as clade BrHc1 isolates.
The relatedness among Brazilian ITS1-5.8S-ITS2 sequences and other sequences obtained from the public data bank revealed that the majority of Rio de Janeiro H. capsulatum isolates (n = 30) were genetically distinct from the isolates from other Brazilian states, as well from isolates from other Latin American countries or other continents. The high level of genetic similarity among these isolates suggests that only one genetic population is present in the Rio de Janeiro's microenvironment. As noted above, we cannot rule out migration of patients to other geographically distinct locations as a possible explanation of the clustering of the ITS1-5.8S-ITS2 sequences of the four clinical H. capsulatum isolates (84502, 84476, 84564, and 9414) from Rio de Janeiro in clade A. Our analyses suggest that H. capsulatum can be divided into different molecular types that are dispersed around the world. Another interesting aspect was the finding that clades B and C exhibited some geographic specificity, which agrees with previous reports (23).
In general, 84.3% concordance was found for the results obtained with the three molecular typing methods used for our 51 isolates, M13 PCR fingerprinting, PCR-RFLP analysis, and analysis of the sequences of four partial genes encoding proteins (Table 1), suggesting that each of these approaches or combinations of these approaches can be used in future analyses of the genetic diversity of H. capsulatum populations.
It has been reported that comparisons of methods should be based on several parameters, such as typeability, reproducibility, and the index of discrimination, in order to assess which typing method is the most efficient (3). The cophenetic correlation coefficient (0.942) and the excellent discriminatory index (0.915) obtained in the M13 PCR fingerprinting analysis indicate the efficiency of this method. The cophenetic correlation for a cluster tree is defined as the linear correlation coefficient for the cophenetic distances obtained from the tree and the original distances (or dissimilarities) used to construct the tree. Thus, it is a measure of how faithfully the tree represents the dissimilarities among observations. This value should be very close to 1 for a high-quality solution. This measure can be used to compare alternative cluster solutions obtained using different algorithms. The bootstrap analysis of the four genes in Brazilian H. capsulatum isolates supported the data obtained with the other methods, and the analysis showed the stability of the individual clades.
The methods produced complementary results and grouped all isolates into three major genetic clusters. In addition, the phylogenetic analysis of the relationship of 70 strains from different regions of Brazil (n = 51) and other countries (n = 19) using DNA sequence analysis of the ITS region provided information useful for understanding fungal dissemination, the genotypes in Latin America, and evidence for autochthonous or imported cases. Finally, this information confirmed that there is not just one specific genotype in distinct regions of Brazil.
The analysis of our results indicates that the three methods used in this study are reliable and reproducible and have similar sensitivities. Hence, these techniques can be used in a standardized approach for typing H. capsulatum. As noted previously, there has not been a consensus concerning the most effective method for molecular typing of H. capsulatum (31). We demonstrated that the M13 PCR fingerprinting method has important advantages compared to the other methods tested in this work. M13 PCR fingerprinting is more rapid, less expensive, and requires less technical expertise than PCR-RFLP and sequencing analyses. Furthermore, these advantages are especially valuable in situations where laboratory facilities are relatively limited.
Acknowledgments
Financial support for this work was provided by PAPES IV Program (Fiocruz/CNPq) grant Proc. 400175/98-3. R.M.Z.-O. was supported in part by CNPq grant 350338/2000-0. This work was also supported by research grant 352303 from the National Health and Research Council of Australia to W.M. M.D.M.M. was supported in part by Fogarty International Center Interhemispheric Research Training Grant in Infectious Diseases NIH D43-TW007129. J.D.N. was supported in part by NIH grant AI056070-01A2 and by the Center for AIDS Research at the Albert Einstein College of Medicine and Montefiore Medical Center (NIH grant AI-51519).
We thank A. L. Q. Torres (IPEC/CNPq) for help with illustrations.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamrick, J. L., R. W. Lichtwardt, and C. Lan. 1986. Levels of isozyme variation within and among Histoplasma capsulatum localities. Trans. Kans. Acad. Sci. 89:49-56. [PubMed] [Google Scholar]
- 3.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasuga, T., J. W. Taylor, and T. J. White. 1999. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J. Clin. Microbiol. 37:653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasuga, T., T. J. White, G. Koenig, J. McEwen, A. Restrepo, E. Castaneda, C. Da Silva Lacaz, E. M. Heins-Vaccari, R. S. De Freitas, R. M. Zancope-Oliveira, Z. Qin, R. Negroni, D. A. Carter, Y. Mikami, M. Tamura, M. L. Taylor, G. F. Miller, N. Poonwan, and J. W. Taylor. 2003. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol. Ecol. 12:3383-3401. [DOI] [PubMed] [Google Scholar]
- 6.Kauffman, C. A. 2007. Histoplasmosis: a clinical and laboratory update. Clin. Microbiol. Rev. 20:115-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keath, E. J., G. S. Kobayashi, and G. Medoff. 1992. Typing of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J. Clin. Microbiol. 30:2104-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keath, E. J., A. A. Painter, G. S. Kobayashi, and G. Medoff. 1989. Variable expression of a yeast-phase-specific gene in Histoplasma capsulatum strains differing in thermotolerance and virulence. Infect. Immun. 57:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer, W., A. Castaneda, S. Jackson, M. Huynh, and E. Castaneda. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer, W., K. Marszewska, M. Amirmostofian, R. P. Igreja, C. Hardtke, K. Methling, M. A. Viviani, A. Chindamporn, S. Sukroongreung, M. A. John, D. H. Ellis, and T. C. Sorrell. 1999. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790-1799. [DOI] [PubMed] [Google Scholar]
- 11.Muniz, M. M., C. V. Pizzini, J. M. Peralta, E. Reiss, and R. M. Zancope-Oliveira. 2001. Genetic diversity of Histoplasma capsulatum strains isolated from soil, animals, and clinical specimens in Rio de Janeiro State, Brazil, by a PCR-based random amplified polymorphic DNA assay. J. Clin. Microbiol. 39:4487-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U. S. A. 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes-Montes, M. R., M. Bobadilla-Del Valle, M. A. Martinez-Rivera, G. Rodriguez-Arellanes, E. Maravilla, J. Sifuentes-Osornio, and M. L. Taylor. 1999. Relatedness analyses of Histoplasma capsulatum isolates from Mexican patients with AIDS-associated histoplasmosis by using histoplasmin electrophoretic profiles and randomly amplified polymorphic DNA patterns. J. Clin. Microbiol. 37:1404-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rios-Fabra, A., A. R. Moreno, and R. E. Isturiz. 1994. Fungal infection in Latin American countries. Infect. Dis. Clin. North Am. 8:129-154. [PubMed] [Google Scholar]
- 15.Rodrigues, C. C. 2004. Avaliação da infecção por Histoplasma capsulatum por meio de reações intradérmicas em moradores da zona urbana e rural do Município de Pratania (SP). Ph.D. thesis. Faculdade de Medicina de Botucatu/UNESP Botucatu, São Paulo, Brazil.
- 16.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 17.Sandhu, G. S., B. C. Kline, L. Stockman, and G. D. Roberts. 1995. Molecular probes for diagnosis of fungal infections. J. Clin. Microbiol. 33:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva-Ribeiro, V. L., M. F. Ferreira-da-Cruz, B. Wanke, and B. Galvao-Castro. 1987. Canine histoplasmosis in Rio de Janeiro: natural and experimental infections. J. Med. Vet. Mycol. 25:319-322. [DOI] [PubMed] [Google Scholar]
- 19.Spitzer, E. D., B. A. Lasker, S. J. Travis, G. S. Kobayashi, and G. Medoff. 1989. Use of mitochondrial and ribosomal DNA polymorphisms to classify clinical and soil isolates of Histoplasma capsulatum. Infect. Immun. 57:1409-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Standard, P. G., and L. Kaufman. 1976. Specific immunological test for the rapid identification of members of the genus Histoplasma. J. Clin. Microbiol. 3:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 22.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura, M., T. Kasuga, K. Watanabe, M. Katsu, Y. Mikami, and K. Nishimura. 2002. Phylogenetic characterization of Histoplasma capsulatum strains based on ITS region sequences, including two new strains from Thai and Chinese patients in Japan. Nippon Ishinkin Gakkai Zasshi 43:11-19. [DOI] [PubMed] [Google Scholar]
- 24.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 25.Vassart, G., M. Georges, R. Monsieur, H. Brocas, A. S. Lequarre, and D. Christophe. 1987. A sequence in M13 phage detects hypervariable minisatellites in human and animal DNA. Science 235:683-684. [DOI] [PubMed] [Google Scholar]
- 26.Vilgalys, R., and M. Hester. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheat, L. J., P. Connolly, M. Smedema, E. Brizendine, and R. Hafner. 2001. Emergence of resistance to fluconazole as a cause of failure during treatment of histoplasmosis in patients with acquired immunodeficiency disease syndrome. Clin. Infect. Dis. 33:1910-1913. [DOI] [PubMed] [Google Scholar]
- 28.Wheat, L. J., and C. A. Kauffman. 2003. Histoplasmosis. Infect. Dis. Clin. North Am. 17:1-19, vii. [DOI] [PubMed] [Google Scholar]
- 29.Woods, J. P., E. L. Heinecke, J. W. Luecke, E. Maldonado, J. Z. Ng, D. M. Retallack, and M. M. Timmerman. 2001. Pathogenesis of Histoplasma capsulatum. Semin. Respir. Infect. 16:91-101. [DOI] [PubMed] [Google Scholar]
- 30.Woods, J. P., D. Kersulyte, W. E. Goldman, and D. E. Berg. 1993. Fast DNA isolation from Histoplasma capsulatum: methodology for arbitrary primer polymerase chain reaction-based epidemiological and clinical studies. J. Clin. Microbiol. 31:463-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, J. 2006. Fundamentals of fungal molecular population genetic analyses. Curr. Issues Mol. Biol. 8:75-89. [PubMed] [Google Scholar]
- 32.Zancope-Oliveira, R. M., P. Morais e Silva Tavares, and M. M. Muniz. 2005. Genetic diversity of Histoplasma capsulatum strains in Brazil. FEMS Immunol. Med. Microbiol. 45:443-449. [DOI] [PubMed] [Google Scholar]
- 33.Zancope-Oliveira, R. M., and B. Wanke. 1987. Distribution of sources of infection of Histoplasma capsulatum var. capsulatum in Rio da Prata, a municipality of Rio de Janeiro (RJ). Rev. Inst. Med. Trop. Sao Paulo 29:243-250. (In Portuguese.) [DOI] [PubMed] [Google Scholar]
- 34.Zarnowski, R., M. Miyazaki, A. Dobrzyn, J. M. Ntambi, and J. P. Woods. 2007. Typing of Histoplasma capsulatum strains by fatty acid profile analysis. J. Med. Microbiol. 56:788-797. [DOI] [PMC free article] [PubMed] [Google Scholar]