Abstract
The stinkbug Parastrachia japonensis (Hemiptera: Parastrachiidae) is known for its prolonged prereproductive nonfeeding period, maternal care of eggs in an underground nest, and maternal collection and provisioning of food (fruits) for nymphs. A previous study suggested that a bacterial symbiont is involved in uric acid recycling in this insect during the nonfeeding period, but the identity of this symbiont has not been determined. Here we characterized a novel bacterial symbiont obtained from P. japonensis. Molecular phylogenetic analyses based on 16S rRNA, gyrB, and groEL gene sequences consistently indicated that this symbiont constituted a distinct lineage in the Gammaproteobacteria that has no close relatives but is allied with gut symbionts of acanthosomatid and plataspid stinkbugs, as well as with endocellular symbionts of sharpshooters, tsetse flies, and aphids. The symbiont genes had a remarkably AT-biased nucleotide composition and exhibited significantly accelerated molecular evolution. The symbiont genome was extremely reduced; its size was estimated to be 0.85 Mb. These results suggest that there has been an intimate host-symbiont association over evolutionary time. The symbiont was localized in swollen crypts in a posterior part of the midgut, which was a specialized symbiotic organ. The possibility that the symbiont is involved in uric acid recycling is discussed. The designation “Candidatus Benitsuchiphilus tojoi” is proposed for the symbiont.
Many insects have mutualistic associations with symbiotic microorganisms inhabiting the gut, body cavity, or cells, in which the symbionts play important roles in the survival and reproduction of their hosts, including food digestion and nutrient provisioning (4, 5). Most, but not all, heteropteran plant-sucking stinkbugs of the infraorder Pentatomomorpha harbor bacterial symbionts in the midgut (5, 10). It has been reported that in some stinkbug species of the families Pentatomidae, Plataspidae, Acanthosomatidae, Cydnidae, and Alydidae, nymphs exhibit retarded growth, reduced body size, and/or high mortality when they are experimentally deprived of their symbionts (1, 5, 8, 16, 22, 23, 27, 30, 37).
Parastrachiidae is a small stinkbug family containing only one genus and two species, Parastrachia japonensis and Parastrachia nagaensis (36, 39), although recent comprehensive phylogenetic analyses suggested that the genera Parastrachia and Dismegistus form a monophyletic group (11). P. japonensis (Fig. 1A) is known for its peculiar ecology and behavior, including a prolonged prereproductive nonfeeding period for newly emerged adults that lasts for 9 months, maternal care of eggs in an underground nest, and maternal collection and provisioning of food (fruits) for nymphs (7, 41, 43). A previous study reported that when adult insects were treated with an antibiotic in the nonfeeding period, the uricase activity in the midgut was significantly reduced and the mortality of the insects increased dramatically (20). Thus, it was suggested that a bacterial symbiont is involved in uric acid recycling in this host insect during the nonfeeding period, like the bacterial symbionts of termites (28) and cockroaches (6, 34) and a yeast-like symbiont of rice planthoppers (35).
FIG. 1.
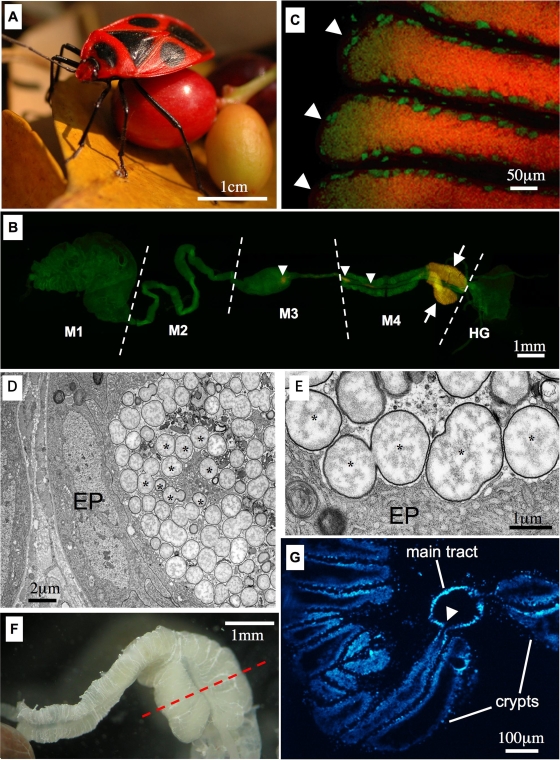
(A) Adult female of P. japonensis carrying fruit. (B and C) In situ hybridization targeting 16S rRNA of the symbiont. Red and green indicate the symbiont and the host nucleus, respectively; yellow is due to superimposed red and green signals. (B) Dissected female alimentary tract. The arrows indicate concentrated symbiont signals in the specialized symbiotic organ, swollen midgut crypts, while the arrowheads indicate sporadic symbiont signals in the midgut main tract. Abbreviations: M1, midgut first section; M2, midgut second section; M3, midgut third section; M4, midgut fourth section with crypts; HG, hindgut. (C) Enlarged image of the swollen midgut crypts. There are strong symbiont signals in the lumen of the crypts. Each arrowhead indicates a crypt. (D and E) Transmission electron microscopy of the swollen midgut crypt. Asterisks indicate symbiont cells. EP, epithelial cells of the crypt. (D) Symbiont cells restricted to the lumen of a crypt. (E) Enlarged image of symbiont cells. (F) Dissected midgut fourth section from an adult female. (G) Section of the midgut fourth section approximately through the plane indicated by the dashed line in panel F. The arrowhead indicates the connection between a crypt and the main tract of the midgut.
Recent microbiological studies have revealed a diverse array of stinkbug symbionts and their lifestyles, including vertically transmitted gammaproteobacterial symbionts in pentatomid, plataspid, acanthosomatid, and scutellerid stinkbugs (8, 16, 18, 23, 29, 30, 31), environmentally acquired betaproteobacterial symbionts (Burkholderia spp.) in alydid stinkbugs (21, 22), and vertically and horizontally transmitted actinobacterial symbionts in pyrrhocorid stinkbugs (19). For the parastrachiid symbiont, the only previous study was performed mainly to examine insect physiology (20), and there was no microbiological characterization of the symbiont. In this study, therefore, we attempted to characterize the bacterial symbiont of P. japonensis.
MATERIALS AND METHODS
Insects.
P. japonensis adults were collected at seven localities in Japan from 2007 to 2009 (see Table S1 in the supplemental material). The collected insects were kept in plastic cases with moistened paper towels and used for experiments within several days.
Insect dissection and DNA extraction.
Adult insects were dissected in sterile phosphate-buffered saline (PBS) (0.8% NaCl, 0.02% KCl, 0.115% Na2HPO4, 0.02% KH2PO4 [pH 7.5]) using fine forceps and microscissors. The alimentary tract (Fig. 1B) was isolated and subjected to either histological analysis or DNA extraction using a NucleoSpin tissue kit (Macherey-Nagel).
Cloning and sequencing.
Three bacterial genes, the 16S rRNA, gyrB, and groEL genes, were cloned and sequenced using DNA samples from the midgut fourth section with crypts. PCR amplification was conducted using AmpliTaq DNA polymerase (Applied Biosystems) and primers 16SA1 (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 16SB1 (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) for the 16S rRNA gene (9), primers gyrBsymF (5′-TTA TCA TGA CWG TAT TAC ATG CWG G-3′) and gyrBsymR (5′-TCC AGC WGA ATC WCC TTC WAC-3′) for the gyrB gene, and primers groEL1F (5′-ATG GCA GCW AAA GAC GTA AAT TYG G-3′) and groEL1R (5′-TTA CAT CAT KCC GCC CAT GC-3′) for the groEL gene (17). The PCR products (1.5 kb for the 16S rRNA gene, 1.0 kb for the gyrB gene, and 1.6 kb for the groEL gene) were cloned and sequenced as described previously (21).
Molecular phylogenetic and evolutionary analyses.
Multiple alignments of the nucleotide sequences were generated using the program Clustal W (42). The alignments were then inspected and corrected manually, and ambiguously aligned sites were removed. Phylogenetic analyses were conducted by using three methods, the maximum parsimony (MP), maximum likelihood (ML), and Bayesian (BA) methods, using the programs PAUP 4.0b10 (40), Phyml (12), and MrBayes v3.0b4 (33), respectively. In the MP analysis, all sites and character changes were weighted equally. In the ML and BA analyses, the GTR+I+G model was selected for the 16S rRNA, gyrB, and groEL gene phylogenies on the basis of the Akaike criterion using the program MrModeltest v2.1 (http://www.ebc.uu.se/systzoo/staff/nylander.html). Bootstrap tests were performed with 100 replications in the MP and ML analyses. In the BA analysis, a total of 11,000 trees were obtained for the 16S rRNA gene (ngen = 160,000, samplefreq = 10, burn in = 5,000), and 8,000 trees were obtained for the gyrB and groEL genes (ngen = 100,000, samplefreq = 10, burn in = 2,000). Based on the data, a 50% majority rule consensus tree was constructed. Relative rate tests were performed using the program RRTree (32) based on genetic distances estimated by using Kimura's two-parameter model (24).
Histology.
Alimentary tracts dissected from female insects were fixed in Carnoy's solution (ethanol-chloroform-acetic acid, 6:3:1) overnight, treated with 6% hydrogen peroxide in 80% ethanol for 3 days to eliminate autofluorescence (25), and then kept in 100% ethanol at room temperature. For tissue sectioning, the preserved samples were cleared using an ethanol-xylene series, embedded in paraffin, and cut into 5-μm serial sections. The tissue sections were mounted on silane-coated glass slides, dewaxed using a xylene-ethanol series, and air dried prior to in situ hybridization. For whole-mount in situ hybridization, the preserved samples were washed twice in PBSTx (PBS containing 0.3% Triton X-100) and directly subjected to the hybridization procedure.
In situ hybridization.
A fluorochrome-labeled oligonucleotide probe, BNTC129-A555 (5′-Alexa555-CCT CTA ATT AGG CAG ATC C-3′) targeting 16S rRNA of the symbiont, was used. Tissue samples were incubated with a hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% SDS, 30% formamide) containing 50 nM probe. For counterstaining of host insect nuclei, 4 μM 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) or 0.5 μM SYTOX green (Invitrogen) was added to the hybridization buffer. After overnight incubation, the samples were thoroughly washed in PBSTx and mounted in Slowfade antifade solution (Molecular Probes). The sectioned and whole-mount samples were observed with an epifluorescence microscope (Axiophot; Carl Zeiss) and/or a laser scanning confocal microscope (LSCM Pascal5; Carl Zeiss). To confirm the specificity of detection, a series of control experiments were conducted as described previously (23).
Electron microscopy.
The midgut fourth section was dissected from adult females in 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2). The tissue was prefixed with the fixative at 4°C for 3 h and postfixed in 2% osmium tetroxide for 60 min at 4°C. After dehydration using an ethanol series, the materials were embedded in Spurr resin and cut into ultrathin sections (thickness, 75 nm), which were double stained with uranyl acetate and lead citrate and observed with a transmission electron microscope (H-7000; Hitachi).
Pulsed-field gel electrophoresis.
The dissected midgut fourth sections were homogenized in phosphate-buffered saline containing 10 mM EDTA and filtered through a 20-μm nylon net filter (Millipore). The symbiont cells obtained were embedded in 1% low-melting-point agarose, and agarose plugs were treated with proteinase K at 50°C overnight. After thorough washing, the symbiont DNA trapped in a plug was digested with a restriction enzyme (I-CeuI) and subjected to pulsed-field gel electrophoresis using a CHEF Mapper XA (Bio-Rad).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this study have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers AB548048 to AB548068.
RESULTS AND DISCUSSION
Phylogenetic placement of the symbiont of P. japonensis.
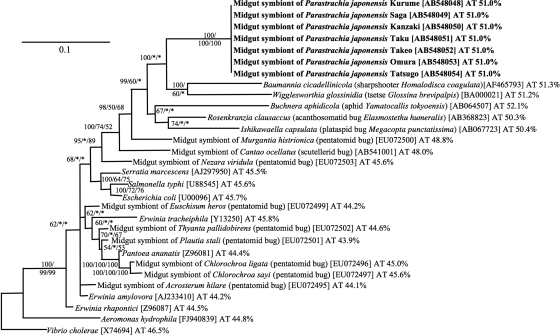
The 16S rRNA gene sequences were identical within and between the local populations of P. japonensis. Phylogenetic analyses showed that the symbiont of P. japonensis formed a distinct lineage in the Gammaproteobacteria, which had no close relatives but was allied with gut symbionts of acanthosomatid and plataspid stinkbugs, as well as with endocellular symbionts of sharpshooters, tsetse flies, and aphids (Fig. 2). Similar results were obtained with the gyrB and groEL gene sequences; the symbiont sequences were identical within and between the local populations and constituted a distinct lineage in the Gammaproteobacteria (see Fig. S1 and S2 in the supplemental material).
FIG. 2.
Molecular phylogenetic analysis of the symbiont of P. japonensis and representatives of the Gammaproteobacteria based on 16S rRNA gene sequences. A Bayesian phylogeny inferred from the results for 1,244 aligned nucleotide sites is shown. At each of the nodes, the posterior probability determined in the Bayesian analysis and bootstrap probabilities determined in the maximum-parsimony and maximum-likelihood analyses are indicated in that order. Branches supported by less than 50% posterior probability were collapsed into polytomies. Asterisks indicate bootstrap probabilities less than 50%. Host insects and sequence accession numbers are indicated in parentheses and brackets, respectively. The AT contents of the nucleotide sequences are also indicated.
Reductive genome evolution in the symbiont of P. japonensis.
The three symbiont gene sequences consistently had a remarkably AT-biased nucleotide composition (Fig. 2; see Fig. S1 and S2 in the supplemental material) and exhibited significantly accelerated molecular evolution compared with free-living gammaproteobacteria (Table 1). The size of the symbiont genome was estimated to be 0.85 Mb (Fig. 3), which was similar to the sizes of the genomes of obligate endocellular symbionts like Buchnera in aphids (∼0.6 Mb) (38) and Wigglesworthia in tsetse flies (∼0.7 Mb) (2), as well as the genomes of obligate gut symbionts like Ishikawaella in plataspid stinkbugs (∼0.8 Mb) (16) and Rosenkranzia in acanthosomatid stinkbugs (∼0.9 Mb) (23), but the symbiont genome was remarkably smaller than the genomes of free-living gammaproteobacteria like Escherichia coli (∼4.6 Mb) (3) and Vibrio cholerae (∼4.0 Mb) (13). This peculiar genetic trait has been identified in ancient endocellular symbiotic bacteria that cospeciated with their hosts (2, 38) and also in some gut symbiotic bacteria of stinkbugs (16, 23). The universal trend toward reductive genome evolution in the obligate mutualistic symbionts has been hypothesized to be related to attenuated purifying selection due to a small population size and substantial bottleneck, which are associated with the lifestyle of vertically transmitted symbionts (16, 26, 44). In this context, it is conjectured that the symbiont of P. japonensis has similarly been subjected to strict vertical transmission from mother to offspring over evolutionary time, presumably establishing an obligate mutualistic association. Recent comprehensive phylogenetic analyses suggested that stinkbugs of the genera Parastrachia and Dismegistus form a monophyletic group (11). Biological and coevolutionary aspects of the host-symbiont association in these stinkbug groups should be investigated.
TABLE 1.
Relative rate test for comparing the rates of molecular evolution of 16S rRNA, gyrB, and groEL gene sequences for the symbiont of P. japonensis and its free-living relatives
| Gene | Lineage 1 (accession no.) | Lineage 2 (accession no.) | Outgroup (accession no.) | K1a | K2b | K1 − K2 | K1/K2 | Pc |
|---|---|---|---|---|---|---|---|---|
| 16S rRNA | Gut symbiont of P. japonensis (AB548050) | Escherichia coli (U00096) and Salmonella enterica serovar Typhi (U88545) | Vibrio cholerae (X74694) | 0.076 | 0.038 | 0.038 | 2.0 | 0.00040 |
| gyrB | Gut symbiont of P. japonensis (AB548057) | Escherichia coli (NC_000913) and Salmonella enterica (NC_003198) | Vibrio cholerae (NC_002505) | 0.156 | 0.054 | 0.102 | 2.9 | 6.0 × 10−6 |
| groEL | Gut symbiont of P. japonensis (AB548064) | Escherichia coli (X07850) and Salmonella enterica serovar Typhi (U01039) | Vibrio cholerae (NC_009456) | 0.065 | 0.031 | 0.034 | 2.1 | 0.0034 |
K1 is the estimated mean distance between lineage 1 and the last common ancestor of lineages 1 and 2.
K2 is the estimated mean distance between lineage 2 and the last common ancestor of lineages 1 and 2.
P values were generated using the program RRTree (32).
FIG. 3.
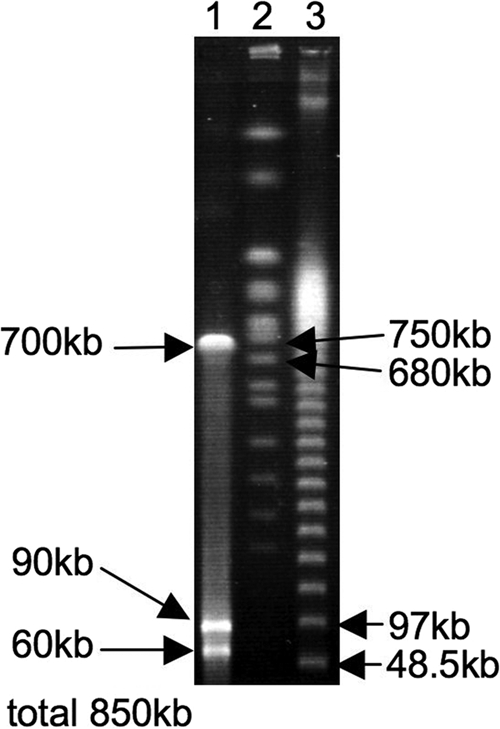
Genome size of the symbiont of P. japonensis estimated by pulsed-field gel electrophoresis. Lane 1, symbiont genomic DNA digested with I-CeuI; lanes 2 and 3, DNA size markers.
In vivo localization of the symbiont in P. japonensis.
In situ hybridization targeting 16S rRNA of the symbiont identified strong signals in the swollen crypts in the posterior part of the midgut fourth section (Fig. 1B and C). In contrast, few symbiont signals were observed in the smaller crypts in the anterior part of the midgut fourth section (Fig. 1B). In the other parts of the alimentary tract there were few symbiont signals, except for sporadic signals occasionally seen in the main tract of the midgut (Fig. 1B). Transmission electron microscopy showed that the lumen of the swollen crypts was filled with globular bacterial cells with a uniform morphotype (Fig. 1D and E), whereas the lumen of the smaller crypts in the anterior part was populated by few bacteria (data not shown). In the midgut fourth section, many crypts were arranged in two rows and fused into a ribbon-shaped structure (Fig. 1F). Histological examination confirmed that the lumen of the crypts was connected to the lumen of the midgut main tract (Fig. 1G). These results indicate that in P. japonensis (i) the midgut fourth section is differentiated into a swollen posterior part and a less-developed anterior part, (ii) the lumen of the crypts in the swollen part specifically harbors the symbiont and is a symbiotic organ, and (iii) the symbiont is also present in the main tract of the midgut, but in much smaller quantities. In other stinkbug groups, including the Pentatomidae, Plataspidae, Acanthosomatidae, Scutelleridae, and Alydidae, the midgut fourth section is uniformly populated by the symbiotic bacteria of the insects (1, 15, 18, 21, 23). To our knowledge, a symbiotic organ restricted to the posterior part of the midgut fourth section is unique to the parastrachiid stinkbug.
Vertical transmission route of the symbiont in P. japonensis.
The structural connection between the midgut crypts and the main tract (Fig. 1G) suggests that there is vertical transmission of the symbiont via anal excretion. However, the mechanism for vertical transmission of the symbiont in P. japonensis is not known and should be studied. Because of the peculiar ecology and behavior of this insect (7, 41), we suggest several possible mechanisms for vertical transmission of the symbiont. The symbiont may be transmitted by contamination of the egg surface with maternal symbiont-containing excrement upon oviposition, as reported for the Pentatomidae, Acanthosomatidae, Scutelleridae, and Pyrrhocoridae (1, 18, 19, 23, 30, 31). It is notable that females of P. japonensis deposit not only fertile eggs but also infertile trophic eggs, which are consumed by newborn nymphs. Experimental removal of the trophic eggs resulted in retarded nymphal growth and elevated mortality (14), suggesting that the trophic eggs may contribute not only nutrients but also the beneficial symbiont to the offspring, as the mother-made symbiont capsules do in the Plataspidae (8, 15, 16). Females of P. japonensis lay an egg mass in the nest, remain near the eggs until they hatch, and collect and provision fruits of Schoepfia jasminodora for the nymphs (43). Hence, it is conceivable that the symbiont is passed directly from the mother to nymphs during maternal care, as reported previously for a cydnid species (37).
Biological role of the symbiont in P. japonensis.
A previous study suggested that a bacterial symbiont is involved in uric acid recycling in the prereproductive nonfeeding period in P. japonensis (20). Considering the highly developed symbiotic system (Fig. 1) and the presumably long-lasting host-symbiont association (Fig. 2 and 3 and Table 1), it appears likely that the gut symbiont characterized in this study plays a biological role in uric acid recycling. Of course, the possibility that other minor bacterial associates that we failed to detect in this study are involved in uric acid recycling cannot be ruled out. Whether the symbiont itself has uricolytic activity or the uricolysis of the host insect is affected by symbiont infection should be examined in future studies.
Proposal of candidate name.
On the basis of the results described above, we propose the name “Candidatus Benitsuchiphilus tojoi” for the novel symbiont lineage. The generic name indicates the association with P. japonensis and refers to the Japanese name of this insect (Benitsuchi-Kamemushi). The specific epithet refers to Sumio Tojo, who, along with colleagues, proposed that there is symbiont-mediated uricolytic activity in P. japonensis during the prereproductive nonfeeding period (20).
Supplementary Material
Acknowledgments
We thank N. Baba, K. Inadomi, H. Mukai, and S. Tojo for providing insect samples.
This study was funded by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and by Grant-in-Aid for Scientific Research (B) 21370011 from the JSPS. T.H., Y.K., and M.H. were supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abe, Y., K. Mishiro, and M. Takanashi. 1995. Symbiont of brown-winged green bug, Plautia stali Scott. Jpn. J. Appl. Entomol. Zool. 39:109-115. [Google Scholar]
- 2.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Bourtzis, K., and T. Miller. 2003. Insect symbiosis. CRC Press, Boca Raton, FL.
- 5.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, New York, NY.
- 6.Cochran, D. G. 1985. Nitrogen excretion in cockroaches. Annu. Rev. Entomol. 30:29-49. [Google Scholar]
- 7.Filippi, L., M. Hironaka, and S. Nomakuchi. 2001. A review of the ecological parameters and implications of subsociality in Parastrachia japonensis (Hemiptera: Cydnidae), a semelparous species that specializes on a poor resource. Popul. Ecol. 43:41-50. [Google Scholar]
- 8.Fukatsu, T., and T. Hosokawa. 2002. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl. Environ. Microbiol. 68:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukatsu, T., and N. Nikoh. 1998. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl. Environ. Microbiol. 64:3599-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glasgow, H. 1914. The gastric caeca and the caecal bacteria of the Heteroptera. Biol. Bull. 3:101-171. [Google Scholar]
- 11.Grazia, J., R. T. Schuh, and W. C. Wheeler. 2008. Phylogenetic relationships of family groups in Pentatomoidea based on morphology and DNA sequences (Insecta: Heteroptera). Cladistics 24:932-976. [DOI] [PubMed] [Google Scholar]
- 12.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 13.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hironaka, M., S. Nomakuchi, S. Iwakuma, and L. Filippi. 2005. Trophic egg production in a subsocial shield bug, Parastrachia japonensis Scott (Heteroptera: Parastrachiidae), and its functional value. Ethology 111:1089-1102. [Google Scholar]
- 15.Hosokawa, T., Y. Kikuchi, X.-Y. Meng, and T. Fukatsu. 2005. The making of symbiont capsule in the plataspid stinkbug Megacopta punctatissima. FEMS Microbiol. Ecol. 54:471-477. [DOI] [PubMed] [Google Scholar]
- 16.Hosokawa, T., Y. Kikuchi, N. Nikoh, M. Shimada, and T. Fukatsu. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosokawa, T., Y. Kikuchi, M. Shimada, and T. Fukatsu. 2007. Obligate symbiont involved in pest status of host insect. Proc. R. Soc. B 274:1979-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiwa, N., T. Hosokawa, Y. Kikuchi, N. Nikoh, X.-Y. Meng, M. Ito, and T. Fukatsu. 16 April 2010, posting date. Primary gut symbiont and secondary Sodalis-allied symbiont in the scutellerid stinkbug Cantao ocellatus. Appl. Environ. Microbiol. doi: 10.1128/AEM.00421-10. [DOI] [PMC free article] [PubMed]
- 19.Kaltenpoth, M., S. A. Winter, and A. Kleinhammer. 2009. Localization and transmission route of Coriobacterium glomerans, the endosymbiont of pyrrhocorid bugs. FEMS Microbiol. Ecol. 69:373-383. [DOI] [PubMed] [Google Scholar]
- 20.Kashima, T., T. Nakamura, and S. Tojo. 2006. Uric acid recycling in the shield bug, Parastrachia japonensis (Hemiptera: Parastrachiidae), during diapause. J. Insect Physiol. 52:816-825. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi, Y., X.-Y. Meng, and T. Fukatsu. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikuchi, Y., T. Hosokawa, and T. Fukatsu. 2007. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 73:4308-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi, Y., T. Hosokawa, N. Nikoh, X.-Y. Meng, Y. Kamagata, and T. Fukatsu. 2009. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 25.Koga, R., T. Tsuchida, and T. Fukatsu. 2009. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl. Entomol. Zool. 44:281-291. [Google Scholar]
- 26.Moran, N. A. 1996. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U. S. A. 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller, H. J. 1956. Experimentelle Studien an der Symbiose von Coptosoma scutellatum Geoffr. (Hem. Heteropt.). Z. Morphol. Ökol. Tiere 44:459-482. [Google Scholar]
- 28.Potrikus, C. J., and J. A. Breznak. 1981. Gut bacteria recycle uric-acid nitrogen in termites—a strategy for nutrient conservation. Proc. Natl. Acad. Sci. U. S. A. 78:4601-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prado, S. S., and R. P. P. Almeida. 2009. Phylogenetic placement of pentatomid stink bug gut symbionts. Curr. Microbiol. 58:64-69. [DOI] [PubMed] [Google Scholar]
- 30.Prado, S. S., and R. P. P. Almeida. 2009. Role of symbiotic gut bacteria in the development of Acrosterum hilare and Murgantia histrionica. Entomol. Exp. Appl. 132:21-29. [Google Scholar]
- 31.Prado, S. S., D. Rubinoff, and R. P. P. Almeida. 2006. Vertical transmission of a pentatomid caeca-associated symbiont. Ann. Entomol. Soc. Am. 99:577-585. [Google Scholar]
- 32.Robinson-Rechavi, M., and D. Huchon. 2000. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics 16:296-297. [DOI] [PubMed] [Google Scholar]
- 33.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 34.Sabree, Z. L., S. Kambhampati, and N. A. Moran. 2009. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc. Natl. Acad. Sci. U. S. A. 106:19521-19526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasaki, T., M. Kawamura, and H. Ishikawa. 1996. Nitrogen recycling in the brown planthopper, Nilaparvata lugens: involvement of yeast-like endosymbionts in uric acid metabolism. J. Insect Physiol. 42:125-129. [Google Scholar]
- 36.Schaefer, C. W., L. Zheng, and S. Tachikawa. 1991. A review of Parastrachia (Hemiptera: Cydnidae: Parastrachiinae). Orient. Insects 25:131-144. [Google Scholar]
- 37.Schorr, H. 1957. Zür Verhaltensbiologie und Symbiose von Brachypelta aterrima Först (Cydnidae, Heteroptera). Z. Morphol. Ökol. Tiere 45:561-602. [Google Scholar]
- 38.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakai, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 39.Sweet, M. H., and C. W. Schaefer. 2002. Parastrachiinae (Hemiptera: Cydnidae) raised to family level. Ann. Entomol. Soc. Am. 95:441-448. [Google Scholar]
- 40.Swofford, D. L. 2001. PAUP* version 4.0b10. Sinauer, Sunderland, MA.
- 41.Tachikawa, S., and C. W. Schaefer. 1985. Biology of Parastrachia japonensis (Hemiptera: Pentatomoidea: ?-idae). Ann. Entomol. Soc. Am. 78:387-397. [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukamoto, L., and S. Tojo. 1992. A report of progressive provisioning in a stink bug, Parastrachia japonensis (Hemiptera: Cydnidae). J. Ethol. 10:21-29. [Google Scholar]
- 44.Wernegreen, J. J. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3:850-861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.