Abstract
Using RNA-based techniques and hot spring samples collected from Yunnan Province, China, we show that the amoA gene of aerobic ammonia-oxidizing archaea can be transcribed at temperatures higher than 74°C and up to 94°C, suggesting that archaeal nitrification can potentially occur at near boiling temperatures.
Aerobic ammonia-oxidizing archaea (AOA) are one major group of microorganisms mediating the autotrophic ammonia oxidation (2) which is central to the global nitrogen cycle (9). AOA possess an ammonia monooxygenase (AMO) which is the enzyme responsible for catalyzing aerobic ammonia oxidation, and its α subunit is encoded by the amoA gene (15). Multiple amoA gene-based molecular studies have demonstrated that AOA can be adapted to a large gradient of environmental variables with respect to temperature (0.2 to 97°C) and pH (2.5 to 9.0) (see the review by Erguder et al. [2] and the references therein). However, so far, only moderately thermophilic “Candidatus Nitrososphaera gargensis” and thermophilic “Candidatus Nitrosocaldus yellowstonii” have been obtained in culture and show the capability of oxidizing ammonia at high temperatures; they can produce nitrite at 46°C and 60 to 74°C at pHs 7 to 8, respectively (1, 4). In addition, Reigstad et al. (12) demonstrated biological ex situ nitrification at 85°C and pH 3.0, using terrestrial hot-spring samples. This indicated that the AMO enzyme is active at temperatures of up to 85°C. In the meantime, with the use of DNA-based molecular techniques, Reigstad et al. (12) and Zhang et al. (16) retrieved AOA amoA gene clone sequences from global terrestrial hot springs with a large gradient of pHs (2.5 to 9.0) and temperatures (38 to 97°C). However, the AOA amoA gene has never been transcribed from environments with temperatures higher than 74°C. In the present study, we performed RNA-based studies investigating the abundance and diversity in hot springs (temperature, 44.5 to 94.0°C; pH, 2.4 to 9.0) of Yunnan Province in southwestern China.
A total of 11 hot-spring samples were selected for field measurements and sample collection (Table 1). Hach kit-based field measurements showed that temperatures of the sampled hot springs ranged from 44.5°C to 94.0°C and pH from 2.4 to 9.0 (Table 1). Mats or mat-containing sinter/sediment samples were collected and subjected to RNA extraction with the use of a FastRNA Pro soil-direct kit (Qbiogene, Inc., CA) according to the manufacturer's protocols. The resulting crude RNA was digested with RNase-free DNase I (Takara, Japan). The DNase-digested RNA samples were verified to be free of genomic DNA contamination by PCR amplification with primer sets specific for total archaea, bacteria, and AOA according to conditions described elsewhere (see Table S1 in the supplemental material and cited references for details). The DNA-free RNA samples were reverse transcribed into cDNA by using the Promega AMV reverse transcription system (Promega Corporation, Madison, WI) as previously described (7). The archaeal amoA gene and total bacterial and archaeal 16S rRNA genes in the synthesized cDNA were quantified by qPCR (see Table S1 in the supplemental material) according our previous studies (6, 7). Bacterial and archaeal 16S rRNA gene abundances were on the magnitude of 108 to 1010 copies per gram of solids, and the AOA amoA gene abundance ranged from 4.5 × 104 to 3.52 × 106 copies per gram of solids in the investigated hot springs (Table 2). The abundance of the transcribed AOA amoA gene in high-temperature hot springs is comparable to those in low-temperature biotopes (7, 8, 11).
TABLE 1.
Water chemistry and temperatures of 11 hot-spring samples in Yunnan Province, China, and description of samples collected from these hot springsa
| Sample (GPS location) | Temp (°C) | pH | Concn of: |
Sample description | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SO42− (mM) | NO2− (μM) | NO3− (μM) | NH4+ (μM) | Fe2+ (μM) | Sulfide (μM) | TDS (g liter−1) | ||||
| DGG (24°57′12.7″/98°26′17.4″) | 84.0 | 7.6 | 3.13 | 1.02 | 8.23 | 1.18 | <0.04 | 9.69 | 5.95 | Gray sinter |
| DGGTYQ (24°57′12.7″/98°26′17.4″) | 87.0 | 2.4 | 5.21 | 0.54 | 0.97 | 69.41 | 303.57 | 6.56 | 4.53 | Black sediment |
| YJQ (24°57′03″/98°26′09.5″) | 94.0 | 9.0 | 0.07 | 0.96 | 9.03 | 0.59 | <0.04 | 150.00 | 5.80 | Brown sediment |
| ZZQ (24°57′03″/98°26′09.5″) | 89.5 | 3.5 | 0.73 | 0.72 | 3.06 | 20.00 | 17.86 | 1.25 | 0.70 | Brown sediment |
| WMXX (24°56′59.6″/98°26′15.7″) | 64.8 | 6.7 | 0.34 | 3.52 | 7.58 | <0.5 | 0.71 | <0.3 | 2.33 | Black microbial mat |
| DHB (24°39′37.2″/98°43′11.9″) | 44.5 | 7.1 | 0.30 | 0.07 | 0.48 | <0.5 | 0.36 | <0.3 | 0.82 | Black microbial mat |
| BLZ1 (24°39′23.3″/98°40′03.4″) | 60.0 | 6.3 | 0.42 | 0.28 | <0.7 | <0.5 | <0.04 | <0.3 | 0.96 | Black sediment |
| BLZ2 (24°39′23.3″/98°40′03.4″) | 55.0 | 6.7 | 0.49 | 0.22 | <0.7 | 1.18 | <0.04 | <0.3 | 1.80 | Black sediment |
| NJYPZT (26°15′01.2″/99°59′22.2″) | 70.0 | 7.9 | 0.31 | 1.28 | 4.68 | 8.24 | 0.54 | <0.3 | 1.94 | Rufous sediment |
| LZT1 (26°14′57.1″/99°59′31.0″) | 62.0 | 6.9 | 1.46 | 2.41 | 5.81 | 2.94 | 0.54 | <0.3 | 2.19 | Black sediment |
| LZT2 (26°14′58.3″/99°59′32.6″) | 80.0 | 7.25 | 1.25 | 1.04 | 1.77 | 12.35 | 0.18 | <0.3 | 2.20 | Black sediment |
Samples from Tengchong, Yunnan Province, are abbreviated as follows: DGG, Dagunguo; DGGTYQ, Dagunguo-Tiyanqu; YJQ, Yanjingquan; ZZQ, Zhenzhuquan; WMXX, Wuming-Xiaoxi. Samples from Longling, Yunnan Province, are abbreviated as follows: DHB, Dahebian; BLZ1, Balazhang 1; BLZ2, Balazhang 2. Samples from Eryuan, Yunnan Province, are abbreviated as follows: NJYPZT, Niujie-Yongping-Zaotang; LZT1, Laizitang 1; LZT2, Laizitang 2. TDS, total dissolved solids.
TABLE 2.
Abundance of 16S rRNA genes and archaeal amoA genes and sequencing information for 11 hot-spring samples collected from Yunnan Province, Chinab
| Sample | No. of copies g−1 of: |
na | Coverage (%) at cutoff of: |
No. of OTUs at cutoff of: |
H′ at cutoff of: |
Chao1 at cutoff of: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA gene |
Archaeal amoA gene |
||||||||||||||
| Bacteria | SD | Archaea | SD | amoA | SD | 2% | 5% | 2% | 5% | 2% | 5% | 2% | 5% | ||
| DGG | 1.58 × 109 | 2.94 × 104 | 7.94 × 108 | 1.76 × 105 | 4.50 × 104 | 2.17 × 102 | 22 | 100.00 | 100.00 | 4 | 3 | 1.0 | 1.0 | 4.0 | 4.0 |
| DGGTYQ | 2.69 × 109 | 1.55 × 104 | 4.63 × 108 | 3.95 × 104 | 3.52 × 106 | 6.53 × 103 | 31 | 93.55 | 93.55 | 5 | 5 | 1.1 | 1.0 | 5.5 | 5.5 |
| YJQ | 2.93 × 1010 | 2.62 × 105 | 3.82 × 1010 | 4.11 × 105 | 2.60 × 106 | 1.98 × 103 | 28 | 100.00 | 100.00 | 5 | 5 | 1.7 | 1.7 | 7.0 | 7.0 |
| ZZQ | 7.42 × 108 | 1.25 × 103 | 1.64 × 108 | 3.68 × 104 | 4.25 × 105 | 5.22 × 102 | 35 | 94.29 | 94.29 | 5 | 5 | 1.1 | 1.0 | 5.5 | 5.5 |
| WMXX | 8.95 × 109 | 2.94 × 104 | 2.28 × 109 | 1.99 × 105 | 1.25 × 106 | 1.91 × 103 | 32 | 96.88 | 96.88 | 4 | 4 | 1.1 | 1.0 | 4.0 | 4.0 |
| DHB | 1.22 × 1011 | 3.27 × 105 | 1.09 × 1010 | 1.60 × 105 | 2.30 × 106 | 2.10 × 103 | 26 | 53.85 | 58.33 | 16 | 13 | 2.8 | 2.4 | 60.0 | 30.0 |
| BLZ1 | 1.04 × 1011 | 3.09 × 106 | 2.22 × 1010 | 1.70 × 105 | 1.28 × 106 | 1.21 × 102 | 25 | 96.00 | 96.00 | 3 | 3 | 0.8 | 0.8 | 3.0 | 3.0 |
| BLZ2 | 3.33 × 109 | 8.16 × 103 | 2.16 × 109 | 4.32 × 104 | 5.34 × 105 | 5.81 × 102 | 31 | 90.32 | 90.32 | 9 | 8 | 1.6 | 1.7 | 14.0 | 8.5 |
| NJYPZT | 2.66 × 1010 | 2.49 × 104 | 4.85 × 109 | 3.27 × 104 | 5.34 × 105 | 9.81 × 101 | 25 | 76.00 | 92.00 | 11 | 8 | 2.1 | 1.7 | 14.8 | 7.3 |
| LZT1 | 1.60 × 109 | 4.32 × 103 | 1.99 × 109 | 2.49 × 104 | 5.62 × 105 | 4.22 × 102 | 43 | 83.72 | 90.70 | 12 | 10 | 2.1 | 1.7 | 20.5 | 12.0 |
| LZT2 | 4.20 × 109 | 1.71 × 104 | 1.80 × 109 | 8.38 × 104 | 9.91 × 104 | 1.53 × 102 | 39 | 92.31 | 92.31 | 6 | 6 | 1.3 | 1.2 | 8.5 | 6.0 |
Number of clones.
The diversity indices were derived from clone libraries.
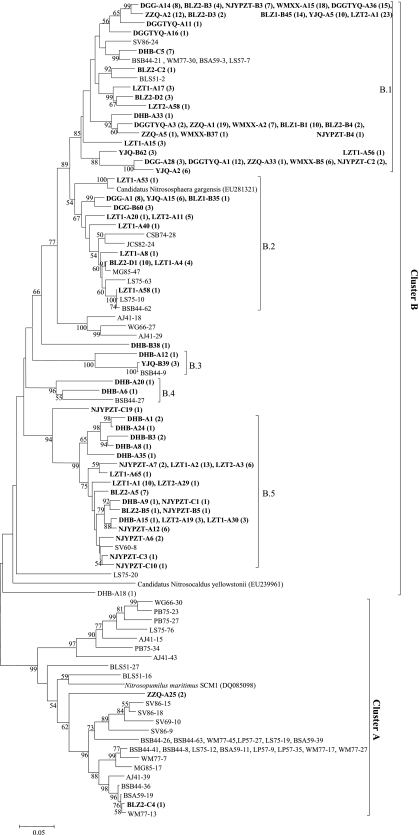
The cDNA samples were PCR amplified using AOA-specific primer sets (see Table S1 in the supplemental material) as described previously (7). The resulting PCR products were used for constructing the amoA gene clone libraries according to established procedures (7). A total of 337 AOA amoA gene clones were randomly selected for sequencing, and the obtained sequences (Table 1) were subjected to operational taxonomic unit (OTU) analysis by using DOTUR 1.53 (13), with cutoffs of 2% and 5% (3). The diversity indices of Shannon (H′) and Chao1 were also calculated using DOTUR. One sequence from each OTU was then selected as a representative for phylogenetic analysis. The number of clones in each sample represented 54 to 100% coverage (at 2% cutoff) for the clone libraries (Table 2). The representative sequences at 2% cutoff, reference sequences from a report by Zhang et al. (16), and amoA gene sequences of “Candidatus Nitrosopumilus maritimus,” “Candidatus Nitrososphaera gargensis,” and “Candidatus Nitrosocaldus yellowstonii” were combined for phylogenetic analysis using the MEGA 4.1 (14). The amoA phylogenetic nomenclature in the report by Zhang et al. (16) was employed in this study (Fig. 1). The phylogenetic analysis showed that only two amoA gene clones retrieved in this study were affiliated with the cluster A named by Zhang et al. (16). In contrast, 99% of clones retrieved in this study were classified into the cluster B and distributed into three groups: B.1, B.2, and B.5 (Fig. 1). The retrieved sequences in the B.1 and B.2 groups were related (90 to 99%) to those from Tengchong hot springs that were determined by Zhang et al. (16) (Fig. 1). The B.2 clones were related (identity 90 to 99%) to moderately thermophilic “Candidatus Nitrososphaera gargensis” (4). In addition, all clone sequences in the cluster B were distantly (<80% identity) related to thermophilic “Candidatus Nitrosocaldus yellowstonii” (1) (Fig. 1).
FIG. 1.
Neighbor-joining tree (partial sequences, ∼635 bp) showing the phylogenetic relationships of archaeal amoA gene sequences cloned from this study and that of Zhang et al. (16) and amoA gene sequences of three AOA isolates or cultures. Clone sequences from this study are shown in boldface type. One representative clone within each OTU is shown, and the number of clones within each OTU is shown in parentheses. The classification system for clusters A and B in the report by Zhang et al. (16) was employed in this study. Scale bars indicate the Jukes-Cantor distances. Bootstrap values of >50% (for 1,000 iterations) are shown.
Previous studies indicated that environmental factors (e.g., ammonium concentration, organic carbon, temperature, salinity, dissolved oxygen [DO], pH, sulfide, and phosphate levels) may affect AOA distributions (2, 10). In order to evaluate the correlation of the measured geochemical variables with amoA gene abundance and diversity in this study, the simple Mantel tests were performed using the zt software (http://www.psb.ugent.be/∼erbon/mantel/) according to established procedures (5). Significant positive correlation (r > 0.5; P < 0.05) was present between the AOA amoA gene abundance (either absolute or relative) and a number of environmental variables but absent (r < 0.5) between the AOA amoA gene abundance and the measured environmental variables (see Table S2 in the supplemental material). Without further investigation, however, it is uncertain whether the observed positive correlations are real or just coincidental.
In summary, our data show that the AOA amoA gene can be transcribed in hot-spring samples with temperatures higher than 74°C and up to 94°C. However, ex situ experiments are required to verify the potential activity of AOA at such high temperatures and to find the reasons for the observed correlations between the AOA amoA gene and measured environmental variables. This will be a major focus of our future research.
Nucleotide sequence accession numbers.
Sequences were deposited in the GenBank database under accession numbers GQ226055 to GQ226135.
Supplementary Material
Acknowledgments
This research was supported by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, China Postdoctoral Science Foundation (20090450457), and the China University of Geosciences (Beijing) Teaching Laboratory funds (H.J.), National Basic Research Program of China (W.L.) (2010CB833800), National Science Foundation of China (40972211) (C.Z.), COMRA fund grant (DYXM-115-02-2-17) (P.W.), and Research Fund of the State Key Laboratory of Geological Processes and Mineral Resources (GPMR2008K08B) of China University of Geosciences—Beijing and the 111 Project (no. B07011) of China (H.D.).
We are grateful to Shicai Deng at China University of Geosciences, Beijing; Yan Li and Xiaoyang Zhi at Yunnan University; and Libo Yu at Third Institute of Oceanography, SOA, for helping with sample collection and qPCR work. We are grateful to three anonymous reviewers whose constructive criticisms significantly improved the quality of the manuscript.
Footnotes
Published ahead of print on 30 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.de la Torre, J. R., C. B. Walker, A. E. Ingalls, M. Konneke, and D. A. Stahl. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810-818. [DOI] [PubMed] [Google Scholar]
- 2.Erguder, T. H., N. Boon, L. Wittebolle, M. Marzorati, and W. Verstraete. 2009. Environmental factors shaping the ecological niches of ammonia oxidizing archaea. FEMS Microbiol. Rev. 33:855-869. [DOI] [PubMed] [Google Scholar]
- 3.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Nat. Acad. Sci. U. S. A. 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatzenpichler, R., E. V. Lebedeva, E. Spieck, K. Stoecker, A. Richter, H. Daims, and M. Wagner. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang, H., H. Dong, S. Deng, B. Yu, Q. Huang, and Q. Wu. 2009. Response of archaeal community structure to environmental changes in lakes on the Tibetan Plateau, northwestern China. Geomicrobiol. J. 26:289-297. [Google Scholar]
- 6.Jiang, H., H. Dong, B. Yu, Y. Li, S. Ji, X. Liu, and C. Zhang. 2007. Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan Plateau. Environ. Microbiol. 9:2603-2621. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, H., H. Dong, B. Yu, G. Lv, S. Deng, N. Berzins, and M. Dai. 2009. Diversity and abundance of ammonia-oxidizing archaea and bacteria in Qinghai Lake, northwestern China. Geomicrobiol. J. 26:199-211. [Google Scholar]
- 8.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 9.Madigan, M. T., J. M. Martinko, P. V. Dunlap, and D. P. Clark. 2010. Brock biology of microorganisms, 12th ed. Benjamin Cummings, Menlo Park, CA.
- 10.Martens-Habbena, W., P. M. Berube, H. Urakawa, J. R. de la Torre, and D. A. Stahl. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976-981. [DOI] [PubMed] [Google Scholar]
- 11.Nicol, G. W., and C. Schleper. 2006. Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol. 14:207-212. [DOI] [PubMed] [Google Scholar]
- 12.Reigstad, L. J., A. Richter, H. Daims, T. Urich, L. Schwark, and C. Schleper. 2008. Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol. Ecol. 64:167-174. [DOI] [PubMed] [Google Scholar]
- 13.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 15.Treusch, A. H., S. Leininger, A. Kletzin, S. C. Schuster, H. P. Klenk, and C. Schleper. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985-1995. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, C. L., Q. Ye, Z. Huang, W. Li, J. Chen, Z. Song, W. Zhao, C. Bagwell, W. P. Inskeep, C. Ross, L. Gao, J. Wiegel, C. S. Romanek, E. L. Shock, and B. P. Hedlund. 2008. Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl. Environ. Microbiol. 74:6417-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.