Abstract
Listeria monocytogenes strains are classified in at least three distinct phylogenetic lineages. There are correlations between lineage classification and source of bacterial isolation; e.g., human clinical and food isolates usually are classified in either lineage I or II. However, human clinical isolates are overrepresented in lineage I, while food isolates are overrepresented in lineage II. σB, a transcriptional regulator previously demonstrated to contribute to environmental stress responses and virulence in L. monocytogenes lineage II strains, was hypothesized to provide differential abilities for L. monocytogenes survival in various niches (e.g., food and human clinical niches). To determine if the contributions of σB to stress response and virulence differ across diverse L. monocytogenes strains, ΔsigB mutations were created in strains belonging to lineages I, II, IIIA, and IIIB. Paired parent and ΔsigB mutant strains were tested for survival under acid and oxidative stress conditions, Caco-2 cell invasion efficiency, and virulence using the guinea pig listeriosis infection model. Parent and ΔsigB mutant strain transcriptomes were compared using whole-genome expression microarrays. σB contributed to virulence in each strain. However, while σB contributed significantly to survival under acid and oxidative stress conditions and Caco-2 cell invasion in lineage I, II, and IIIB strains, the contributions of σB were not significant for these phenotypes in the lineage IIIA strain. A core set of 63 genes was positively regulated by σB in all four strains; different total numbers of genes were positively regulated by σB in the strains. Our results suggest that σB universally contributes to L. monocytogenes virulence but specific σB-regulated stress response phenotypes vary among strains.
Listeria monocytogenes is a Gram-positive, rod-shaped bacterium that can cause listeriosis, a life-threatening invasive disease in humans and animals. While listeriosis is rarely diagnosed in healthy individuals, the elderly, the immunocompromised, and pregnant women and their fetuses are particularly at risk of infection. The vast majority (99%) of listeriosis infections are food borne, and approximately 20% of diagnosed infections result in death (36).
Increasing evidence indicates that L. monocytogenes strains belong to multiple lineages that appear to differ in their abilities to be transmitted to humans (20). Characterization of L. monocytogenes isolates from a variety of hosts and environments by multiple subtyping methods (35, 63), including multilocus enzyme electrophoresis (MLEE) (47), has shown that strains comprising the species L. monocytogenes belong to two major divisions designated lineages I and II and at least one additional distinct genetic lineage. The third lineage, lineage III, can be subdivided into lineages IIIA/C and IIIB (lineage IIIB has recently been classified as lineage IV [65]), based on results obtained with a number of molecular subtyping strategies, including pulsed-field gel electrophoresis (PFGE) (20) and virulence gene sequencing (20, 68). While different nomenclatures have been used for these L. monocytogenes lineages (47, 53), the main groups described in multiple studies consistently include the same L. monocytogenes serotypes (20, 53, 66, 68). Based on the most commonly applied lineage designations (20), lineage I includes predominantly strains with the 1/2b, 3b, 3c, and 4b serotypes and lineage II includes primarily strains with the 1/2a, 1/2c, and 3a serotypes (38). Strains associated with human clinical listeriosis cases are significantly overrepresented in lineage I compared to strains associated with animal listeriosis cases or contaminated foods (20). Lineage I isolates appear to have significantly greater pathogenic potential than lineage II strains, as suggested by their relatively enhanced ability to spread to neighboring host cells in a cell culture plaque assay (20, 68). On the other hand, lineage II strains are significantly more common among food isolates than among isolates from human listeriosis cases (20). However, preferential recovery of lineage II strains over lineage I strains may occur with at least some selective media and isolation protocols (5), which may have contributed to this observation. Lineage III includes predominantly strains with the 4a and 4c serotypes, as well as some serotype 4b strains that are distinct from the strains grouped in lineage I (20, 26). Strains classified in lineage III appear to be associated with isolation from animals, occasionally with isolation from human listeriosis cases, and only rarely with isolation from foods (20).
L. monocytogenes survival and persistence in diverse environments, including food processing plants, are facilitated by its ability to survive and grow at wide ranges of temperatures (0 to 45°C) (14), pH values (pH 4.4 to 9.4) (14), and other environmental conditions. The σB general stress regulator, which is conserved across many Gram-positive bacteria, including Staphylococcus aureus (69), Bacillus anthracis (16), and Bacillus licheniformis (3), has been shown to contribute to bacterial survival following exposure to environmental stresses, including acid, oxidative, and energy stresses (9, 15, 67). In the L. monocytogenes strains evaluated to date, σB positively regulates at least 160 genes directly and as many as 200 genes both directly and indirectly (48), including transcription of a number of virulence genes (e.g., prfA, bsh, inlA, and inlB) (30, 31, 33, 44, 48, 49). An L. monocytogenes 10403S ΔsigB null mutant has reduced invasiveness in human intestinal epithelial cells (32) and reduced virulence in intragastrically inoculated guinea pigs (17).
The contributions of σB to L. monocytogenes stress responses and virulence have been characterized most thoroughly for lineage II strains, including strains 10403S (44, 48) and EGD-e (21, 53). For example, the relative virulence characteristics of the lineage II strain 10403S and its isogenic ΔsigB mutant have been evaluated in the guinea pig model, but contributions of σB to gastrointestinal infection have not been reported for other L. monocytogenes lineages. Information about the contributions of σB to regulation of gene expression in strains belonging to lineages I, IIIA, and IIIB is just emerging. A recent comparative transcriptomic study found that sigB (lmo0895) was expressed at higher levels in lineage II strains than in lineage I strains (53). Also, a number of previously identified σB-regulated genes (30, 48) were expressed at higher levels in lineage II strains than in lineage I strains; these genes include opuCA and lmo1421, which encode a known compatible solute transporter protein and a putative compatible solute transporter protein, respectively. With respect to the stress response phenotype, Moorhead and Dykes (37) reported that the relative importance of σB in stress responses was not the same in all strains of L. monocytogenes, as assessed by comparison of the survival of two ΔsigB mutants having different serotypes under various stress conditions. Specifically, there were greater differences between a serotype 1/2a strain and its ΔsigB mutant than between a serotype 4a wild-type strain and its ΔsigB mutant.
We hypothesized that the inherent differences in the abilities of L. monocytogenes strains to survive under stress conditions and subsequently cause disease may be at least partially attributable to differences in the contributions of σB among strains. To determine if the contributions of σB to stress responses and virulence differ across diverse L. monocytogenes strains, we evaluated L. monocytogenes lineage I, II, IIIA, and IIIB strains using a combination of phenotypic and transcriptomic analyses, including a multigenome microarray to characterize differences in the σB regulons. Each pair of isogenic parent and ΔsigB strains was also assayed for survival under acid and oxidative stress conditions, invasiveness in a Caco-2 cell model, and virulence in the guinea pig listeriosis infection model.
MATERIALS AND METHODS
Bacterial strains and storage.
The strains selected for this study included L. monocytogenes lineage I, II, IIIA, and IIIB isolates (Table 1). FSL J1-194 (lineage I) was isolated from a sporadic human listeriosis case (50). It is serotype 1/2b, which is commonly associated with human disease (29), and ribotype DUP-1042B, which is considered a ribotype with enhanced virulence characteristics (20), as demonstrated by plaques in tissue culture plaque assays that are larger than the plaques formed by strains belonging to other ribotypes. 10403S is a laboratory type strain (2) and was selected (i) because it is a lineage II strain and (ii) to facilitate comparisons between this study and previous studies that also used this strain (10, 15, 17). FSL J2-071 and FSL J1-208 were selected from available lineage IIIA and IIIB strains, respectively. Both of these strains were isolated from clinical animal cases, and they are serotypes 4c and 4a, respectively, which are common in lineage III strains (38). While the strains selected for this study represent L. monocytogenes diversity, additional strains belonging to each lineage would have to be evaluated to determine if the strains used here are representative of the lineages (12). FSL A1-254, a 10403S isogenic ΔsigB mutant, was used in previous studies (9, 15, 67). Isogenic sigB null (ΔsigB) mutations were created in the other three strains using allelic exchange mutagenesis, as described previously (25, 67). Primers used for creation of mutants are shown in Table S1 in the supplemental material. Stock cultures were stored at −80°C in brain heart infusion (BHI) containing 15% glycerol. Cultures were streaked onto BHI agar (Difco, Detroit, MI) and incubated at 37°C for 24 h to obtain isolated colonies for inoculation of overnight cultures. The growth conditions for each experiment below are described.
TABLE 1.
Strains used in this study
| Lineage | Strain | Serotype | Ribotype | Origin | Reference(s) |
|---|---|---|---|---|---|
| I | FSL J1-194 | 1/2b | DUP-1042B | Human clinical isolate | 50 |
| I | FSL C6-001 (ΔsigB) | 1/2b | DUP-1042B | FSL J1-194 | This study |
| II | 10403S | 1/2a | DUP-1030A | Laboratory type strain | 2 |
| II | FSL A1-254 (ΔsigB) | 1/2a | DUP-1030A | 10403S | 15, 67 |
| IIIA | FSL J2-071 | 4c | DUP-1061A | Bovine clinical isolate | 68 |
| IIIA | FSL O1-006 (ΔsigB) | 4c | DUP-1061A | FSL J2-071 | This study |
| IIIB | FSL J1-208 | 4a | DUP-10142 | Caprine clinical isolate | 68 |
| IIIB | FSL O1-005 (ΔsigB) | 4a | DUP-10142 | FSL J1-208 | This study |
Acid and oxidative stress survival assays.
For acid and oxidative stress survival assays, strains were grown in 5 ml of BHI broth at 37°C with shaking (230 rpm) for 12 h. A 1% inoculum was transferred to 5 ml of preheated BHI broth and grown to an optical density at 600 nm (OD600) of ∼0.4. After growth to an OD600 ∼0.4, a ∼1% (vol/vol) inoculum was transferred to 50 ml of prewarmed (37°C) BHI broth in a 300-ml nephelo flask (Bellco, Vineland, NJ) to obtain a calculated OD600 of 0.004 (the volume transferred was adjusted based on the actual OD600 of the starting inoculum). Following the second passage, strains were grown to stationary phase (defined as 10 h after inoculation of the flask). Two 5-ml aliquots of stationary-phase cells were transferred to sterile 16-mm test tubes. For acid stress tests, one aliquot was used to determine the volume (in μl) of 12 N HCl (VBR, Westchester, PA) needed to reduce the pH of the culture to 2.5, as determined by direct measurement with a pH meter (Beckman, Coulter Inc., Fullerton, CA); the second aliquot was used for an experiment. Following addition of 12 N HCl to the experimental cultures, they were gently vortexed and immediately returned to incubation at 37°C with shaking. Aliquots of the acidified cultures were removed at 10, 30, and 60 min after acidification. Bacteria were quantified by serial dilution and standard plate counting. For oxidative stress tests, cumene hydroperoxide (CHP) (Sigma-Aldrich) that had been dissolved in dimethyl sulfoxide (DMSO) was added to 900-μl portions of 10-h cultures in 1.5-ml Eppendorf tubes to obtain a final CHP concentration of 13.0 mM. The tubes were incubated for 15 min at 37°C with shaking. An equal volume of DMSO was added to nonstressed control cultures; survival was assessed by serial dilution and standard plate counting. At least three independent replicates were performed for each L. monocytogenes strain tested under each condition.
Caco-2 cell invasion assay.
Caco-2 invasion assays were performed as previously described (40). Confluent Caco-2 monolayers were inoculated with 10 μl of a stationary-phase culture (approximately 4.8 × 107 cells/well) grown as described above for the acid and oxidative stress survival assays. Intracellular L. monocytogenes cells were enumerated by spiral plating (Autoplate 4000; Spiral Biotech, Norwood, MA) 10-fold dilutions of lysed Caco-2 cell suspensions in phosphate-buffered saline (PBS) on BHI agar plates. The invasion efficiency was expressed as the log ratio of the L. monocytogenes cells recovered to the cells in the initial inoculum. Three independent invasion assays were performed for each L. monocytogenes strain tested.
Statistical analyses of stress and invasion assays.
Statistical analyses were performed with Statistical Analysis Software (SAS) 9.0 (SAS Institute, Inc., Cary, NC). Regression analysis was used to calculate the death rate for cells exposed to pH 2.5, which was expressed as average log number of CFU that died per hour for each strain. Repeated-measures analysis of variance (ANOVA) was used to test if there was a significant difference in the death rate between a wild-type strain and its isogenic ΔsigB mutant. Two-sided t tests were used to test for differences in cell death due to oxidative stress and in invasion efficiency between wild-type and ΔsigB strains. For all statistical analyses a P value of < 0.05 was considered significant. ANOVA was also used to test for differences in stress survival and in invasion efficiency among the four wild-type strains.
Microarray, cDNA labeling, and microarray hybridization.
Bacteria were initially grown in 5 ml of BHI broth at 37°C with shaking (230 rpm) for 15 h. A 1% inoculum was transferred to 5 ml of prewarmed BHI broth and grown to an OD600 of ∼0.4. After growth to an OD600 of ∼0.4, a ∼1% (vol/vol) inoculum was transferred to 50 ml of prewarmed (37°C) BHI broth in a 300-ml nephelo flask (Bellco, Vineland, NJ) to obtain a calculated OD600 of 0.004 (the volume transferred was adjusted based on the actual OD600 of the starting inoculum). Following the second passage, cells were collected at stationary phase (defined as growth to an OD600 of 1.0, followed by incubation for an additional 3 h). Prior to centrifugation, RNAProtect bacterial reagent (Qiagen, Valencia, CA) was added to the cultures according to the manufacturer's instructions to stabilize the mRNA; pellets were stored at −80°C prior to RNA isolation. RNA was isolated as previously described (48). Briefly, bacterial cells were lysed enzymatically with lysozyme (Fisher Scientific, Pittsburg, PA) and mechanically using six bursts of sonication (Misonix, Farmingdale, NY) at 18 W on ice for 30 s. Total RNA was isolated and purified using an RNeasy midi kit (Qiagen) according to the manufacturer's protocol. RNA was eluted from the column using RNase-free water. Total RNA was incubated with RNasin (Promega, Madison, WI) to inhibit RNases and with RQ1 DNase (Promega) to remove contaminating DNA. After phenol-chloroform extraction, UV spectrophotometry (Nanodrop, Wilmington, DE) was used to quantify and assess the purity of the RNA. RNA integrity was assessed by agarose gel electrophoresis. Purified RNA was stored in RNase-free water at −80°C prior to reverse transcription (RT).
The Pathogen Functional Genomics Resource Center (PFGRC)/J. Craig Venter Institute (JCVI) L. monocytogenes microarray (version 2) was used to identify mRNA transcript level differences between wild-type strains and their ΔsigB mutants. PFGRC/JCVI microbial RNA aminoallyl labeling for microarray standard operating procedure M007 (http://pfgrc.jcvi.org/index.php/microarray/protocols.html) was used to reverse transcribe and label the total RNA. Spectrophotometry was used to quantify cDNA and the total numbers of picomoles of Cy3 or Cy5 (Amersham Biosciences, Piscataway, NJ) incorporated. Cy3-labeled cDNA and Cy5-labeled cDNA were combined and thoroughly dehydrated prior to hybridization. PFGRC/JCVI standard operating procedure M008 (http://pfgrc.jcvi.org/index.php/microarray/protocols.html) was used to hybridize the Cy3 and Cy5 dye-labeled cDNA to the microarray. Briefly, microarray slides were blocked in a prehybridization buffer supplemented with bovine serum albumen (BSA) (Sigma-Aldrich) and washed with deionized water and then with isopropyl alcohol. Dried Cy3- and Cy5-labeled cDNA was hydrated with hybridization buffer containing 0.6 mg/ml sheared salmon sperm DNA (Invitrogen), denatured twice at 95°C for 5 min, and briefly centrifuged. cDNA was hybridized to the microarray at 42°C for 16 h. Following hybridization, slides were washed using the PFGRC/JCVI M008 protocol and dried by centrifugation.
Microarray image processing, replicates, and statistical analysis.
Image processing and analyses were performed as previously described (48). Data preprocessing and statistical analyses were performed using the LIMMA package available from the BioConductor software project for the R programming environment (18, 54). Background correction was performed using the “normexp” method to produce more robust ratios for low-intensity spots, and print tip normalization was used to correct for spatial effects and dye intensity bias (56). The empirical Bayes approach was used to assess differential expression (55). Three replicates were performed for each comparison of wild-type and isogenic ΔsigB strains. For each probe, fold changes, moderated t-statistics, and P values (adjusted for multiple tests by controlling for the false discovery rate) were calculated. Raw and normalized microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE21427.
Hybridization index, identification of differentially expressed genes, and association of σB-dependent genes with JCVI role categories.
The PFGRC/JCVI L. monocytogenes microarray contained multiple probes for certain loci that lacked a 70-mer consensus sequence for a given locus for the four strains represented on the microarray. When multiple probes for a single locus were present, the probe that best matched the sequence for a given strain used in the present study was selected as the representative probe for subsequent analyses of that strain. BLASTN was used to calculate the hybridization index (HI), which was the level of identity (expressed as a percentage) between each probe and each strain; BLASTN results were parsed to compare levels of identity between each strain and the probes. EGD-e locus names were used to describe the corresponding open reading frames (ORFs) in the lineage I, IIIA, and IIIB strains as the genomes of these strains have not been annotated yet; EGD-e locus names were also used in lieu of 10403S locus names to facilitate comparisons among all of the strains in this study. The locus names of genes unique to F2365, F6852, and H7858 (i.e., genes with no EGD-e homologue) were used in this study. Genes whose transcript levels were higher in a wild-type strain than in the isogenic ΔsigB mutant (≥1.5-fold change) and whose adjusted P values were <0.05 were considered positively differentially expressed and thus σB dependent. A χ2 test for trend was used to determine if there was an association between σB-dependent expression and JCVI role category. Subsequent Fisher's exact tests were used to determine which role categories were significantly associated with σB-dependent genes. P values of <0.05 were considered significant. The odds ratio (OR) for the presence of a σB-dependent gene in a given category rather than the other categories was determined.
TaqMan qRT-PCR and putative σB promoter comparisons.
TaqMan quantitative reverse transcription-PCR (qRT-PCR) was used to confirm that there were lineage-specific σB-dependent genes. Total RNA was extracted from cells grown as described above for microarray analyses using an Ambion MicrobExpress kit (Ambion). RNA quality and integrity were assessed with an Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA), and the total RNA concentration was checked by spectrophotometry (Nanodrop). qRT-PCR was performed using an ABI Prism 7000 sequence detection system (Applied Biosystems) essentially as previously described (58), except that RNA was reverse transcribed to cDNA with cDNA reverse transcription reagents (Applied Biosystems) using random hexamers prior to quantification of transcript levels. To evaluate residual genomic DNA levels, the same reaction was performed with each sample without reverse transcriptase. In previous studies by our group (31, 58), the rpoB primers and probe were used to calculate the residual DNA copy number and to normalize transcript levels as there is minimal variation in the transcript level of this housekeeping gene under different conditions. Primers (IDT DNA, Coralville, IA) and probes (Applied Biosystems) used in this study (see Table S2 in the supplemental material) were designed with Primer Express (Applied Biosystems) based on a consensus sequence for the gene in the four wild-type strains. For some strains, the DNA sequence of a gene, including at least 200 nucleotides upstream of the coding region, was not available. To PCR amplify and sequence the region of interest for these strains, primers were designed based on the sequences of other wild-type strains. TaqMan primers and probes were designed based on the conserved sequence for all strains used in this study.
Guinea pig listeriosis infection model.
The guinea pig listeriosis infection model was used to assess the virulence of L. monocytogenes parent and ΔsigB strains as previously described by Garner et al. (17). Bacteria were grown as described above for RNA isolation, except that cells were grown to early stationary phase (defined as growth to an OD600 of 0.8, followed by incubation for an additional 1 h) for consistency with previous guinea pig listeriosis infection model studies (17). Aliquots of early-stationary-phase bacterial cultures were concentrated by centrifugation and resuspended in 1 ml PBS (pH 7.4) containing 15% glycerol. Cells were frozen and stored in multiple aliquots at −80°C for use in three replicates; an aliquot of each strain was thawed, and the cells were enumerated by serial dilution and spiral plating to determine cell viability 1 day prior to inoculation of animals. An aliquot was thawed immediately before inoculation; the concentration of the inoculum was adjusted to 1.0 × 1010 CFU/ml, which was confirmed by serial dilution and plating immediately after inoculation of the guinea pig.
Animal protocols (protocol 2002-0060) were approved by the Cornell University Institutional Animal Care and Use Committee prior to initiation of the experiments. Male Hartley guinea pigs (Elm Hill, Chelmsford, MA) weighing 348 ± 43 g that were 3 weeks old were housed individually, which allowed collection of each animal's feces. Animals were provided feed and water ad libitum. Cages were changed daily, and animal health and weight were monitored and recorded daily. Animals were acclimated for 5 days prior to inoculation; experiments were performed in triplicate using three animals per bacterial strain tested.
Intragastric inoculation of guinea pigs was performed as described previously (1). Briefly, feed was withheld for 12 h prior to inoculation. Animals were anesthetized with isoflurane administered via inhalation using oxygen as a carrier gas (17). L. monocytogenes was administered intragastrically by gavage. The stomach pH was neutralized with 1 ml of PBS containing 125 mg calcium carbonate (pH 7.4) prior to inoculation of L. monocytogenes (1.0 × 1010 CFU/ml). Each guinea pig was weighed daily and immediately prior to euthanasia by CO2 at 72 h after inoculation. The brain, liver, spleen, mesenteric lymph nodes, and small intestine were harvested and evaluated individually for L. monocytogenes as previously described (17). Recovered organs were kept on ice until they were processed. All organs were weighed and visually inspected for lesions. A 20-cm portion of the small intestine distal to the cecum was harvested. After the contents were removed, the small intestine segments were rinsed twice in 20 ml PBS, incubated at room temperature for 90 min in 20 ml of Dulbecco modified Eagle medium (DMEM) (Invitrogen) containing 100 mg/ml gentamicin to kill extracellular bacteria, and then rinsed three times in PBS prior to homogenization. The liver was homogenized in 60 ml of sterile PBS in a small autoclaved blender unit for 30 s; the brain, spleen, mesenteric lymph nodes, and small intestine were homogenized in 30 ml of PBS for 30 s. Following homogenization, homogenates were directly spread plated in duplicate on BHI media; homogenates and dilutions of homogenates (in PBS) were also spiral plated on BHI agar. All samples were also enriched to enable detection of L. monocytogenes in all organs, as follows: 10 ml homogenate was added to 90 ml Listeria enrichment broth (LEB) (Difco, Sparks, MD), incubated at 30°C, and then plated (50 μl) on Oxford medium (ThermoFisher, Waltham, MA) after 24 and 48 h.
After inoculation, feces were collected from all guinea pigs daily and processed as described by Garner et al. (17). A total of 0.5 g of feces was homogenized in 4.5 ml of PBS. Homogenized samples were serially diluted in PBS; homogenates were spread plated on Oxford medium (Oxoid, Basingstoke, United Kingdom), and subsequently dilutions in PBS were spiral plated on Oxford medium and grown for 24 h at 35°C. L. monocytogenes colonies were enumerated and confirmed on L. monocytogenes plating medium (LMPM) (Biosynth, Naperville, IL).
Statistical analyses were performed with Statistical Analysis Software (SAS) 9.0 (SAS Institute, Inc., Cary, NC). The levels of recovery (in log CFU/g) of L. monocytogenes from organs (i.e., liver, spleen, mesenteric lymph nodes, and small intestine) were used as the main measure of virulence. For specimens that were negative as determined by direct plating and positive as determined by enrichment, numbers of bacteria were conservatively estimated to represent the mean between the direct plating and enrichment detection limits; for specimens that were negative by both direct plating and enrichment, bacterial numbers were conservatively estimated to represent the mean between the enrichment detection limit and 0 CFU/g. One-sided t tests were used to determine whether the number L. monocytogenes bacteria (log CFU/g) recovered from a given organ or feces was higher for the wild-type strain than for the isogenic ΔsigB mutant. A P value of <0.05 was considered significant.
RESULTS
Contributions of σB to survival of stationary-phase cells at pH 2.5 and under oxidative stress conditions are different for different strains.
For both survival under acid stress conditions and survival under oxidative stress conditions, there were no significant differences in the death rates among the four wild-type strains. While the death rates of the wild-type lineage I, lineage II, and lineage IIIB strains under acid stress conditions differed significantly from those of their isogenic ΔsigB mutants (P = 0.0004, P < 0.0001, and P = 0.0047, respectively) (Table 2), the death rate of the lineage IIIA wild-type strain and the death rate of its isogenic ΔsigB mutant did not differ significantly (P = 0.2920). The largest difference in death rates (∼5 log CFU/h) was observed for the lineage II wild-type strain and its ΔsigB mutant, suggesting that σB is more important for survival under acid stress conditions in the lineage II strain than in the other strains. Further, while oxidative stress assays showed that there were significant differences in the numbers of cells killed between lineage I, lineage II, and lineage IIIB wild-type strains and their ΔsigB mutants (P = 0.0225, P < 0.0001, and P = 0.0291, respectively) (Table 2), σB did not contribute significantly to the survival of the lineage IIIA strain under these conditions (P = 0.0827). σB apparently played the largest role in survival under oxidative stress conditions in the lineage II strain, as shown by the largest difference (∼1.4 log CFU) between the lineage II wild-type strain and its ΔsigB mutant. σB contributed significantly to the survival of the lineage I, II, and IIIB strains at pH 2.5 and under oxidative stress conditions but apparently contributed little to the survival of the lineage IIIA strain under the same conditions.
TABLE 2.
Contributions of σB to survival under acid and oxidative stress conditions for all four L. monocytogenes lineages
| Lineage | Death rate (log CFU/h) at pH 2.5 |
Death (log CFU) with 13 mM CHP |
||||
|---|---|---|---|---|---|---|
| Wild typea | ΔsigB strainb | P valuec | Wild typed | ΔsigB straine | P valuef | |
| I | 3.95 ± 0.93 | 6.62 ± 0.66 | 0.0004 | 1.27 ± 0.04 | 2.10 ± 0.23 | 0.0225 |
| II | 1.75 ± 0.49 | 6.76 ± 0.79 | <0.0001 | 1.10 ± 0.08 | 2.57 ± 0.12 | <0.0001 |
| IIIA | 4.02 ± 2.23 | 5.74 ± 1.89 | 0.2920 | 1.14 ± 0.22 | 1.74 ± 0.40 | 0.0827 |
| IIIB | 3.65 ± 0.77 | 6.37 ± 0.73 | 0.0047 | 1.38 ± 0.30 | 2.09 ± 0.22 | 0.0291 |
Average death rate (± standard deviation) for wild-type parent strain exposed to pH 2.5 for 1 h.
Average death rate (± standard deviation) (log CFU/h) for isogenic ΔsigB strain exposed to pH 2.5 for 1 h.
P value for the time-strain interaction. A P value of <0.05 indicates there that was a significant difference in the average death rate between the wild-type and isogenic ΔsigB strains.
Average death value (± standard deviation) (log CFU) for wild-type parent strain after 15 min of exposure to 13 mM CHP.
Average death value (± standard deviation) (log CFU) for isogenic ΔsigB strain after 15 min of exposure to 13 mM CHP.
P value for one-sided t test. A P value of <0.05 indicates that there was a significant difference in the average death value between the wild-type and isogenic ΔsigB strains.
Contributions of σB to invasion of Caco-2 cells by stationary-phase L. monocytogenes cells are different for strains belonging to different lineages.
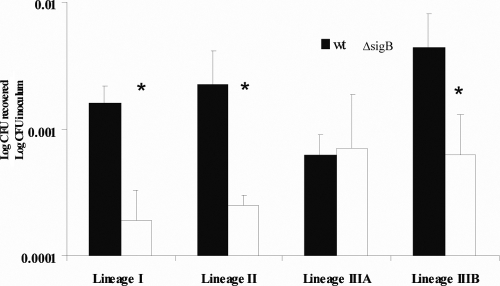
Assays of invasion of Caco-2 epithelial cells by stationary-phase L. monocytogenes cells (Fig. 1) showed that there were significant differences in invasion efficiency between wild-type strains and their isogenic ΔsigB mutants for lineage I (P = 0.0004), lineage II (P = 0.0319), and lineage IIIB (P = 0.0192) strains. Specifically, lineage I, II, and IIIB wild-type strains had higher invasion efficiencies than their isogenic ΔsigB mutants. No significant difference in invasion efficiency was found between the lineage IIIA wild-type strain and its ΔsigB mutant (P = 0.1991). The lineage IIIA parent strain showed the lowest invasion efficiency of the four parent strains; ANOVA showed that there was a significant effect (P = 0.0126) of the factor “strain” on the invasion efficiencies of the four parent strains, and a Tukey's post hoc test showed that the lineage IIIA wild-type strain was significantly less invasive than the lineage IIIB wild-type strain (P = 0.0097), while no other wild-type strains differed significantly in their invasion efficiencies. Overall, σB contributed significantly to invasion of Caco-2 cells by stationary-phase lineage I, II, and IIIB wild-type strains but did not appear to contribute to invasion by the lineage IIIA strain under the same conditions.
FIG. 1.
Invasion efficiencies (log CFU recovered/log CFU initial inoculum) for stationary-phase L. monocytogenes lineage I, II, IIIA, and IIIB strains in Caco-2 cells. The bars indicate means for at least three independent experiments; the error bars indicate 1 standard deviation from the mean. Each wild-type strain (wt) is paired with its isogenic ΔsigB mutant, as indicated on the x axis. The invasion efficiencies for each pair of strains were assessed by using two-sample t tests (*, P < 0.05).
Core σB regulon consists of at least 63 genes.
The transcript levels of a core set of 63 genes were higher in all four wild-type strains than in their ΔsigB mutants (≥1.5-fold change), and these genes were significantly positively differentially expressed (adjusted P value, <0.05) in all strains (Table 3 ). Putative σB-dependent promoters were located upstream of 50 of these 63 genes or predicted operons (79.4%); the putative σB promoters were identified using a hidden Markov model (HMM) previously developed for identification of σB promoters in L. monocytogenes strain 10403S (48). The core regulon included genes previously reported to be σB regulated, including inlA, opuCA, and rsbX, which encode internalin A, a glycine betaine-carnitine-choline ABC transporter, and a negative regulator of σB-dependent gene expression, respectively. Interestingly, 12 of the 63 core genes have no known function, and 7 of them encode conserved hypothetical proteins, indicating that over 30% of the genes in the core σB regulon, which may make important contributions to L. monocytogenes physiology, remain to be characterized.
TABLE 3.
Positively regulated σB-dependent genes present in all four L. monocytogenes lineages
| Genea | Productb | Lineage I |
Lineage II |
Lineage IIIA |
Lineage IIIB |
||||
|---|---|---|---|---|---|---|---|---|---|
| Fold changec | Adjusted P valued | Fold changec | Adjusted P valued | Fold changec | Adjusted P valued | Fold changec | Adjusted P valued | ||
| lmo0133 | Conserved hypothetical protein | 4.5 | 0.0003 | 5.1 | <0.0001 | 9 | <0.0001 | 14.5 | <0.0001 |
| lmo0134 | Acetyltransferase, GNAT family | 6.9 | 0.0001 | 5.6 | <0.0001 | 14.3 | <0.0001 | 7.7 | <0.0001 |
| lmo0169 | Similar to a glucose uptake protein | 5.5 | 0.0002 | 3.4 | <0.0001 | 5.1 | <0.0001 | 6.9 | <0.0001 |
| lmo0170 | Conserved hypothetical protein | 3.6 | 0.0006 | 3 | 0.0008 | 5.6 | <0.0001 | 7.4 | <0.0001 |
| lmo0210 | Similar to l-lactate dehydrogenase | 4.5 | <0.0001 | 2.6 | <0.0001 | 3.8 | 0.0001 | 4.6 | 0.0058 |
| lmo0211 | Similar to B. subtilis general stress protein | 1.7 | 0.0335 | 1.5 | 0.0001 | 2.1 | 0.0021 | 2.4 | <0.0001 |
| lmo0405 | Phosphate transporter family protein | 2.7 | 0.0410 | 1.7 | 0.0106 | 2.1 | 0.0053 | 2.1 | 0.0036 |
| lmo0433 | Internalin A | 3.6 | 0.0004 | 3.1 | <0.0001 | 4.8 | <0.0001 | 7 | <0.0001 |
| lmo0515 | Conserved hypothetical protein | 3.3 | 0.0116 | 3.4 | 0.0002 | 3.5 | <0.0001 | 5.4 | <0.0001 |
| lmo0539 | Similar to tagatose-1,6-diphosphate aldolase | 14.4 | <0.0001 | 7.5 | <0.0001 | 19.5 | <0.0001 | 24.2 | <0.0001 |
| lmo0555 | Similar to di-tripeptide transporter | 7.2 | 0.0002 | 4.1 | <0.0001 | 7.1 | <0.0001 | 9.3 | <0.0001 |
| lmo0593 | Similar to transport proteins (formate) | 6 | 0.0032 | 5.7 | <0.0001 | 9 | 0.0003 | 18.2 | <0.0001 |
| lmo0596 | Similar to unknown proteins | 14.2 | 0.0001 | 22.7 | <0.0001 | 33.8 | <0.0001 | 36 | <0.0001 |
| lmo0602 | Weakly similar to transcription regulator | 2.6 | 0.0023 | 3.7 | <0.0001 | 2.8 | 0.0003 | 2.5 | 0.0121 |
| lmo0610 | Similar to internalin proteins, putative peptidoglycan-bound protein (LPXTG motif) | 1.9 | 0.0380 | 3.7 | <0.0001 | 5.5 | <0.0001 | 8.2 | <0.0001 |
| lmo0642 | Putative membrane protein | 3.4 | 0.0028 | 2 | 0.0002 | 2 | 0.0029 | 3.1 | 0.0005 |
| lmo0655 | Similar to phosphoprotein phosphatases | 4.4 | <0.0001 | 2.9 | <0.0001 | 2.5 | 0.0014 | 3.1 | 0.0007 |
| lmo0722 | Similar to pyruvate oxidase | 4.6 | 0.0002 | 5.4 | <0.0001 | 8.8 | <0.0001 | 3.8 | 0.0070 |
| lmo0781 | Similar to mannose-specific PTS component IID | 10 | <0.0001 | 15.6 | <0.0001 | 18.2 | <0.0001 | 25.4 | <0.0001 |
| lmo0782 | Similar to mannose-specific PTS component IIC | 12.9 | <0.0001 | 13.5 | <0.0001 | 20.3 | <0.0001 | 22 | <0.0001 |
| lmo0783 | Similar to mannose-specific PTS component IIB | 6.4 | <0.0001 | 12 | <0.0001 | 14.8 | <0.0001 | 18 | <0.0001 |
| lmo0784 | PTS system, IIAB component, authentic frameshift | 2 | 0.0341 | 5.7 | <0.0001 | 5.5 | <0.0001 | 6.4 | <0.0001 |
| lmo0794 | Similar to B. subtilis YwnB protein | 6.1 | 0.0007 | 12.8 | <0.0001 | 10.3 | <0.0001 | 25.4 | <0.0001 |
| lmo0796 | Conserved hypothetical protein | 1.9 | 0.0029 | 4 | <0.0001 | 8.3 | <0.0001 | 12.3 | <0.0001 |
| lmo0880 | LysM domain protein | 7.4 | <0.0001 | 6.7 | <0.0001 | 16.4 | <0.0001 | 6.8 | 0.0006 |
| lmo0896 | Indirect negative regulation of σB-dependent gene expression (serine phosphatase) | 1.5 | 0.0244 | 2.3 | <0.0001 | 1.7 | 0.0197 | 2.7 | 0.0017 |
| lmo0911 | Unknown | 2.1 | 0.0152 | 2.1 | <0.0001 | 9.2 | <0.0001 | 1.8 | 0.0200 |
| lmo0913 | Succinate semialdehyde dehydrogenase | 6.6 | <0.0001 | 13.4 | <0.0001 | 16 | <0.0001 | 22.3 | <0.0001 |
| lmo0937 | Unknown | 6.4 | <0.0001 | 10.4 | <0.0001 | 16.2 | <0.0001 | 18.9 | <0.0001 |
| lmo0953 | Unknown | 3.2 | 0.0156 | 6.5 | <0.0001 | 12 | <0.0001 | 16.3 | <0.0001 |
| lmo0956 | Similar to N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25) | 2.6 | 0.0073 | 2.1 | <0.0001 | 4.4 | <0.0001 | 4.3 | <0.0001 |
| lmo0957 | Glucosamine-6-phosphate isomerase | 2.3 | 0.0006 | 1.6 | 0.0027 | 3.1 | 0.0027 | 2.4 | 0.0441 |
| lmo0994 | Unknown | 24.4 | <0.0001 | 14.1 | <0.0001 | 54.2 | <0.0001 | 79.2 | <0.0001 |
| lmo1140 | Unknown | 4 | 0.0084 | 3.5 | <0.0001 | 4.9 | 0.0001 | 4.9 | <0.0001 |
| lmo1241 | Conserved hypothetical protein | 2.7 | 0.0012 | 2.2 | 0.0002 | 4.2 | <0.0001 | 5 | <0.0001 |
| lmo1295 | Similar to host factor 1 protein | 1.8 | 0.0120 | 3.4 | <0.0001 | 5.4 | <0.0001 | 8 | <0.0001 |
| lmo1375 | Peptidase, M20/M25/M40 family | 2.4 | 0.0168 | 3 | <0.0001 | 2.4 | 0.0025 | 2.1 | 0.0381 |
| lmo1425 | Similar to betaine-carnitine-choline ABC transporter (membrane protein) | 1.9 | 0.0062 | 3.4 | <0.0001 | 3.5 | <0.0001 | 4.7 | <0.0001 |
| lmo1428 | Similar to glycine betaine-carnitine-choline ABC transporter (ATP-binding protein) | 2.2 | 0.0008 | 2.9 | <0.0001 | 2.7 | 0.0001 | 3.5 | 0.0004 |
| lmo1433 | Pyridine nucleotide disulfide oxidoreductase family protein | 4.7 | 0.0006 | 4.2 | <0.0001 | 3 | 0.0378 | 3.7 | 0.0309 |
| lmo1602 | Similar to unknown proteins | 4.2 | <0.0001 | 4.6 | <0.0001 | 1.7 | 0.0417 | 2.7 | <0.0001 |
| lmo1605 | UDP-N-acetylmuramate-alanine ligase | 7.4 | 0.0001 | 2.2 | <0.0001 | 2.4 | 0.0047 | 2.8 | <0.0001 |
| lmo1606 | FtsK/SpoIIIE family protein | 9.1 | <0.0001 | 5.6 | <0.0001 | 3.5 | <0.0001 | 4.1 | <0.0001 |
| lmo1694 | Similar to CDP-abequose synthase | 8 | <0.0001 | 8 | <0.0001 | 26.2 | <0.0001 | 1.8 | 0.0162 |
| lmo1799 | Putative peptidoglycan-bound protein (LPXTG motif) | 2.8 | 0.0017 | 1.7 | 0.0002 | 4.8 | <0.0001 | 4.7 | <0.0001 |
| lmo2085 | Cell wall surface anchor family protein | 11 | <0.0001 | 12.2 | <0.0001 | 14.1 | 0.0007 | 16.8 | 0.0067 |
| lmo2130 | Similar to unknown protein | 2.1 | 0.0112 | 2.6 | <0.0001 | 1.9 | 0.0104 | 2.3 | 0.0006 |
| lmo2191 | Similar to unknown proteins | 2.2 | 0.0066 | 3 | <0.0001 | 2.3 | <0.0001 | 2.3 | <0.0001 |
| lmo2269 | Unknown | 4.3 | 0.0096 | 5.7 | <0.0001 | 5.6 | <0.0001 | 6.2 | <0.0001 |
| lmo2391 | Conserved hypothetical protein similar to B. subtilis YhfK protein | 6.2 | <0.0001 | 9.1 | <0.0001 | 21.6 | <0.0001 | 28.5 | <0.0001 |
| lmo2434 | Glutamate decarboxylase gamma | 3.2 | 0.0193 | 2.7 | <0.0001 | 4 | 0.0097 | 3.8 | <0.0001 |
| lmo2454 | Unknown | 3.6 | 0.0003 | 4.6 | <0.0001 | 6.6 | <0.0001 | 8.2 | <0.0001 |
| lmo2463 | Similar to transport protein | 3 | 0.0426 | 3.9 | <0.0001 | 6.2 | <0.0001 | 4.3 | 0.0266 |
| lmo2485 | PspC domain protein, truncated | 1.9 | 0.0113 | 4.4 | <0.0001 | 1.8 | 0.0006 | 2 | 0.0021 |
| lmo2570 | Putative membrane protein | 5.5 | <0.0001 | 4.5 | <0.0001 | 7.3 | <0.0001 | 8.9 | <0.0001 |
| lmo2571 | Similar to nicotinamidase | 4.4 | <0.0001 | 5.8 | <0.0001 | 7.3 | <0.0001 | 8.3 | <0.0001 |
| lmo2572 | Similar to chain A, dihydrofolate reductase | 4.4 | 0.0002 | 1.6 | 0.0018 | 7.5 | <0.0001 | 3.5 | <0.0001 |
| lmo2573 | Alcohol dehydrogenase, zinc dependent | 3.4 | 0.0002 | 4.6 | <0.0001 | 5.6 | <0.0001 | 7.5 | <0.0001 |
| lmo2673 | Conserved hypothetical protein | 6.9 | 0.0009 | 9.7 | <0.0001 | 26 | <0.0001 | 37.2 | <0.0001 |
| lmo2674 | Similar to ribose-5-phosphate epimerase | 9.8 | <0.0001 | 3.8 | <0.0001 | 10.1 | <0.0001 | 13.7 | <0.0001 |
| lmo2724 | Similar to unknown proteins | 2.8 | 0.0030 | 2.9 | <0.0001 | 4.7 | <0.0001 | 4.9 | <0.0001 |
| lmo2748 | Similar to B. subtilis stress protein YdaG | 11.6 | <0.0001 | 10 | <0.0001 | 15.3 | <0.0001 | 5.1 | 0.0063 |
| lmo-f2365_0703 | Conserved hypothetical protein | 2.2 | 0.0277 | 3.3 | <0.0001 | 2.2 | 0.0038 | 2.4 | 0.0186 |
Gene names are based on the L. monocytogenes EGDe locus.
Common names of products are based on the EGDe annotation.
Changes in the transcript level in the parent strain compared to the transcript level in the ΔsigB strain. A ≥1.5-fold change was considered significant.
An adjusted P value of <0.05 was considered significant.
Of the 63 genes identified here as σB-dependent genes in all four strains, 56 and 59 were previously identified as genes positively regulated by σB by Raengpradub et al. (48) and Ollinger et al. (44), respectively (see Table S3 in the supplemental material). In addition, 45 of the 63 core σB-dependent genes identified here were previously identified as σB-dependent genes in L. monocytogenes EGD-e cells grown to early stationary phase (21) (see Table S3 in the supplemental material). As cells were grown to stationary phase prior to RNA isolation in each of these studies, genes with functions that contribute to recovery of cells from stationary phase may be overrepresented among the genes identified as σB-dependent genes.
σB-dependent genes found to be significantly positively differentially expressed in at least one strain.
A total of 425 genes were found to be positively differentially expressed in at least one of the four L. monocytogenes strains (>1.5-fold change; adjusted P value, <0.05) and thus constitute the σB pan-regulon (see Table S3 in the supplemental material). Of these 425 genes, 170 were positively differentially expressed in the lineage I strain, 252 were positively differentially expressed in the lineage II strain, 201 were positively differentially expressed in the lineage IIIA strain, and 207 were positively differentially expressed in the lineage IIIB strain. For 83 of the 425 positively differentially expressed genes there was a putative σB promoter upstream of the transcriptional start site or upstream of the first gene in a putative operon.
To broadly assess the functional roles of σB-dependent genes across all strains, the gene classification in JCVI role categories (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi) was examined. A χ2 test for trend indicated that σB-dependent genes (i.e., genes that were σB dependent in at least one strain) were not randomly distributed among JCVI role categories (P < 0.0001). Results of subsequent Fisher's exact tests indicated that σB-dependent genes were significantly overrepresented (P < 0.05) in two JCVI role categories (“cellular processes” and “energy metabolism”) and significantly underrepresented in three categories (“DNA metabolism,” “regulatory function,” and “unknown function”) (Table 4; see Table S4 in the supplemental material).
TABLE 4.
JCVI role categories with over- or underrepresentation for σB-dependent genes
| JCVI role categorya | No. of annotated genes | No. of σB-dependent genesb | Fisher's exact test P valuec | Odds ratiod |
|---|---|---|---|---|
| Cellular processes | 165 | 35 | 0.0463 | 1.5 |
| Energy metabolism | 380 | 103 | <0.0001 | 2.3 |
| DNA metabolism | 102 | 8 | 0.0263 | 0.5 |
| Regulatory functions | 226 | 24 | 0.0361 | 0.6 |
| Unknown function | 109 | 9 | 0.0312 | 0.5 |
JCVI L. monocytogenes EGD-e role categories.
Number of σB-dependent genes in all lineage representatives for each role category.
Fisher's exact test was used to identify significant associations between σB-dependent genes and the total number of genes in a role category; two-sided P values of <0.05 were considered significant.
Odds ratios were calculated to characterize significant associations between σB-dependent genes and role category.
Odds ratios (OR) were calculated to identify associations between σB-dependent regulation and role category. σB-dependent genes were overrepresented in the cellular processes role category, with an OR of 1.5 (P = 0.0462); the σB-dependent genes in this role category included several genes that are associated with pathogenesis (e.g., inlA, inlB, inlC2, and inlD). In the energy metabolism role category, σB-dependent genes were overrepresented, with an OR of 2.5 (P < 0.0001). Of the σB-dependent genes in the energy metabolism role category, 17 are classified in the pyruvate dehydrogenase subrole category, and 11 of these genes encode phosphotransferase system (PTS) subunits, suggesting that σB is important for modulating metabolism. This observation suggests that σB-dependent PTSs have a role in survival during stationary phase and under other environmental stress conditions. While the DNA metabolism role category was one-half as likely as other categories to contain σB-dependent genes (P = 0.0263; OR, 0.5), five of the six σB-dependent genes in this role category were associated with DNA replication, recombination, and repair, while one gene (lmo1361) was similar to an exodeoxyribonuclease gene, suggesting that σB modulates genes that may be necessary to maintain DNA integrity, particularly in stationary-phase cells.
Despite evidence that σB is involved in a number of transcriptional regulatory networks (11), σB-dependent genes were underrepresented in the regulatory functions role category (P = 0.0361; OR, 0.6). Of the 24 σB-dependent genes in this category, 15 were involved in transcriptional regulation, including genes encoding 4 GntR family transcriptional regulators (lmo0958, lmo1725, lmo2003, lmo2004) and 2 MerR family transcriptional regulators (lmo1788, lmo2593). The members of the GntR family have been characterized as global regulators of primary metabolism in a number of bacteria (8, 23, 42), and MerR-like regulators have been shown to play a role in optimizing transcription from σ70-dependent promoters with atypical distances between the −35 and −10 elements (4). Finally, 9 σB-dependent genes were classified in the unknown role category. While σB-dependent genes were underrepresented in the unknown role category (p = 0.0312; OR, 0.5), 109 of 425 genes (25.5%) identified as σB-dependent genes in stationary-phase cells (irrespective of role category) are described as (i) similar to unknown, (ii) unknown, or (iii) conserved hypothetical protein according to their common name designations.
Genes were identified as exclusively σB-dependent genes in lineage I or lineage II, suggesting that there are differences in σB regulons among strains.
We hypothesized that identification of differences in σB regulons between lineage I and II strains could provide insight into differences in pathogenic potential; therefore, we compared the σB regulons identified in the strains used in this study. To be considered exclusively σB dependent in one lineage but not in the other lineages and to reduce false-negative results, genes that were differentially expressed in the lineages were required to have a hybridization index (HI) of ≥95%. For 106 of the 170 lineage I genes and 252 lineage II genes with higher transcript levels in the wild-type strain than in the ΔsigB mutant, the transcript levels were significantly higher in both the lineage I and II wild-type strains.
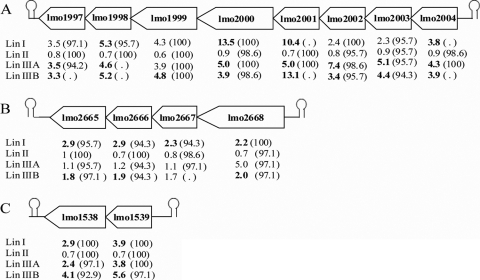
Of the genes whose transcript levels were determined to be higher in the wild-type strain than in the ΔsigB mutant strain, 55 σB-dependent genes were found in the lineage I strains and not in the lineage II strains using the HI criterion (Table 5). lmo1997 to lmo2004, lmo2665 to lmo2668, and lmo1538 and lmo1539 are putative operons comprised of a number of genes whose transcript levels are higher in the lineage I wild-type strain than in the ΔsigB mutant (Fig. 2). For four of the eight genes in the putative lmo1997-lmo1998-lmo1999-lmo2000-lmo2001-lmo2002-lmo2003-lmo2004 operon the transcript levels were significantly higher in the lineage I strain than in its ΔsigB mutant, while the transcript levels of none of the genes in this operon were significantly higher in the lineage II wild-type strain (Table 5 and Fig. 2A). Specifically, the transcript levels of lmo2001, lmo2000, and lmo1998 were 10.4-, 13.5-, and 5.3-fold higher, respectively, in the lineage I wild-type strain (Fig. 2A) than in its ΔsigB mutant. lmo2003 and lmo2004 encode GntR family transcriptional regulators and are upstream of six genes encoding components of a PTS. Similarly, lmo2665, lmo2666, and lmo2667 encode components of a PTS and are preceded by lmo2668 (Fig. 2B), which is similar to a BglG family transcriptional antiterminator gene. These results suggest that these PTSs are important in stationary phase in at least some strains. lmo1538 and lmo1539 (Fig. 2C) encode a glycerol kinase and a glycerol uptake facilitator, respectively, and the transcript levels of both of these genes were significantly higher in the lineage I wild-type strain than in the isogenic ΔsigB mutant. Differences in transcript levels were not significant in the lineage II strain (HI, 100%). Further, lmo2507, which encodes a protein highly similar to the cell division ATP-binding protein FtsE, was σB dependent in the lineage I strain and not in the lineage II strain. Modulated expression of FtsE has been proposed to be used by L. monocytogenes (60) and other species (52) to suppress cell division under stress conditions. Finally, transcription of lmo0315, which encodes a protein with a possible function in thiamine biosynthesis, was σB dependent in the lineage I strain but not in the lineage II strain.
TABLE 5.
Positively regulated σB-dependent genes present in lineage I but not in lineage II
| Genea | Productb | Fold changec |
Adjusted P valued |
||
|---|---|---|---|---|---|
| Lineage I | Lineage II | Lineage I | Lineage II | ||
| lmo0130 | Ser/Thr protein phosphatase family protein | 2.1 | 1.3 | 0.0046 | 0.0764 |
| lmo0188 | Dimethyladenosine transferase (16S rRNA dimethylase) | 1.7 | 1.1 | 0.0348 | 0.3556 |
| lmo0217 | Similar to B. subtilis DivIC protein | 1.8 | 1.2 | 0.0167 | 0.0230 |
| lmo0239 | Hypothetical protein | 1.9 | 1.1 | 0.0244 | 0.4018 |
| lmo0315 | Similar to thiamine biosynthesis protein | 2.5 | 1.0 | 0.0395 | 0.9741 |
| lmo0640 | Similar to oxidoreductase | 1.8 | 1.2 | 0.0081 | 0.0748 |
| lmo0958 | Transcriptional regulator, GntR family | 1.7 | 1.4 | 0.0082 | 0.0100 |
| lmo0959 | Llm protein | 1.8 | 1.1 | 0.0398 | 0.3736 |
| lmo1076 | N-Acetylmuramoyl-l-alanine amidase, family 4 | 1.6 | 1.2 | 0.0264 | 0.2002 |
| lmo1237 | Similar to glutamate racemase | 1.7 | 1.0 | 0.0324 | 0.8658 |
| lmo1255 | PTS, trehalose-specific, IIBC component | 2.7 | 1.3 | 0.0227 | 0.0495 |
| lmo1293 | Similar to glycerol-3-phosphate dehydrogenase | 1.8 | 0.5 | 0.0200 | 0.0026 |
| lmo1348 | Similar to aminomethyltransferase | 1.7 | 0.9 | 0.0447 | 0.1346 |
| lmo1357 | Acetyl-coenzyme A carboxylase, biotin carboxylase | 1.9 | 1.3 | 0.0290 | 0.0092 |
| lmo1389 | Similar to sugar ABC transporter, ATP-binding protein | 2.2 | 1.2 | 0.0020 | 0.0868 |
| lmo1390 | Similar to ABC transporter (permease proteins) | 2.2 | 1.2 | 0.0041 | 0.0399 |
| lmo1391 | Putative ABC transporter, permease protein | 1.8 | 1.2 | 0.0187 | 0.2869 |
| lmo1538 | Similar to glycerol kinase | 2.9 | 0.7 | 0.0001 | 0.0195 |
| lmo1539 | Similar to glycerol uptake facilitator | 3.9 | 0.7 | 0.0000 | 0.0746 |
| lmo1542 | Ribosomal protein L21 | 1.5 | 1.4 | 0.0354 | 0.0355 |
| lmo1570 | Highly similar to pyruvate kinases | 1.8 | 1.1 | 0.0227 | 0.7807 |
| lmo1658 | 30S ribosomal protein S2 | 2.2 | 1.3 | 0.0029 | 0.0530 |
| lmo1849 | Similar to metal cation ABC transporter, ATP-binding protein | 2.3 | 1.0 | 0.0224 | 0.8362 |
| lmo1956 | Similar to transcriptional regulator (Fur family) | 2.0 | 1.0 | 0.0148 | 0.9788 |
| lmo1998 | Similar to opine catabolism protein | 5.3 | 0.7 | 0.0467 | 0.0471 |
| lmo2000 | PTS, mannose/fructose/sorbose family, IID component | 13.5 | 0.9 | 0.0019 | 0.7744 |
| lmo2001 | PTS, IIC component | 10.4 | 0.7 | 0.0089 | 0.0102 |
| lmo2004 | Transcriptional regulator, GntR family | 3.8 | 0.9 | 0.0379 | 0.4411 |
| lmo2020 | Similar to cell division initiation protein (septum placement) | 2.5 | 1.4 | 0.0096 | 0.0014 |
| lmo2038 | UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase | 2.9 | 1.2 | 0.0124 | 0.0404 |
| lmo2058 | Similar to heme O oxygenase | 2.1 | 1.2 | 0.0462 | 0.1411 |
| lmo2101 | Pyridoxine biosynthesis protein | 2.4 | 1.1 | 0.0011 | 0.5643 |
| lmo2102 | Unknown | 1.9 | 1.2 | 0.0066 | 0.0845 |
| lmo2118 | Similar to phosphoglucomutase | 1.5 | 0.9 | 0.0367 | 0.6917 |
| lmo2167 | Metallo-beta-lactamase family protein | 2.0 | 1.2 | 0.0201 | 0.1227 |
| lmo2208 | Hydrolase, haloacid dehalogenase-like family | 2.0 | 1.0 | 0.0153 | 0.9761 |
| lmo2216 | Similar to histidine triad (HIT) protein | 1.7 | 1.3 | 0.0444 | 0.0018 |
| lmo2217 | Similar to unknown protein | 1.5 | 1.4 | 0.0329 | 0.0010 |
| lmo2223 | Conserved hypothetical protein | 2.0 | 1.4 | 0.0136 | 0.0056 |
| lmo2232 | CBS domain protein | 2.5 | 1.2 | 0.0123 | 0.1146 |
| lmo2240 | Similar to ABC transporter (ATP-binding protein) | 1.7 | 1.1 | 0.0137 | 0.4621 |
| lmo2397 | Similar to NifU protein | 1.8 | 1.2 | 0.0244 | 0.3112 |
| lmo2415 | Similar to ABC transporter, ATP-binding protein | 2.0 | 1.0 | 0.0254 | 0.8350 |
| lmo2507 | Highly similar to the cell division ATP-binding protein FtsE | 2.7 | 1.3 | 0.0290 | 0.0181 |
| lmo2547 | Highly similar to homoserine dehydrogenase | 1.5 | 0.9 | 0.0341 | 0.6523 |
| lmo2633 | Ribosomal protein S10 | 1.6 | 1.0 | 0.0187 | 0.8983 |
| lmo2660 | Similar to transketolase | 1.7 | 0.9 | 0.0305 | 0.1802 |
| lmo2664 | Similar to sorbitol dehydrogenase | 2.2 | 1.1 | 0.0027 | 0.7538 |
| lmo2665 | Similar to PTS galactitol-specific enzyme IIC component | 2.9 | 1.0 | 0.0064 | 0.9200 |
| lmo2666 | Similar to PTS galactitol-specific enzyme IIB component | 2.9 | 0.7 | 0.0004 | 0.0005 |
| lmo2667 | Similar to PTS galactitol-specific enzyme IIA component | 2.3 | 0.8 | 0.0015 | 0.0316 |
| lmo2668 | Similar to transcriptional antiterminator (BglG family) | 2.2 | 0.7 | 0.0014 | 0.0058 |
| lmo2758 | Similar to inosine monophosphate dehydrogenase | 1.7 | 1.1 | 0.0239 | 0.5999 |
| lmo2791 | Partition protein, ParA homolog | 1.9 | 1.0 | 0.0260 | 0.7694 |
| lmoh7858_0080.5 | Hypothetical protein | 3.2 | 1.0 | 0.0002 | 0.9174 |
Gene names are based on the L. monocytogenes EGDe locus. Bold type indicates putative operons.
Common names of the products are based on the EGDe annotation.
Changes in the transcript level in the parent strain compared to the transcript level in the ΔsigB strain. A ≥1.5-fold change was considered significant.
An adjusted P value of <0.05 was considered significant.
FIG. 2.
σB-dependent operons in lineage I but not in lineage II. The diagrams show three putative operons that appear to be σB dependent in the lineage I wild-type strain but not in the lineage II wild-type strain. (A) Operon consisting of lmo2004, lmo2003, lmo2002, lmo2001, lmo2000, lmo1999, lmo1998, and lmo1997. Genes in this operon were flanked by Rho-independent terminators, which are indicated by stem-loop structures. The numbers below each gene indicate the fold change in the transcript level in the parent strain compared to that in the ΔsigB strain for each lineage; bold type indicates values that are significantly different (≥1.5-fold change; adjusted P value, <0.05). The values in parentheses are the hybridization index (expressed as a percentage) for each gene; some hybridization indices could not be calculated as BLAST results found no match because the genomes are not complete yet. (B) Fold changes (significantly differentially expressed genes indicated by boldface type) and hybridization indices for the σB-dependent putative operon comprised of lmo2668, lmo2667, lmo2666, and lmo2665. (C) Fold changes (significantly differentially expressed genes indicated by boldface type) and hybridization indices for the σB-dependent putative operon comprised of lmo1539 and lmo1538.
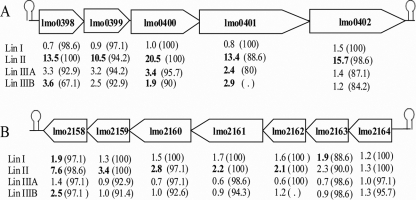
The transcript levels of 108 genes that were not σB dependent in the lineage I strain were higher in the lineage II wild-type strain than in its ΔsigB mutant (Table 6 ). In particular, the transcript levels of the putative lmo0398-lmo0399-lmo0400- lmo0401-lmo0402 operon, which encodes components of a PTS, a glycosyl hydrolase, and a BglG family transcriptional terminator, were at least 10.5-fold higher in the lineage II wild-type strain than in the ΔsigB mutant (Fig. 3 A). There was not a significant change for any of the genes in this operon when the lineage I strain was compared to its ΔsigB mutant (HI for each gene, >95%). Other genes in this category include (i) lmo2159 and lmo2160 and (ii) lmo0043, lmo2159, and lmo2160, which are in the putative lmo2164-lmo2163-lmo2162-lmo2161-lmo2160-lmo2159-lmo2158 operon (Fig. 3B), encode an oxidoreductase and an endonuclease, respectively, and may play a role in stationary-phase survival (15). lmo0043, which was also previously identified as a σB-dependent gene in the lineage II strain EGD-e (21), encodes a protein that is predicted to catalyze the conversion of l-arginine to l-citrulline and ammonia, which may aid in survival under acid stress conditions (13).
TABLE 6.
Genes positively regulated by σB in lineage II but not in lineage I
| Genea | Productb | Fold changec |
Adjusted P valued |
||
|---|---|---|---|---|---|
| Lineage I | Lineage II | Lineage I | Lineage II | ||
| lmo0019 | Conserved hypothetical protein | 1.4 | 2.9 | 0.1649 | 0.0000 |
| lmo0043 | Similar to arginine deiminase | 1.9 | 3.7 | 0.3567 | 0.0000 |
| lmo0135 | Oligopeptide ABC transporter, oligopeptide-binding protein | 0.8 | 2.0 | 0.2953 | 0.0000 |
| lmo0136 | Oligopeptide ABC transporter, permease protein | 0.6 | 1.5 | 0.1425 | 0.0000 |
| lmo0264 | Internalin C2 | 1.6 | 5.3 | 0.3567 | 0.0000 |
| lmo0265 | Peptidase, M20/M25/M40 family | 1.5 | 8.2 | 0.7760 | 0.0000 |
| lmo0292 | Similar to heat shock protein HtrA serine protease | 1.4 | 1.6 | 0.1117 | 0.0017 |
| lmo0321 | Similar to unknown proteins | 3.5 | 5.3 | 0.1010 | 0.0000 |
| lmo0342 | Similar to transketolase | 1.5 | 1.6 | 0.5712 | 0.0185 |
| lmo0343 | Similar to transaldolase | 1.1 | 2.0 | 0.9782 | 0.0011 |
| lmo0344 | Similar to dehydrogenase/reductase | 1.3 | 1.8 | 0.8013 | 0.0083 |
| lmo0345 | Similar to sugar phosphate isomerase | 1.3 | 1.5 | 0.8159 | 0.0002 |
| lmo0346 | Similar to triosephosphate isomerase | 1.3 | 1.9 | 0.8578 | 0.0062 |
| lmo0348 | Dihydroxyacetone kinase | 1.2 | 1.8 | 0.9272 | 0.0098 |
| lmo0398 | Similar to phosphotransferase system enzyme IIA | 0.7 | 13.5 | 0.1649 | 0.0000 |
| lmo0399 | PTS IIABC component, degenerate | 0.9 | 10.5 | 0.8049 | 0.0000 |
| lmo0400 | PTS IIABC component, degenerate | 1.0 | 20.5 | 0.9922 | 0.0000 |
| lmo0401 | Glycosyl hydrolase, family 38 | 0.8 | 13.4 | 0.3601 | 0.0000 |
| lmo0402 | Similar to transcriptional antiterminator (BglG family) | 1.5 | 15.7 | 0.1404 | 0.0000 |
| lmo0439 | Conserved hypothetical protein | 3.8 | 5.6 | 0.1291 | 0.0000 |
| lmo0449 | Unknown | 1.7 | 1.5 | 0.4564 | 0.0238 |
| lmo0584 | Conserved hypothetical membrane protein | 1.2 | 1.7 | 0.3709 | 0.0001 |
| lmo0589 | Unknown | 1.7 | 1.7 | 0.1977 | 0.0079 |
| lmo0590 | Similar to a fusion of two types of conserved hypothetical proteins, conserved hypothetical | 1.1 | 1.7 | 0.9228 | 0.0003 |
| lmo0591 | Membrane protein, putative | 1.2 | 1.5 | 0.4479 | 0.0166 |
| lmo0626 | Similar to unknown protein | 1.4 | 2.0 | 0.8736 | 0.0005 |
| lmo0628 | Unknown | 2.9 | 3.3 | 0.2091 | 0.0000 |
| lmo0647 | Unknown | 1.4 | 2.0 | 0.1241 | 0.0003 |
| lmo0648 | Magnesium transporter, CorA family | 1.7 | 1.7 | 0.2015 | 0.0014 |
| lmo0649 | Transcriptional regulator, GntR family | 2.7 | 1.7 | 0.1298 | 0.0004 |
| lmo0650 | Conserved membrane protein | 1.8 | 1.9 | 0.0763 | 0.0000 |
| lmo0759 | Glyoxalase family protein | 0.8 | 1.6 | 0.5762 | 0.0049 |
| lmo0760 | Unknown | 0.9 | 1.6 | 0.5681 | 0.0016 |
| lmo0811 | Carbonic anhydrase | 1.7 | 1.7 | 0.1107 | 0.0002 |
| lmo0818 | Cation transport ATPase, E1-E2 family | 0.7 | 1.7 | 0.4525 | 0.0023 |
| lmo0819 | Unknown | 1.5 | 1.7 | 0.0833 | 0.0010 |
| lmo0928 | Similar to 3-methyladenine DNA glycosylase | 2.0 | 1.5 | 0.1498 | 0.0150 |
| lmo0929 | Sortase family protein | 1.8 | 1.6 | 0.0663 | 0.0032 |
| lmo0995 | Membrane protein, putative | 1.1 | 3.4 | 0.9752 | 0.0001 |
| lmo1037 | B. subtilis YoaT protein homolog lmo1037 (imported) | 1.0 | 1.6 | 0.9873 | 0.0064 |
| lmo1064 | Hypothetical protein | 1.4 | 1.7 | 0.7066 | 0.0002 |
| lmo1072 | Highly similar to pyruvate carboxylase | 1.2 | 1.5 | 0.5247 | 0.0062 |
| lmo1121 | Unknown | 1.9 | 2.4 | 0.0539 | 0.0000 |
| lmo1226 | Similar to transporter (to B. subtilis YdgH protein) | 1.2 | 1.5 | 0.5341 | 0.0023 |
| lmo1242 | B. subtilis YdeI protein homolog lin1206 | 1.1 | 1.6 | 0.8038 | 0.0004 |
| lmo1243 | Uncharacterized conserved protein, PhnB family CAC3689 | 0.9 | 1.5 | 0.8989 | 0.0117 |
| lmo1360 | Highly similar to methylene tetrahydrofolate dehydrogenase and methenyl tetrahydrofolate cyclohydrolase | 1.6 | 1.6 | 0.1421 | 0.0017 |
| lmo1388 | CD4 T-cell-stimulating antigen, lipoprotein | 1.6 | 1.7 | 0.1146 | 0.0066 |
| lmo1421 | Similar to glycine betaine/carnitine/choline ABC transporter (ATP-binding protein) | 1.7 | 2.2 | 0.1477 | 0.0003 |
| lmo1426 | Similar to glycine betaine/carnitine/choline ABC transporter (osmoprotectant-binding protein) | 1.8 | 3.1 | 0.0541 | 0.0000 |
| lmo1427 | Glycine betaine/l-proline ABC transporter, permease protein | 1.3 | 3.0 | 0.4386 | 0.0000 |
| lmo1527 | Similar to protein export membrane protein SecDF | 1.8 | 1.5 | 0.1750 | 0.0006 |
| lmo1534 | l-Lactate dehydrogenase | 1.5 | 1.6 | 0.1649 | 0.0005 |
| lmo1571 | 6-Phosphofructokinase | 1.4 | 1.5 | 0.3590 | 0.0001 |
| lmo1580 | Universal stress protein family | 1.3 | 2.0 | 0.2022 | 0.0000 |
| lmo1622 | Conserved hypothetical protein | 1.2 | 1.6 | 0.5714 | 0.0011 |
| lmo1635 | Conserved hypothetical protein | 2.1 | 1.7 | 0.1056 | 0.0037 |
| lmo1637 | Putative ABC transporter, permease protein | 1.4 | 1.6 | 0.2716 | 0.0002 |
| lmo1666 | Conserved hypothetical protein | 1.4 | 1.6 | 0.7464 | 0.0002 |
| lmo1681 | Similar to cobalamin-independent methionine synthase | 1.2 | 1.5 | 0.7274 | 0.0061 |
| lmo1636 | Similar to ABC transporter (ATP-binding protein) | 2.0 | 1.9 | 0.0936 | 0.0001 |
| lmo1696 | Putative membrane protein | 1.2 | 1.5 | 0.4402 | 0.0073 |
| lmo1698 | Acetyltransferase, GNAT family | 1.1 | 3.5 | 0.8156 | 0.0001 |
| lmo1702 | Glyoxalase family protein | 1.0 | 1.6 | 0.9402 | 0.0369 |
| lmo1713 | Cell shape-determining protein | 1.5 | 1.5 | 0.4402 | 0.0244 |
| lmo1749 | Similar to shikimate kinase | 1.5 | 1.9 | 0.5157 | 0.0003 |
| lmo1790 | Metallo-beta-lactamase family protein | 1.5 | 1.7 | 0.2399 | 0.0001 |
| lmo1806 | Highly similar to acyl carrier proteins | 1.2 | 1.9 | 0.7644 | 0.0000 |
| lmo1883 | Chitinase | 2.6 | 3.2 | 0.3466 | 0.0000 |
| lmo1929 | Similar to nucleoside diphosphate kinase | 1.1 | 1.8 | 0.9873 | 0.0001 |
| lmo1930 | Heptaprenyl diphosphate synthase component II (imported) | 2.2 | 1.6 | 0.1056 | 0.0003 |
| lmo1931 | 2-Heptaprenyl-1,4-naphthoquinone methyltransferase | 1.9 | 1.8 | 0.0763 | 0.0002 |
| lmo1932 | Heptaprenyl diphosphate synthase component I, putative | 1.8 | 1.5 | 0.1616 | 0.0272 |
| lmo1933 | Similar to GTP cyclohydrolase I | 2.2 | 1.9 | 0.5761 | 0.0002 |
| lmo2031 | Conserved hypothetical protein TIGR00044 | 1.2 | 1.6 | 0.4204 | 0.0111 |
| lmo2033 | Highly similar to cell division protein FtsA | 1.4 | 1.6 | 0.2091 | 0.0015 |
| lmo2159 | Oxidoreductase, Gfo/Idh/MocA family | 1.3 | 3.4 | 0.3295 | 0.0000 |
| lmo2160 | AP endonuclease family 2 C terminus family | 1.5 | 2.8 | 0.0988 | 0.0000 |
| lmo2161 | ThuA protein | 1.7 | 2.2 | 0.1642 | 0.0000 |
| lmo2162 | Conserved hypothetical protein | 1.6 | 2.1 | 0.1513 | 0.0001 |
| lmo2169 | Unknown | 1.4 | 1.7 | 0.2512 | 0.0000 |
| lmo2196 | Similar to pheromone ABC transporter (binding protein) | 0.9 | 1.6 | 0.9436 | 0.0006 |
| lmo2230 | Similar to arsenate reductase | 10.5 | 18.7 | 0.0947 | 0.0000 |
| lmo2231 | Similar to unknown proteins | 0.9 | 3.1 | 0.9720 | 0.0009 |
| lmo2368 | MutT/nudix family protein | 1.6 | 1.5 | 0.0577 | 0.0054 |
| lmo2386 | Similar to B. subtilis YuiD protein | 1.1 | 1.9 | 0.9436 | 0.0000 |
| lmo2387 | Conserved hypothetical protein | 1.4 | 4.0 | 0.3857 | 0.0000 |
| lmo2389 | Similar to NADH dehydrogenase | 1.9 | 1.8 | 0.1056 | 0.0000 |
| lmo2399 | Similar to conserved hypothetical proteins | 1.5 | 1.8 | 0.2577 | 0.0004 |
| lmo2437 | Conserved hypothetical protein | 1.0 | 2.0 | 0.9866 | 0.0000 |
| lmo2465 | Conserved hypothetical protein | 1.5 | 1.8 | 0.3884 | 0.0000 |
| lmo2511 | Similar to conserved hypothetical proteins like B. subtilis YvyD protein | 1.2 | 1.9 | 0.5123 | 0.0001 |
| lmo2520 | N-Acylamino acid racemase | 0.8 | 1.8 | 0.6061 | 0.0000 |
| lmo2522 | Similar to hypothetical cell wall-binding protein from B. subtilis | 1.0 | 1.6 | 0.9825 | 0.0028 |
| lmo2534 | ATP synthase F0, C subunit | 1.4 | 1.5 | 0.1426 | 0.0006 |
| lmo2536 | Highly similar to ATP synthase subunit i | 1.4 | 1.6 | 0.2900 | 0.0009 |
| lmo2568 | Unknown | 1.4 | 1.6 | 0.6145 | 0.0236 |
| lmo2611 | Adenylate kinase | 1.0 | 1.5 | 0.9866 | 0.0143 |
| lmo2638 | Pyridine nucleotide-disulfide oxidoreductase family protein | 1.0 | 1.5 | 0.9914 | 0.0025 |
| lmo2670 | Hypothetical protein | . | 2.3 | NA | 0.0000 |
| lmo2695 | Dihydroxyacetone kinase, Dak1 subunit, putative | 1.6 | 1.6 | 0.1042 | 0.0003 |
| lmo2696 | Dihydroxyacetone kinase family protein | 1.5 | 1.8 | 0.1100 | 0.0001 |
| lmo2720 | Acetyl-coenzyme A synthetase | 1.0 | 1.5 | 0.9951 | 0.0022 |
| lmo2739 | Transcriptional regulator, Sir2 family | 1.7 | 1.5 | 0.0538 | 0.0038 |
| lmo2741 | Major facilitator family transporter | 1.7 | 1.5 | 0.0698 | 0.0010 |
| lmo2742 | SH3 domain protein | 0.7 | 1.5 | 0.1447 | 0.0015 |
| lmo2832 | Glycerate kinase 2 | 1.2 | 1.5 | 0.8437 | 0.0083 |
| lmof2365_1394 | Hypothetical protein | 1.9 | 1.5 | 0.1498 | 0.0068 |
Gene names are based on the L. monocytogenes EGDe locus. Bold type indicates putative operons.
Common names of the products are based on the EGDe annotation.
Changes in the transcript level in the parent strain compared to the transcript level in the ΔsigB strain. A ≥1.5-fold change was considered significant.
An adjusted P value of <0.05 was considered significant.
FIG. 3.
σB-dependent operons in lineage II but not in lineage I. The diagrams show two putative operons that appear to be σB dependent in the lineage II wild-type strain but not in the lineage I wild-type strain. (A) Operon consisting of lmo0398, lmo0399, lmo0400, lmo0401, and lmo0402. Genes in this operon were flanked by Rho-independent terminators, which are indicated by stem-loop structures. The numbers below each gene indicate the fold change in the transcript level in the parent strain compared to that in the ΔsigB strain for each lineage; bold type indicates values that are significantly different (≥1.5-fold change; adjusted P value, <0.05). The values in parentheses are the hybridization index (expressed as a percentage) for each strain; some hybridization indices could not be calculated as BLAST results found no match because the genomes are not complete yet. (B) Fold changes (significantly differentially expressed genes indicated by boldface type) and hybridization indices for the σB-dependent putative operon comprised of lmo2164, lmo2163, lmo2162, lmo2161, lmo2160, lmo2159, and lmo2158.
Confirmation of select differentially expressed genes in lineage I and II by TaqMan qRT-PCR.
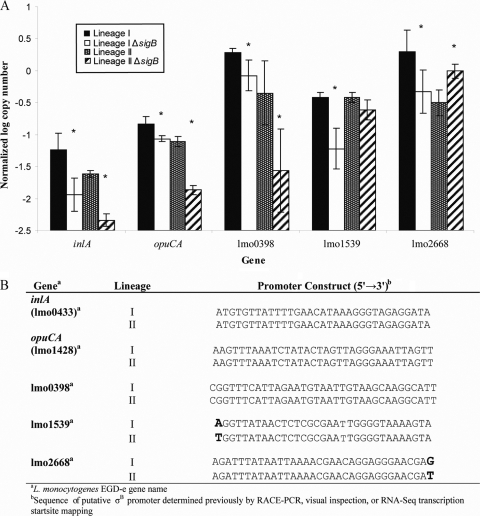
We used TaqMan qRT-PCR to confirm differences in transcript levels between wild-type and ΔsigB mutants for (i) select genes that were σB dependent in all four strains and (ii) select genes that were σB dependent in either lineage I or II strains but not in both lineage I and II strains, based on microarray analyses. Previous studies (33, 58) and microarray analyses in this study showed that transcription of inlA and opuCA is σB dependent. Using TaqMan qRT-PCR for all four strains, we confirmed that the inlA transcript copy numbers were significantly higher in the wild-type strains than in their isogenic ΔsigB mutants (P < 0.05) (Fig. 4; see Table S5 in the supplemental material). We confirmed that transcription of opuCA was σB dependent in lineages I and II (P < 0.05) (Fig. 4). Microarray analyses suggested that σB-dependent transcription of lmo0398, which is similar to a gene encoding a phosphotransferase system (PTS) enzyme IIA component, was different in different lineages. Using TaqMan qRT-PCR, we found that transcription of lmo0398 was σB dependent in both lineages I and II, as shown by significantly higher transcript levels in the wild-type parent strains than in their isogenic ΔsigB mutants (P < 0.05). While microarray analyses did not identify lmo0398 as a σB-dependent gene in the lineage I strain, the differences in transcript levels between the lineage I wild-type and ΔsigB mutant strains were statistically significant when we used TaqMan qRT-PCR, which is a more sensitive approach than microarray analysis and thus is able to detect smaller differences in transcript levels, as appeared to be the case with the lineage I strain. The σB-dependent nature of lmo0398 is further supported by the observation that the putative σB promoter region upstream of this gene (determined by RNA-Seq transcriptional start site mapping [43] and visual inspection) was 100% conserved in all strains. Lineage-specific σB-dependent transcription of lmo1539, which is similar to a glycerol uptake facilitator gene, was confirmed by qRT-PCR. Specifically, the transcript levels were significantly higher in the lineage I wild-type strain (P < 0.01) than in its ΔsigB mutant, while there were no significant differences in transcript levels between the lineage II wild-type strain and its ΔsigB mutant. The coding sequence of lmo1539 was highly conserved in all strains. The differences in σB-dependent transcription of lmo1539 between lineage I and II strains may reflect differences in the σB promoter sequences identified by HMM and confirmed by RNA-Seq transcription start site mapping (43). Similarly, differences in σB-dependent transcription of lmo2668, which encodes a protein similar to a BglG family transcriptional antiterminator, among lineages were found when microarray analyses were used. qRT-PCR confirmed that the transcript levels of lmo2668 were higher in the lineage I wild-type strain than in the ΔsigB mutant, suggesting that there is direct σB-dependent transcription of lmo2668. Interestingly, the transcript levels were significantly higher in the lineage II ΔsigB mutant than in the parent strain (P = 0.0104), supporting the hypothesis that lmo2668 is not positively regulated by σB in this lineage II strain and may be indirectly downregulated by σB. Similar to the findings for lmo1539, differences in σB-dependent transcription among strains belonging to different lineages may reflect a single-nucleotide polymorphism (SNP) in the putative σB promoters. This preliminary evidence suggests that diversification of σB promoter sequences among lineages may modulate transcription of some genes in the σB regulon, which may affect stress response systems in L. monocytogenes strains.
FIG. 4.
Confirmation of lineage I or lineage II σB-dependent genes by TaqMan qRT-PCR and comparison of promoter regions. (A) Normalized log copy numbers of transcripts for five genes (inlA, opuCA, lmo0398, lmo1539, and lmo2668) in lineage I and II strains and their isogenic ΔsigB mutants. The bars indicate the average log-transformed levels of transcripts (normalized to rpoB) for three independent replicates (i.e., three RNA isolations performed on different days); the error bars indicate one standard deviation from the mean. For each gene, one-sided t tests were used to determine if the transcript levels in the ΔsigB mutants were significantly different from those in the wild-type strains (*, P < 0.05). (B) Putative σB promoter sequences for five genes determined to be σB dependent by microarray analyses in either lineage I or lineage II or in both lineages. Bold type indicates nucleotide differences between lineage I and II putative σB promoter sequences. RACE, random amplification of cDNA ends.
Guinea pig listeriosis infection model.
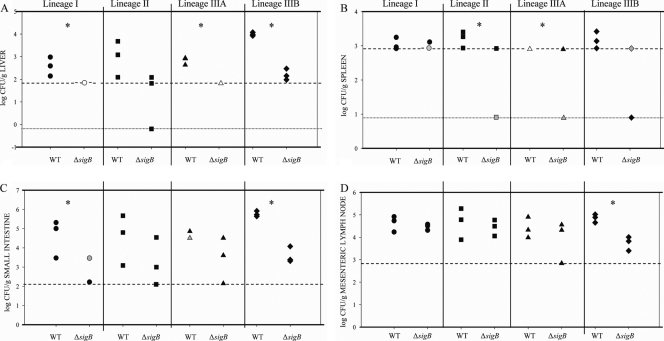
We used the guinea pig intragastric infection model to determine if σB contributed to virulence in strains belonging to different L. monocytogenes lineages. The numbers of bacteria (in log CFU/g) were determined for five organs (brain, liver, mesenteric lymph nodes, spleen, and small intestine) harvested from each animal that was intragastrically inoculated with an L. monocytogenes mutant or wild-type strain at 72 h postinfection (Fig. 5); three animals were inoculated with each strain. At 72 h after intragastric inoculation, the numbers of the ΔsigB mutant bacteria in the organs tested were generally lower than the numbers of the corresponding parent strain bacteria (Fig. 5). Significantly lower numbers of ΔsigB strain bacteria than of parent strain bacteria were recovered for lineage I (liver and small intestine [P < 0.05 for both]), lineage II (spleen [P < 0.05]), lineage IIIA (liver and spleen [P < 0.01 for both]), and lineage IIIB (small intestine [P < 0.01], as well as liver and mesenteric lymph nodes [P < 0.001 for both]). L. monocytogenes was not recovered from the brain of any inoculated animal, consistent with the results of a previous study of guinea pigs (39), possibly indicating that spread to the brain may occur only at later times after inoculation. While the differences between the recovery of the wild-type strain and the recovery of the ΔsigB mutant from organs in this study are not as large as those reported in a previous study (17), the variation is likely attributable to (i) our use of fewer replicates for each comparison in an effort to reduce animal usage, which resulted in a decrease in the power to detect differences, and (ii) the inherent variation in a complex biological system. Overall, σB contributed to virulence in all L. monocytogenes strains tested.
FIG. 5.
Log CFU/g L. monocytogenes recovered from organs: scatter plots of L. monocytogenes recovered from the organs of guinea pigs at 72 h after intragastric inoculation. The strains (a wild-type strain [WT] and the corresponding ΔsigB strain belonging to each lineage) are indicated on the x axis. The numbers of bacteria in the liver (A), spleen (B), small intestine (C), and mesenteric lymph nodes (D) are indicated on the y axis. Data were obtained from three guinea pigs that were intragastrically inoculated with each strain. Black symbols indicate a single data point, gray symbols indicate two overlapping data points, and open symbols indicate three overlapping data points. The detection limits, which were different for different organs due to different organ weights, are indicated by horizontal dashed and dotted lines in each panel. The dashed horizontal lines indicate the detection limit for direct plating; the dotted lines in panels A and B indicate the detection limits for enrichment procedures. The data reported for the plating detection limit were positive for L. monocytogenes after enrichment, but the bacterial counts were below the counts detectable by standard plate counting. For the data reported for the enrichment detection limit there was no recovery of L. monocytogenes after enrichment. An asterisk indicates that significantly (P < 0.05, one-sided t test) higher numbers of bacteria were recovered from organs from animals inoculated with the wild-type strain than from organs from animals inoculated with the isogenic ΔsigB mutant.
Significantly smaller numbers of ΔsigB mutant cells than of wild-type cells were recovered from feces at 72 h postinoculation for the lineage I (P = 0.0163), II (P = 0.0163), and IIIA (P = 0.0472) strains (Table 7). Fewer lineage IIIB ΔsigB cells than lineage IIIB wild-type cells were recovered from feces, but the difference was not statistically significant (P = 0.0532) (Table 7). The reduced numbers of ΔsigB cells in fecal material suggest that strains lacking σB may have a decreased ability to (i) attach to intestinal epithelial cells and/or (ii) survive in the intestinal tract.
TABLE 7.
Guinea pig weight and fecal shedding of L. monocytogenes at 72 h postinoculation
| Lineage | Weight (%) |
Fecal shedding (log CFU/g) |
||||
|---|---|---|---|---|---|---|
| Wild typea | ΔsigB mutantb | P valuec | Wild typed | ΔsigB mutante | P valuef | |
| I | 101.1 ± 8.9 | 106.5 ± 5.6 | 0.2129 | 5.3 ± 0.9 | 3.1 ± 0.7 | 0.0163 |
| II | 99.0 ± 8.4 | 106.6 ± 4.9 | 0.123 | 4.8 ± 1.5 | 0.0 ± 0.0 | 0.0163 |
| IIIA | 99.4 ± 3.7 | 105.7 ± 3.3 | 0.0465 | 6.1 ± 1.4 | 2.6 ± 2.4 | 0.0472 |
| IIIB | 91.6 ± 2.6 | 105.9 ± 9.2 | 0.0305 | 6.1 ± 0.1 | 3.3 ± 2.9 | 0.0532 |
Average weight (± standard deviation) of guinea pig infected with wild-type strain at 72 h postinoculation (after euthanasia), expressed as a percentage of the weight at zero time (inoculation), which was defined as 100%.
Average weight (± standard deviation) of guinea pig infected with the ΔsigB strain at 72 h postinoculation, expressed as a percentage of the weight at zero time, which was defined as 100%.
P value for t test for the weight of guinea pigs inoculated with the wild-type strain and the weight of guinea pigs inoculated with the ΔsigB strain. A P value of <0.05 indicates that there was a significant difference in the weight of animals at 72 h postinoculation.
Average fecal shedding (± standard deviation) of wild-type parent strain at 72 h postinoculation.
Average fecal shedding (± standard deviation) of ΔsigB strain at 72 h postinoculation.
P value for t test. A P value of <0.05 indicates that there is a significant difference in the average fecal shedding of L. monocytogenes between the wild-type and isogenic ΔsigB strains.
Finally, we tested whether there was a significant difference at 72 h postinoculation between the weights of animals that had been infected with the wild-type strains and the weights of animals that had been infected with the ΔsigB strains. The weight at 72 h postinoculation was expressed as a percentage of the weight of the animal at the time of infection, which was defined as 100%. While there were no significant differences between the weights of animals inoculated with the wild-type strain and the weights of animals inoculated with the ΔsigB strain for lineage I (P = 0.2129) or lineage II (P = 0.1230), animals inoculated with the wild-type lineage IIIA (P = 0.0465) and lineage IIIB (P = 0.0305) strains weighed significantly less than animals inoculated with the corresponding isogenic ΔsigB strains (Table 7).
DISCUSSION
In this study, we used phenotypic and transcriptomic approaches to characterize contributions of σB to the stress response and virulence in L. monocytogenes lineage I, II, IIIA, and IIIB strains. The data generated using these approaches showed that (i) the contributions of σB to acid and oxidative stress resistance are different in different strains; (ii) the σB-dependent genes in L. monocytogenes include a pan-regulon consisting of approximately 400 genes that are σB dependent in at least one strain and a core regulon consisting of at least 60 genes that are σB dependent in all of the strains characterized here, and (iii) while σB contributes to in vitro intestinal epithelial cell invasion only in some strains, it contributes to virulence in the guinea pig in all L. monocytogenes strains tested, supporting the hypothesis that σB makes strain-specific contributions to gene regulation in L. monocytogenes and universal contributions to virulence.
Phenotypic characterization reveals that the contributions of σB to survival under stress conditions are different for strains belonging to different lineages.
In this study, we tested the ability of a wild-type strain and its isogenic ΔsigB mutant belonging to each lineage to resist acid and oxidative stress in an initial effort to screen for differences in the contributions of σB to stress responses. We found that σB played a significant role in resistance to acid and oxidative stresses in the strains belonging to lineages I, II, and IIIB tested here. These results are consistent with the results of previous studies of lineage II strains that demonstrated that σB contributes to survival under environmental stress conditions (e.g., acid, oxidative, and energy stresses), as shown by reduced survival of an isogenic ΔsigB mutant under the same conditions (9, 15, 67). Further, σB plays a significant role in resistance to acid and salt stresses in Listeria innocua (48), as well as in resistance to acid, oxidative, and ethanol stresses in S. aureus (6), heat, ethanol, and osmotic stresses in Bacillus cereus (61), and heat, osmotic, ethanol, acid, freezing, and desiccation stresses in Bacillus subtilis (24, 64). To our knowledge, this study is the first study demonstrating that σB contributes to survival under stress conditions in a lineage I strain, and importantly, our results support the hypothesis that σB may play an important role in the survival and transmission of the L. monocytogenes strains that are the most significant risk for human health (i.e., lineage I strains).
We found that σB played a limited role in resistance of the L. monocytogenes lineage IIIA strain to acid and oxidative stresses. Therefore, despite significant evidence that there was σB-dependent differential gene expression and virulence and despite the absence of premature stop codons in rsb genes in the lmo0889-lmo0888-lmo0887-lmo0896 operon, which encode regulators of σB (data not shown), the L. monocytogenes lineage IIIA strain showed a limited σB-dependent phenotype under the conditions tested. The apparent σB-independent phenotype of the lineage IIIA strain (serotype 4c) observed under these conditions is consistent with the results of a previous study (37) which identified differences in the contributions of σB to environmental stress between serotype 1/2a and 4c strains. Specifically, σB played a minimal role in resistance to acid, oxidative, and heat stresses in the serotype 4c strain as there were no significant differences in survival between the wild-type strain and its isogenic ΔsigB mutant (37). Phenotypic diversification of response regulation has also been observed in other bacteria, including Escherichia coli (41) and Salmonella enterica serovar Typhimurium (27).
σB-dependent genes in L. monocytogenes include a pan-regulon consisting of approximately 400 genes and a core regulon consisting of at least 60 genes.
Whole-genome microarray approaches using two well-characterized lineage II laboratory strains (EGD-e and 10403S) previously identified more than 200 σB-dependent genes in L. monocytogenes (21, 48), and up to 168 genes were reported to be positively regulated by σB (21, 30, 44, 48). Using four strains, one each belonging to lineages I, II, IIIA, and IIIB, we identified a total of ∼400 genes as genes that are positively regulated by σB in at least one strain; these genes, therefore, represent an initial description of the L. monocytogenes σB pan-regulon. In comparison, the reported σB regulons in other Gram-positive organisms (e.g., S. aureus and B. subtilis) contain from ∼100 to 150 genes (45, 46, 62); each of these regulons was determined using a single strain. Over 60 of the L. monocytogenes pan-regulon genes were σB dependent in all four strains; thus, these genes constitute the core L. monocytogenes σB regulon. The majority of the genes comprising the core σB regulon were also identified as genes positively regulated by σB in previous studies using the lineage II strains EGD-e and 10403S (21, 44, 48). While it is likely that the core σB regulon was underestimated in the present study due to (i) low hybridization indexes (HI) for some genes in some strains and (ii) use of a single growth condition with a single microarray platform, it is not surprising that the L. monocytogenes core regulon is small compared to the pan-regulon across a diverse set of strains. For example, while σB is conserved in many different Gram-positive organisms, the L. monocytogenes, S. aureus, B. cereus, and B. subtilis σB regulons share only three genes (rsbV, rsbW, and sigB), which are members of the sigB operon (62). This observation supports the notion that the σB regulon has evolved to perform niche-specific functions in different organisms.
Overall, σB-dependent genes in Gram-positive bacteria contribute to a wide variety of functions, including energy metabolism, regulatory functions, and pathogenesis (21, 22, 45, 48, 62). In this study, L. monocytogenes σB-dependent genes were significantly overrepresented in the cellular processes and energy metabolism L. monocytogenes JCVI role categories, which indicates that σB contributes to multiple mechanisms important for cell function, survival, and pathogenesis. Likewise, the σB regulon in L. monocytogenes EGD-e included genes encoding proteins involved in a wide range of metabolic functions, genes encoding general stress proteins, and genes having unknown functions (21). While the regulatory functions role category was underrepresented among the σB-dependent genes identified here, the σB-dependent genes in this role category encode potentially important transcriptional regulators and include genes encoding GntR family and MerR-like regulators. These proteins act as global regulators of primary metabolism (8, 23, 42) and contribute to optimization of σ70-dependent promoters in Gram-negative bacteria (4), respectively. Combined with the results of previous studies that have shown that there are regulatory networks for σB and PrfA in L. monocytogenes (19, 59), as well as for σB and other regulators in L. monocytogenes (11) and in other bacteria (7, 57), our findings contribute to an emerging body of evidence supporting the hypothesis that σB plays an important role in regulatory networks and contributes to the stress response and virulence across L. monocytogenes lineages, even though the specific role of σB is clearly different in different strains.
L. monocytogenes strains classified in different lineages appear to differ in the likelihood that they are associated with human illnesses and in overall pathogenic potential (20, 68). Interestingly, a number of differences between σB regulons were observed in the lineage I and II strains examined in this study. For example, genes encoding a glycerol kinase and glycerol uptake facilitator were σB dependent in the lineage I strain but not in the lineage II strain. The putative σB-dependent promoters for lmo1539, which encodes the glycerol uptake facilitator, were different in the lineage I and II strains, which may be responsible for the differences in σB-dependent transcription between these strains. A recent study demonstrated that L. monocytogenes growth in the presence of glycerol results in upregulation of a number of PrfA-controlled genes (28, 68). Furthermore, lmo0315, which encodes a protein with a possible role in thiamine biosynthesis, was σB dependent in the lineage I strain but not in the lineage II strain. A recent study (51) concluded that L. monocytogenes employs thiamine uptake and biosynthesis of thiamine precursors to grow in host cytoplasm. Therefore, based on these findings in combination with our data, it is tempting to speculate that differences in glycerol-dependent regulation of virulence genes and a potentially enhanced ability to grow intracellularly may affect the pathogenic potential of strains that are classified in different lineages.
Transcriptional profiles have been reported for wild-type strains representing L. monocytogenes lineages I and II (53), and evidence has indicated that the transcript levels of a number of σB-dependent genes were higher in the lineage II strains than in the lineage I strains (based on lineage designations that are consistent with the first description of L. monocytogenes lineages by Piffaretti et al. in 1989 [47]). Of the σB-related genes reported to be expressed more highly in lineage II (53), five were found here to be σB dependent in lineage II strains but not in lineage I strains, including two genes encoding ABC transport system components (lmo1421 and lmo1426), further supporting the hypothesis that there has been diversification of the σB regulon in lineage I and II strains. As a considerable number of genes that were found to be σB dependent in either lineage I or II strains encode proteins with unknown functions or hypothetical proteins, further investigation of these genes and their regulation may provide insight into niche adaption and phenotypic differences among L. monocytogenes strains and lineages. The ability of microarray analyses to sensitively detect differences in the contributions of σB to expression of various genes in the σB core regulon undoubtedly affects relative identification of genes as σB-dependent genes in strains and lineages. Emerging transcriptome profiling techniques, such as RNA-Seq, should provide powerful and sensitive new tools for regulon identification.
While σB does not contribute to in vitro intestinal epithelial cell invasion in all L. monocytogenes strains, it contributes to virulence in the guinea pig in all strains tested.
Previous studies (17, 32) demonstrated that in L. monocytogenes 10403S (a lineage II strain), σB is important for invasion of Caco-2 cells, as shown by the reduced recovery of ΔsigB mutant cells compared to the recovery of cells of the parent strain. Kim et al. (33) demonstrated that σB contributed significantly to L. monocytogenes 10403S invasion of human enterocytes and hepatocytes, predominantly through InlA- and InlB-mediated pathways as both inlA and inlB are transcribed from σB-dependent promoters (32, 33). In the present study, wild-type lineage I, II, and IIIB strains had significantly higher invasion efficiencies than their isogenic ΔsigB mutants, indicating that σB contributes to invasion of Caco-2 cells. Interestingly, we found that σB did not contribute to the efficiency of invasion of Caco-2 cells in the lineage IIIA strain despite the fact that (i) the L. monocytogenes lineage IIIA strain has a full-length inlA transcript and (ii) microarray analyses identified over 200 genes whose transcript levels were higher in the wild-type strain than in the ΔsigB mutant, indicating that there is a large σB regulon that includes well-characterized σB-dependent genes important for virulence (e.g., bsh, opuCA, inlA, and gadB [30]). The possible explanations for this finding include (i) the possibility that σB activity is regulated differently in the lineage IIIA strain than in other strains and (ii) the possibility that genes critical for invasion efficiency, acid resistance, and oxidative stress resistance show limited or no σB dependence in the lineage IIIA strain tested, even though our microarray analyses did not identify clear candidate genes that might explain these differences. It is tempting to speculate that the phenotypic diversification of the lineage IIIA strain may be attributable in part to the selective pressures of its environment. In recent work Kvitek et al. (34) found significant phenotypic variation in stress sensitivity and gene expression among Saccharomyces cerevisiae strains isolated from distinct environments but similar phenotypic and transcriptional profiles for strains isolated from similar niches.
We used the guinea pig listeriosis infection model as an in vivo system to quantify and characterize the contributions of σB to virulence in L. monocytogenes strains from lineages I, II, IIIA, and IIIB. All L. monocytogenes ΔsigB strains had reduced virulence in the guinea pig listeriosis infection model, as shown by the reduced recovery of the ΔsigB strains compared to the recovery of their isogenic parents from at least one organ. A recent study by Ollinger et al. (44) provided a possible mechanistic explanation for the importance of σB in the guinea pig listeriosis infection model. σB regulates activity of the major L. monocytogenes virulence regulator PrfA, ranging from transcriptional activation of prfA at the P2prfA promoter to posttranscriptional downregulation of PrfA regulon expression under conditions when PrfA is highly active (e.g., under intracellular conditions). This rheostat-like role for σB in controlling PrfA activity appears to be important for the tightly regulated control of virulence gene expression in L. monocytogenes. Thus, contributions of σB to virulence gene expression in the lineage IIIA strain appear to be essential for full virulence of this strain. While σB contributes to invasion of intestinal epithelial cells in vitro and survival under acid and stress conditions in some strains, it universally contributed to virulence in guinea pigs in the L. monocytogenes strains tested, further underscoring the central importance of contributions of σB to gene regulation and virulence in L. monocytogenes.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health award RO1-AI052151-01A1 (to K.J.B.)
We thank J. Corron, S. Chaturongakul, M. E. Palmer, E. D. Fortes, and Y. Soyer for their assistance with the guinea pig gastrointestinal model for infection and T. M. Bergholz for assistance with microarray data analyses.
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bakardjiev, A. I., B. A. Stacy, S. J. Fisher, and D. A. Portnoy. 2004. Listeriosis in the pregnant guinea pig: a model of vertical transmission. Infect. Immun. 72:489-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 3.Brody, M. S., and C. W. Price. 1998. Bacillus licheniformis sigB operon encoding the general stress transcription factor sigma B. Gene 212:111-118. [DOI] [PubMed] [Google Scholar]
- 4.Brown, N. J., J. V. Soyanov, S. P. Kidd, and J. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 5.Bruhn, J. B., B. F. Vogel, and L. Gram. 2005. Bias in the Listeria monocytogenes enrichment procedure: lineage 2 strains outcompete lineage 1 strains in University of Vermont selective enrichments. Appl. Environ. Microbiol. 71:961-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cebrián, G., N. Sagarzazu, A. Aertsen, R. Pagán, S. Condón, and P. Mañas. 2009. Role of the alternative sigma factor σB on Staphylococcus aureus resistance to stresses of relevance to food preservation. J. Appl. Microbiol. 107:187-196. [DOI] [PubMed] [Google Scholar]
- 7.Cerca, N., J. L. Brooks, and K. K. Jefferson. 2008. Regulation of the intercellular adhesin locus regulator (icaR) by SarA, σB, and IcaR in Staphylococcus aureus. J. Bacteriol. 190:6530-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chai, Y., R. Kolter, and R. Losick. 2009. A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J. Bacteriol. 191:2423-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaturongakul, S., and K. J. Boor. 2006. σB Activation under environmental and energy stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 72:5197-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaturongakul, S., S. Raengpradub, M. Wiedmann, and K. J. Boor. 2008. Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol. 16:388-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, P. D., S. Ryan, C. G. Gahan, and C. Hill. 2005. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71:2832-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curran, T. M., J. Lieou, and R. E. Marquis. 1995. Arginine deiminase system and acid adaptation of oral streptococci. Appl. Environ. Microbiol. 61:4494-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FAO/WHO. 2004. Microbiological risk assessment series 5: risk assessment of Listeria monocytogenes in ready-to-eat foods. FAO/WHO, Geneva, Switzerland. http://www.who.int/foodsafety/publications/micro/en/mra5_contents.pdf.
- 15.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouet, A., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garner, M. R., B. L. Njaa, M. Wiedmann, and K. J. Boor. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentleman, R., V. Carey, D. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray, M. J., N. E. Freitag, and K. J. Boor. 2006. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect. Immun. 74:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hain, T., H. Hossain, S. S. Chatterjee, S. Machata, U. Volk, S. Wagner, B. Brors, S. Haas, C. T. Kuenne, A. Billion, S. Otten, J. Pane-Farre, S. Engelmann, and T. Chakraborty. 2008. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e σB regulon. BMC Microbiol. 8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecker, M., A. Reder, S. Fuchs, M. Pagels, and S. Engelmann. 2009. Physiological proteomics and stress/starvation responses in Bacillus subtilis and Staphylococcus aureus. Res. Microbiol. 160:245-258. [DOI] [PubMed] [Google Scholar]
- 23.Hillerich, B., and J. Westpheling. 2006. A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J. Bacteriol. 188:7477-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holtmann, G., M. Brigulla, L. Steil, A. Schutz, K. Barnekow, U. Volker, and E. Bremer. 2004. RsbV-independent induction of the SigB-dependent general stress regulon of Bacillus subtilis during growth at high temperature. J. Bacteriol. 186:6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528-535. [PubMed] [Google Scholar]
- 26.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen, F., S. Leach, S. J. Wilde, A. Davies, G. S. A. B. Stewart, and T. Humphrey. 2000. Invasiveness in chickens, stress resistance and RpoS status of wild-type Salmonella enterica subsp. enterica serovar Typhimurium definitive type 104 and serovar Enteritidis phage type 4 strains. Microbiology. 146:3227-3235. [DOI] [PubMed] [Google Scholar]
- 28.Joseph, B., S. Mertins, R. Stoll, J. Schar, K. R. Umesha, Q. Luo, S. Muller-Altrock, and W. Goebel. 2008. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 190:5412-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 30.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2006. Contributions of Listeria monocytogenes σB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152:1827-1838. [DOI] [PubMed] [Google Scholar]
- 32.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, H., H. Marquis, and K. J. Boor. 2005. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kvitek, D. J., J. L. Will, and A. P. Gasch. 2008. Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet. 4:e1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lozniewski, A., A. Humbert, D. Corsaro, J. Schwartzbrod, M. Weber, and A. Le Faou. 2001. Comparison of sludge and clinical isolates of Listeria monocytogenes. Lett. Appl. Microbiol. 32:336-339. [DOI] [PubMed] [Google Scholar]
- 36.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moorhead, S. M., and G. A. Dykes. 2003. The role of the sigB gene in the general stress response of Listeria monocytogenes varies between a strain of serotype 1/2a and a strain of serotype 4c. Curr. Microbiol. 46:461-466. [DOI] [PubMed] [Google Scholar]
- 38.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nightingale, K. K., R. A. Ivy, A. J. Ho, E. D. Fortes, B. L. Njaa, R. M. Peters, and M. Wiedmann. 2008. inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 74:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nightingale, K. K., K. Windham, K. E. Martin, M. Yeung, and M. Wiedmann. 2005. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71:8764-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notley-McRobb, L., T. King, and T. Ferenci. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogasawara, H., Y. Ishida, K. Yamada, K. Yamamoto, and A. Ishihama. 2007. PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J. Bacteriol. 189:5534-5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliver, H. F., R. H. Orsi, L. Ponnala, U. Keich, W. Wang, Q. Sun, S. W. Cartinhour, M. J. Filiatrault, M. Wiedmann, and K. J. Boor. 2009. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ollinger, J., B. Bowen, M. Wiedmann, K. J. Boor, and T. M. Bergholtz. 2009. Listeria monocytogenes σB modulates PrfA-mediated virulence factor expression. Infect. Immun. 77:2113-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pané-Farré, J., B. Jonas, K. Förstner, S. Engelmann, and M. Hecker. 2006. The σB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296:237-258. [DOI] [PubMed] [Google Scholar]
- 46.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. U. S. A. 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raengpradub, S., M. Wiedmann, and K. J. Boor. 2008. Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74:158-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rauch, M., Q. Luo, S. Muller-Altrock, and W. Goebel. 2005. SigB-dependent in vitro transcription of prfA and some newly identified genes of Listeria monocytogenes whose expression is affected by PrfA in vivo. J. Bacteriol. 187:800-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauders, B. D., E. D. Fortes, D. L. Morse, N. Dumas, J. A. Kiehlbauch, Y. Schukken, J. R. Hibbs, and M. Wiedmann. 2003. Molecular subtyping to detect human listeriosis clusters. Emerg. Infect. Dis. 9:672-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schauer, K., J. Stolz, S. Scherer, and T. M. Fuchs. 2009. Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J. Bacteriol. 191:2218-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt, K. L., N. D. Peterson, R. J. Kustusch, M. C. Wissel, B. Graham, G. J. Phillips, and D. S. Weiss. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Severino, P., O. Dussurget, R. Z. Vencio, E. Dumas, P. Garrido, G. Padilla, P. Piveteau, J. P. Lemaitre, F. Kunst, P. Glaser, and C. Buchrieser. 2007. Comparative transcriptome analysis of Listeria monocytogenes strains of the two major lineages reveals differences in virulence, cell wall, and stress response. Appl. Environ. Microbiol. 73:6078-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth, G. K. 2005. Limma: linear models for microarray data, p. 397-420. In R. V. Carey, S. Dudoit, R. Irizarry, and W. Huber (ed.), Bioinformatics and computational biology solutions using R and bioconductor. Springer, New York, NY.
- 55.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- 56.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31:265-273. [DOI] [PubMed] [Google Scholar]
- 57.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 58.Sue, D., D. Fink, M. Wiedmann, and K. J. Boor. 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843-3855. [DOI] [PubMed] [Google Scholar]
- 59.Toledo-Arana, A., O. Dussurget, G. Nikitas, N. Sesto, H. Guet-Revillet, D. Balestrino, E. Loh, J. Gripenland, T. Tiensuu, K. Vaitkevicius, M. Barthelemy, M. Vergassola, M.-A. Nahori, G. Soubigou, B. Regnault, J.-Y. Coppee, M. Lecuit, J. Johansson, and P. Cossart. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950-956. [DOI] [PubMed] [Google Scholar]
- 60.van der Veen, S., T. Hain, J. A. Wouters, H. Hossain, W. M. de Vos, T. Abee, T. Chakraborty, and M. H. J. Wells-Bennik. 2007. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiology 153:3593-3607. [DOI] [PubMed] [Google Scholar]
- 61.van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. de Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Schaik, W., M. van der Voort, D. Molenaar, R. Moezelaar, W. M. de Vos, and T. Abee. 2007. Identification of the σB regulon of Bacillus cereus and conservation of σB-regulated genes in low-GC-content Gram-positive bacteria. J. Bacteriol. 189:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vela, A. I., J. F. Fernandez-Garayzabal, J. A. Vazquez, M. V. Latre, M. M. Blanco, M. A. Moreno, L. de La Fuente, J. Marco, C. Franco, A. Cepeda, A. A. Rodriguez Moure, G. Suarez, and L. Dominguez. 2001. Molecular typing by pulsed-field gel electrophoresis of Spanish animal and human Listeria monocytogenes isolates. Appl. Environ. Microbiol. 67:5840-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volker, U., B. Maul, and M. Hecker. 1999. Expression of the sigma B-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ward, T. J., T. F. Ducey, T. Usgaard, K. A. Dunn, and J. P. Bielawski. 2008. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 74:7629-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wiedmann, M., J. Bruce, C. Keating, A. Johnson, P. McDonough, and C. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.