Abstract
To gain insight into the diversity and origins of antibiotic resistance genes, we identified resistance genes in the soil in an apple orchard using functional metagenomics, which involves inserting large fragments of foreign DNA into Escherichia coli and assaying the resulting clones for expressed functions. Among 13 antibiotic-resistant clones, we found two genes that encode bifunctional proteins. One predicted bifunctional protein confers resistance to ceftazidime and contains a natural fusion between a predicted transcriptional regulator and a β-lactamase. Sequence analysis of the entire metagenomic clone encoding the predicted bifunctional β-lactamase revealed a gene potentially involved in chloramphenicol resistance as well as a predicted transposase. A second clone that encodes a predicted bifunctional protein confers resistance to kanamycin and contains an aminoglycoside acetyltransferase domain fused to a second acetyltransferase domain that, based on nucleotide sequence, was predicted not to be involved in antibiotic resistance. This is the first report of a transcriptional regulator fused to a β-lactamase and of an aminoglycoside acetyltransferase fused to an acetyltransferase not involved in antibiotic resistance.
In the middle of the 20th century, the discovery of antibiotics dramatically changed the tools available to cure infectious disease. Today, antibiotics control many diseases, but use, overuse, and misuse have led to an alarming increase in the frequency of human pathogens that do not respond to antibiotic therapy (4, 14, 16, 31), underscoring the need for new antibiotics (reviewed in reference 10) and a better understanding of the origins of antibiotic resistance.
Soil microbial communities are potential reservoirs of antibiotic resistance genes, but these reservoirs have rarely been studied. Bacteria living in soil are attractive for this investigation because of their inherent genetic diversity and the abundance of antibiotic-producing soil bacteria, which predicts the presence of resistance genes in producers and susceptible target bacteria (1, 27). A study of Streptomyces isolates from diverse soils found that all of the 480 strains tested were resistant to multiple antibiotics (15).
Analysis of antibiotic-resistant bacteria typically involves culturing bacteria from an environmental sample on a medium that contains the antibiotic of interest. However, fewer than 1% of all microbes are readily culturable; thus, culture-based analysis excludes most of the microbial diversity (7-9, 18, 20, 24). To access a broad suite of resistance genes, we previously used functional metagenomics, a technique that enables discovery of antibiotic resistance genes from organisms that may or may not be readily culturable. Additionally, selections for functional enzymes can interrogate all expressed proteins in the library and not just those with sequences homologous to known proteins. This approach has revealed genes that would have been missed by sequence analysis alone (19). Since the overwhelming majority of known antibiotic resistance genes originated from culturable bacteria, there are likely many more resistance elements yet to be found. In addition, many of the known resistance genes are from clinically important bacteria. Recent work from our laboratory demonstrated that certain resistance genes found in soil bacteria are related only distantly to those found in clinical isolates, indicating that more work needs to be done to understand the relationship between resistance genes present in soil and those in clinical isolates (1, 27).
In the current study, we constructed a metagenomic library from soil bacterial communities and screened the library for clones that confer antibiotic resistance to tetracycline, β-lactams, or aminoglycoside antibiotics. We identified 13 clones that conferred resistance to at least one of the antibiotics tested. Among these clones we found genes encoding two putative novel bifunctional enzymes.
MATERIALS AND METHODS
Soil sampling.
Soil samples were taken from below apple trees located in an apple orchard in southern Wisconsin (42°58′N, 89°28′W). Two samples, one on the northern side and one on the southern side, from each of two trees were collected on 26 April 2007. After visible vegetation had been removed, a core of soil approximately 5 cm deep and 10 cm in diameter was removed. Each sample was stored on ice for transport to the laboratory, where it was kept frozen at −20°C until DNA was extracted.
Library construction.
Metagenomic libraries were constructed as previously described (21), with the following modifications. Agarose plugs containing lysed bacterial cells were embedded in a 1% low-melt agarose gel and electrophoresed to size select the DNA. The region of the gel corresponding to 40 to 100 kb was excised, and the DNA was extracted using the GELase enzyme according to the manufacturer's protocol (Epicentre Biotechnologies, Madison, WI). The resulting DNA was ligated into the pCC1FOS fosmid vector, packaged into phage, and introduced into the EPI300 strain of Escherichia coli using the CopyControl fosmid library production kit (Epicentre Biotechnologies).
Selections.
Antibiotic-resistant clones were selected as previously described (1). Libraries were grown in 5 ml LB broth amended with 20 μg/ml chloramphenicol and 1× CopyControl induction solution (Epicentre Biotechnologies) for 1 or 2 h, diluted in sterile water, and plated on LB agar containing one of the following antibiotics: 16 μg/ml amoxicillin, 50 μg/ml ampicillin, 50 μg/ml carbenicillin, 16 μg/ml cefamandole, 1 μg/ml ceftazidime, 50 μg/ml cephalexin, 10 μg/ml gentamicin, 10 μg/ml kanamycin, 12.5 μg/ml piperacillin, or 10 μg/ml tetracycline. Plates were incubated at 28°C for 2 days, after which single colonies were streaked on LB agar plates supplemented with the appropriate antibiotic and LB agar plates with 20 μg/ml chloramphenicol. Clones that grew on both media were further analyzed.
strA-strB cloning.
The strA gene was amplified using purified DNA templates, representing each of five library subpools, with the following primers: strAf, 5′-CTGGTTGCCTGTCAGAGG-3′, and strAr, 5′-GTCAGAGGGTCCAATCGC-3′ (modified from reference 28). The strB gene was amplified using the following primers: strBf, 5′-GACTCCTGCAATCGTCAAGG-3′, and strBr, 5′-GCAATGCGTCTAGGATCGAG-3′ (30). The expected sizes for the strA and strB fragments are 753 bp and 561 bp, respectively. The resulting PCR products were precipitated with isopropanol using standard procedures. Each PCR product was then ligated into pGEM-5zf and transformed into E. coli. The plasmid DNA from 10 random colonies from each transformation was isolated; inserts were sequenced using the M13 forward and M13 reverse primers.
Mutagenesis and sequence analysis.
The gene responsible for the observed resistance of a clone was identified in one of two ways. In some cases, using the GPS-1 kit (New England Biolabs, Beverly, MA), we introduced random insertions into the target metagenomic clone. Fosmids with transposon insertions were transformed into strain EPI300 of E. coli. The transformants were screened for the loss of the resistance phenotype, and the DNA flanking the transposon was sequenced using primers supplied in the kit. Sequences were assembled and manipulated using the Lasergene software package (DNAStar, Madison, WI) and either annotated using the RAST program (5) or subjected to BLAST analysis against the NCBI's nr database (2). The BLAST results were accurate according to the NCBI database in April 2010. BLAST results with expected values better than e-20 were considered to be significant.
Alternatively, the gene responsible for resistance was identified by subcloning. Active clones were partially digested with Sau3AI (Promega, Madison, WI), and fragments were purified using the PCR purification and gel extraction kit (Qiagen, Valencia, CA) and ligated to pUC19 (New England Biolabs) digested with BamHI (Promega). The resulting clones were introduced into EPI300 and selected on LB containing antibiotics of interest as described above. Cells harboring clones conferring resistance to the appropriate antibiotic and lacking chloramphenicol resistance associated with the original fosmid backbone were sequenced using the following primers: pucseqF, 5′-TTCCCAGTCACGACGTTGTAAAAC-3′, and pucseqR, 5′- ATGCTTCCGGCTCGTATGTTGTGT-3′.
Cloning resistance genes.
cft2, the ceftazidime resistance gene from clone AOCefta2, was PCR amplified using the following primers: PciIcefta2, 5′-AAAACATGTCAATGCACCAATGGCCTGCGAAAAG-3′, and EcoICRIcefta2, 5′-AAAGAGCTCGGTGGACCGGGTTGACAT-3′. To generate an inducible construct containing the full-length gene, the resulting product was digested with PciI (New England Biolabs) and EcoICRI (Promega) and ligated into pET-28b (Novagen, Madison, WI) that had been digested with EcoICRI and NcoI (Promega). To generate an inducible construct containing just the β-lactamase domain, primers EcoICRIcefta2 and NcoIcefta2lactamaseF (5′-ACAGCCATGGTCAGCCAGCGCCTTGCGGAGATCG-3′) were used to amplify the appropriate region of the open reading frame (ORF). The resulting PCR product was digested with NcoI and EcoICRI and ligated into pET-28b digested with NcoI and EcoICRI.
kan4, the kanamycin resistance gene from clone AOKan4, was amplified using the following primers: BamHIpet44Kan4, 5′-TTCGGATCCCATACGATGGAGTCGCAAGTTGAAGTTT-3′, and HindIIIKan4r, 5′-TAGAAGCTTAAGTCATGATTGATTGTAAGTTTTCC-3′. The N-terminal domain of kan4 was amplified using the BamHIpet44Kan4 primer and HindIIIkan4Nterm, 5′-CTGAAGCTTATTACATTCGGATTTCGATTTCGTCG-3′. The C-terminal domain of kan4 was amplified using primer HindIIIKan4r with BamHIpet44kan4Cterm, 5′-TAAGGGATCCCCTGGTGTAATGCCTGACGCCAATG-3′. Each resulting PCR product was cut with BamHI and HindIII (Promega) and ligated into the BamHI and HindIII sites of pET44a (Novagen).
Antibiotic susceptibility analysis.
Antibiotic susceptibility assays were conducted according to the following protocol. Briefly, each row of a 96-well plate contained Mueller-Hinton broth (Beckton Dickinson and Co., Sparks, MD) and the specified antibiotic present at concentrations according to a 2-fold serial dilution from the first well. The first well contained sufficient antibiotic so that the final concentration would be 512 μg/ml, and the last well contained no antibiotic, to serve as a control for growth. To each well in the assay, ∼105 CFU of a strain carrying a metagenomic clone were added, bringing the volume to 100 μl. The wells were scored for visible growth after incubation for 18 h at 28°C. The susceptibility corresponds to the concentration of antibiotic in the last well that showed no cell growth. Each clone was subjected to three assays. In almost all cases, the three values were identical; when they were not, the lowest value is reported. All values obtained for cells bearing each clone were within one dilution of each other.
Nucleotide sequence accession numbers.
Sequence data obtained from this work were deposited in GenBank with the following accession numbers: AOAmox2, GQ244488; AOCarb3, GQ244489; AOCefta2, GQ244490; AOAmox1, GQ244491; AOCarb11, Q244492; AOCefta11, GQ244493; AOPip4, GQ244494; AOCarb2, GQ244495; AOCefta1, GQ244496; AOKan4, GQ244497; AOKan6, GQ244498; AOKan8, GQ244499; and AOTet43, GQ244501.
RESULTS
Construction of the metagenomic library.
We sampled soil from a commercial apple orchard because the aminoglycoside antibiotic streptomycin is applied in this orchard two to four times per year to prevent the bacterial disease fire blight. The soil was used to construct a metagenomic library that contained approximately 446,000 clones. The average insert size was approximately 30 kb, which produced a library containing over 13 Gb of metagenomic DNA. Assuming an average genome size of 5 Mb, this library captures more than 2,500 bacterial genome equivalents.
Streptomycin resistance.
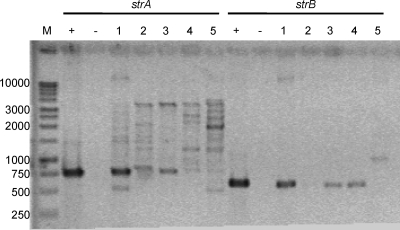
The orchard from which the soil was taken has a history of streptomycin exposure, and consequently, we selected for streptomycin-resistant clones and found none. Since the strA-strB gene pair from Tn5393, which confers streptomycin resistance (12, 13), had been found previously in culturable bacteria isolated from the soil in this orchard (A. Klimowicz, personal communication), we investigated the possibility that the strA-strB gene pair was present in our library but not expressed in the E. coli host. PCR amplification with primers specific to strA and to strB revealed bands corresponding to 753 bp and 561 bp, the expected sizes for the strA and strB fragments, respectively (Fig. 1). The identity of the genes was confirmed by sequencing, which revealed 100% identity at the nucleotide level with the expected strA gene. The same analysis indicated that all of the nine plasmids from library subpool 1 that contained inserts matched the expected strB gene.
FIG. 1.
Detection of strA and strB streptomycin phosphotransferases in the metagenomic library. Each of five subpools of the metagenomic library, as well as a positive and a negative control, was used as a template to amplify the strA and strB genes. A total of 5 μl of each PCR was separated on a 1% agarose gel in 1× Tris-borate-EDTA (TBE). Lane M contains the 1-kb DNA ladder (Promega, Madison, WI). Primers specific to strA were used in lanes labeled “strA” and should result in a band at 753 bp. Primers specific to strB were used in lanes labeled “strB” and should result in a band at 540 bp. Lanes labeled “+” are positive-control PCRs using the strA and strB genes as templates. Lanes labeled “−” contain products of PCR with no DNA template as a negative control. The sizes of the molecular weight markers are indicated on the left.
Clones conferring antibiotic resistance.
To detect genes that confer resistance to other antibiotics, we subjected the metagenomic library to selection on the following 10 antibiotics: the β-lactams ampicillin, amoxicillin, carbenicillin, cefamandole, ceftazidime, cephalexin, and piperacillin; the aminoglycosides gentamicin and kanamycin; and tetracycline. We identified 13 clones that confer resistance to at least one of the antibiotics tested (Table 1). Among the 13 active clones, 11 contained homology to previously reported enzymes involved in antibiotic resistance, including eight β-lactamases, two from each of the four Ambler classes of β-lactamases (3). Although the functions of the enzymes encoded by these clones were assigned based on sequence homology, the sequences of these genes indicate their novelty. The sequences are different enough from those in the NCBI nr database that a nucleotide BLAST analysis of these clones was unable to identify significant matches. A protein BLAST analysis identified matches, although the similarities were still low. Selection on kanamycin, an aminoglycoside antibiotic, yielded two clones that contain aminoglycoside acetyltransferase homologues (aac6′ gene products). Selection for tetracycline resistance yielded the AOTet43 clone encoding an efflux pump. In addition, we identified two clones, AOCefta2 and AOKan4, that both encoded bifunctional enzymes.
TABLE 1.
Clones that confer antibiotic resistancea
| Clone | Resistanceb | Gene function | Best-match organismc |
|---|---|---|---|
| AOAmox2 | Amo, Crb, Cft, Pip | Class A β-lactamase | Chitinophaga (54) (YP_003121133) |
| AOCarb3 | Amo, Cft, Crb | Class A β-lactamase | Pedobacter (49) (ZP_01883167) |
| AOCefta2 | Cft | Class A β-lactamase | USB (49) (ACH59002) |
| AOAmox1 | Amo, Crb, Cft, Pip | Metallo-β-lactamase | Pseudomonas (46) (CAQ53840) |
| AOCarb11 | Amo, Crb, Pip | Metallo-β-lactamase | USB (48) (ACH58985) |
| AOCefta11 | Cft | Class C β-lactamase | Pseudomonas (74) (YP_349452) |
| AOPip4 | Crb, Pip | Class C β-lactamase | Parachlamydia (29) (ZP_06299750) |
| AOCarb2 | Crb | Class D β-lactamase | Chitinophaga (40) (YP_003120606) |
| AOCefta1 | Amo, Crb, Cft | Class D β-lactamase | Legionella (81) (YP_123907) |
| AOKan4 | Kan | AG acetyltransferase | USB (59) (AAS90627) |
| AOKan6 | Kan | AG acetyltransferase | USB (59) (AAS90609) |
| AOKan8 | Kan | AG acetyltransferase | USB (59) (AAS90609) |
| AOTet43 | Tet | Efflux pump | Gordonia (71) (YP_003273675) |
The abbreviations are as follows: Amo, amoxicillin; Cft, ceftazidime; Crb, carbenicillin; Kan, kanamycin; Pip, piperacillin; Tet, tetracycline; USB, uncultured soil bacterium; AG, aminoglycoside.
Resistance refers to the antibiotics in the media on which E. coli cells bearing each fosmid clone were isolated.
BLAST analysis was performed as described previously (2). The values in parentheses are the percent amino acid identity of the best match followed by the GenBank accession number of the best match.
Antibiotic susceptibility assays.
E. coli was the host used for the identification of genes conferring resistance to antibiotics. Therefore, the gene products encoded by these clones are functional in E. coli, and since E. coli is a human commensal, it is important to evaluate the spectrum of antibiotic resistance of E. coli cells bearing each of these genes. We conducted antibiotic susceptibility assays on all clones and have referred to any clone that confers a reproducible decrease in susceptibility to antibiotics as conferring resistance to that antibiotic. For those clones conferring resistance to β-lactam antibiotics, the antibiotic susceptibility value was determined for each of four β-lactams, amoxicillin, carbenicillin, ceftazidime, and piperacillin (Table 2). These were chosen because each clone was isolated on media containing at least one of these antibiotics. These antibiotics also were useful in determining the spectrum of resistance since they represent members of both the penicillin and cephalosporin structural subclasses. Two clones, AOCefta2 and AOCefta11, increased resistance to ceftazidime only. This is consistent with the fact that the clones were found in bacteria cultured with ceftazidime but not with other antibiotics. All other clones containing β-lactamases conferred enhanced resistance to at least one of the other β-lactams tested.
TABLE 2.
MIC assay data
| Clone name | MIC (μg/ml)a |
|||||
|---|---|---|---|---|---|---|
| Amo | Crb | Cft | Pip | Kan | Tet | |
| AOAmox2 | 16 | 16 | 4 | 8 | NT | NT |
| AOCarb3 | 16 | 64 | 2 | 16 | NT | NT |
| AOCefta2 | 4 | 16 | 4 | 4 | NT | NT |
| AOAmox1 | >512 | >512 | 16 | >512 | NT | NT |
| AOCarb11 | 128 | 128 | <0.5 | >512 | NT | NT |
| AOCefta11 | 4 | 16 | 2 | 8 | NT | NT |
| AOPip4 | 4 | 32 | 2 | 32 | NT | NT |
| AOCarb2 | 4 | 32 | 1 | 8 | NT | NT |
| AOCefta1 | 8 | 256 | 4 | 64 | NT | NT |
| AOKan4 | NT | NT | NT | NT | 16 | NT |
| AOKan6 | NT | NT | NT | NT | 8 | NT |
| AOKan8 | NT | NT | NT | NT | 32 | NT |
| AOTet43 | NT | 16 | <0.5 | NT | 2 | 8 |
| Empty vector | 4 | 16 | <0.5 | 4 | 2 | <0.5 |
The abbreviations are as follows: Amo, amoxicillin; Cft, ceftazidime; Crb, carbenicillin; Kan, kanamycin; Pip, piperacillin; Tet, tetracycline; NT, not tested.
For those clones conferring resistance to kanamycin, AOKan4, AOKan6, and AOKan8, the antibiotic susceptibility values were determined for kanamycin only. Each of these clones increased resistance to kanamycin compared to the vector-only control. Because the active gene from the AOTet43 metagenomic insert encodes a putative efflux pump, antibiotic susceptibility values were determined for tetracycline, kanamycin, carbenicillin, and ceftazidime to assess a broad resistance profile. However, AOTet43 conferred resistance only to tetracycline.
Clone AOCefta2.
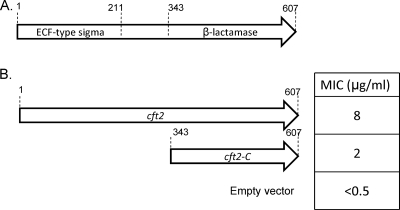
In addition to the eight clones in which the active gene encodes only the β-lactamase, we identified one β-lactamase gene, cft2 in clone AOCefta2, that has a unique genetic arrangement. The protein encoded by the entire 1,821-bp ORF was submitted to a BLAST search, which yielded Lra5 (49% identity), a class A β-lactamase isolated from an uncultured bacterium found in Alaskan soil, as the best match. The β-lactamase homology was restricted to the C-terminal 265 amino acids (Fig. 2A). Although the approximate size expected for a full-length β-lactamase is 265 amino acids, the cft2 ORF encodes a protein of 607 amino acids. To identify the potential function of the rest of the protein, the C terminus (amino acids 343 to 607) was excluded from our analysis and the BLAST search was repeated. We found that the N terminus of this ORF, encoding amino acids 40 to 211, has homology (35% amino acid identity) with an extracytoplasmic function (ECF)-type sigma factor from Rhodoferax ferrireducens T118. Exclusion of the N terminus (amino acids 1 to 211) left only the central region of the ORF, which encodes a 133-amino-acid peptide. BLAST analysis of the central region resulted in a match that had an expected value of 0.63, indicating that the central region shows no significant homology to any proteins in the NCBI nr database.
FIG. 2.
The domains of Cft2. (A) The cft2 ORF is predicted to encode a bifunctional protein. The N-terminal domain, amino acids 1 to 211, encodes a predicted ECF-type sigma factor. The C-terminal domain, amino acids 343 to 607, encodes a class A β-lactamase. (B) The β-lactamase domain of cft2 was cloned into pET28b and expressed in E. coli. The numbers above each ORF reflect amino acid numbering. MIC values are for the full-length and truncated cft2 constructs.
To ensure that the predicted β-lactamase domain is responsible for the observed resistance, we cloned the full-length gene as well as the portion of the gene encoding amino acids 343 to 607 into pET28b (Fig. 2B). The antibiotic susceptibility of E. coli carrying the original fosmid clone was 4 μg/ml of ceftazidime. The cft2 ORF and the part of cft2 encoding just the β-lactamase domain that were each cloned into the pET28b expression vector conferred antibiotic susceptibility values of 8 μg/ml and 2 μg/ml, respectively. These values are all greater than the value observed for the host E. coli carrying empty pET28(b) (<0.5 μg/ml).
To our knowledge, there is only one other report of a bifunctional enzyme that confers resistance to β-lactam antibiotics (1). Therefore, we sequenced the entire insert of the AOCefta2 clone to determine the genomic context of cft2. The resulting 34,867 bp of DNA had a mean GC content of 56%. The GC content is consistent across the entire insert, suggesting that cft2 is a natural fusion of multiple domains. The insert contains 25 predicted genes on the metagenomic DNA fragment, nearly half of which were predicted to code for hypothetical proteins (Table 3). Of the genes with similarity to known genes, two had similarity to genes involved in antibiotic resistance or its dissemination. Approximately 15 kb upstream of the β-lactamase is an ORF predicted to encode a chloramphenicol 3-O-phosphotransferase. In addition, approximately 5 kb upstream of the β-lactamase is an ORF predicted to encode a transposase.
TABLE 3.
Predicted genes encoded by the AOCefta2 metagenomic clone
| nt start | nt stop | Predicted gene functiona | Best-match organismb |
|---|---|---|---|
| 411 | 1 | Endonuclease IV | Sphaerobacter (45) (YP_003319913) |
| 1312 | 470 | Uracil-DNA glycosylase superfamily | Anaeromyxobacter (55) (YP_002494162) |
| 2333 | 1617 | LSU ribosomal protein L1p (L10Ae) | Brevibacillus (61) (YP_002769688) |
| 3465 | 2479 | Hypothetical protein | No matches |
| 5224 | 3593 | Lipase maturation factor 1 | Chthoniobacter (60) (ZP_03131605) |
| 6401 | 7672 | Hypothetical protein | No matches |
| 8033 | 9277 | Hypothetical protein | No matches |
| 10027 | 9380 | Chloramphenicol 3-O-phosphotransferase | Legionella (44) (YP_003454190) |
| 11657 | 10110 | Hypothetical protein | No matches |
| 11964 | 13241 | Hypothetical protein | Deinococcus (31) (NP_285530) |
| 15037 | 13457 | Parallel beta-helix repeat | Bacterium Ellin514 (35) (ZP_03630385) |
| 15233 | 15583 | Hypothetical protein | Dethiobacter (78) (ZP_03728276) |
| 17560 | 15929 | Parallel beta-helix repeat precursor | Bacterium Ellin514 (36) (ZP_03627387) |
| 19631 | 18123 | Hypothetical protein | “Candidatus Kuenenia” (31) (CAJ71036) |
| 20099 | 19740 | Transposase IS116/IS110/IS902 family protein | Solibacter (50) (YP_824442) |
| 21967 | 20546 | Kelch-like ECH-associated protein 1 | Clostridium (35) (ZP_04804093) |
| 23276 | 22227 | Alpha/beta hydrolase fold | Cyanothece (31) (YP_002481203) |
| 23765 | 24982 | Hypothetical protein | Planctomyces (37) (ZP_01855161) |
| 25177 | 26997 | Bifunctional beta-lactamase | Uncultured bacterium (49) (ACH59002) |
| 29196 | 27083 | Cell wall/surface repeat protein | Bacterium Ellin514 (32) (ZP_03632172) |
| 29479 | 30924 | Hypothetical protein | Trichodesmium (27) (YP_721418) |
| 31432 | 30953 | Hypothetical protein | Flavobacteriales (49) (ZP_01105379) |
| 31529 | 32632 | Peptide chain release factor 2 | Geobacter (54) (YP_002538537) |
| 32690 | 33703 | Dipeptidyl-peptidase VI | Bacteroides (26) (ZP_03015828) |
| 33700 | 34737 | Chloromuconate cycloisomerase YkfB1 | Stigmatella (45) (ZP_01463557) |
LSU, large subunit.
BLAST analysis was performed as previously described (2). The values in parentheses are the percent amino acid identity to the best match followed by the GenBank accession number of the best match.
Clone AOKan4.
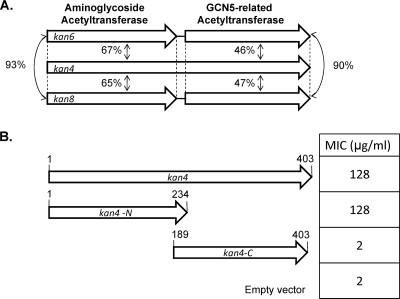
Selection for kanamycin resistance yielded three unique clones. Two of these clones, AOKan6 and AOKan8, each contain a gene that encodes an aminoglycoside acetyltransferase (aac6′). Each also contains a gene encoding an acetyltransferase immediately downstream of the aminoglycoside acetyltransferase (Fig. 3A). The third clone conferring kanamycin resistance, AOKan4, also encodes these two acetyltransferases, but in this case, in a single ORF.
FIG. 3.
Clones conferring kanamycin resistance. (A) The kan4 ORF is predicted to encode a bifunctional protein. The N-terminal domain is predicted to be an aminoglycoside acetyltransferase. The C-terminal domain encodes a predicted GCN5-related acetyltransferase. Proteins encoded by AOKan6 and AOKan8 were aligned to Kan4. The percentages represent the pairwise amino acid identities from each alignment. (B) The parts of kan4 encoding the two domains were cloned separately into pET44a and expressed in E. coli. The numbers above each ORF reflect amino acid numbering. The MICs represent the levels of resistance to kanamycin conferred by each of the clones.
The two acetyltransferase domains in Kan4 were subcloned (Fig. 3B). MIC analysis demonstrated that the N-terminal 233 amino acids alone conferred the same level of kanamycin resistance (128 μg/ml) as the full-length protein. This is consistent with the homology of the N-terminal domain to known aminoglycoside acetyltransferases. The C-terminal domain did not confer resistance to kanamycin.
DISCUSSION
In this study, we used functional metagenomics to identify 13 antibiotic resistance genes in soil from an apple orchard that has been treated with streptomycin. These genes included β-lactamases, aminoglycoside acetyltransferases, a multidrug efflux pump, and a bifunctional protein containing a natural fusion of a β-lactamase and a sigma factor.
Our initial expectation was to identify streptomycin resistance genes from a community of soil bacteria that had experienced repeated treatment with streptomycin. When we did not detect streptomycin resistance in our functional screens, we used PCR to verify that streptomycin resistance genes are indeed present in the metagenomic library. Since our analysis included only the coding region of the strA and strB genes, it is possible that their regulatory elements contain mutations that would prevent their expression. However, the most likely explanation for this discrepancy is that the streptomycin resistance genes are regulated by genetic elements that are not recognized by the E. coli gene expression machinery.
The low sequence identities of the enzymes identified in this study highlight the novelty of these proteins. Many of the best matches in the NCBI nr database share less than 60% amino acid identity with the enzymes that we found in soil metagenomic libraries. Furthermore, nucleotide BLAST analysis of the NCBI nr database consistently failed to identify genes with significant matches. This lack of sequence conservation with previously described enzymes suggests that functional assays would identify genes that would likely be missed in a sequence-only analysis.
The structure and function of the β-lactamase fused to a putative transcriptional regulator are intriguing. Transcriptional regulators are functional inside the cell membrane, whereas β-lactamases cleave extracellular molecules. Further experimentation will address the localization of this protein as well as its role in the life of the microorganism.
Additional analysis of the sequence upstream of the cft2 gene provides evidence that this predicted bifunctional gene was not created as an artifact of the library cloning process. The C terminus of the Cft2 protein shows homology to other β-lactamases in the PER-1 subclass of the class A β-lactamases (29). The region of DNA upstream of enzymes in this subclass contains a transposase as well as a conserved inverted repeat element (22, 23, 25, 26). Similarly, the region upstream of the cft2 gene contains a putative transposase gene as well as a 12 out of 17 bp match to the inverted repeat element consensus sequence.
To our knowledge, the kan4 gene encodes the first reported bifunctional aminoglycoside acetyltransferase in which only one domain is involved in antibiotic resistance. There are four reported bifunctional enzymes in which an aminoglycoside acetyltransferase is fused to another domain that also functions in the detoxification of aminoglycoside antibiotics (6, 11, 17). In the case of Kan4, the N-terminal domain confers resistance to kanamycin, while the C-terminal domain is predicted to encode an acetyltransferase that does not utilize kanamycin as a substrate.
This study, coupled with previous work from our laboratory (1), has uncovered three novel bifunctional enzymes involved in antibiotic resistance. Their presence suggests that these enzymes are more common than previously thought. The findings highlight the benefit of functional metagenomics: without limiting discovery to those genes that can be recognized as resistance determinants by sequence, we uncovered novel enzymes.
Acknowledgments
We thank Lynn Williamson for assistance with library construction and Heather Allen for critical comments on the manuscript.
This work was funded by USDA grant 2006-35319-17466, NIH fellowship GM876101 to J.J.D., and USDA CSREES project number WISR-2006-02619 to L.A.M.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Allen, H. K., L. A. Moe, J. Rodbumrer, A. Gaarder, and J. Handelsman. 2009. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 3:243-251. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambler, R. P. 1980. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289:321-331. [DOI] [PubMed] [Google Scholar]
- 4.Austin, D. J., K. G. Kristinsson, and R. M. Anderson. 1999. The relationship between the volume of antimicrobial consumption in human communities and the frequency of resistance. Proc. Natl. Acad. Sci. U. S. A. 96:1152-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, and O. Zagnitko. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azucena, E., I. Grapsas, and S. Mobashery. 1997. Properties of a bifunctional bacterial antibiotic resistance enzyme that catalyzes ATP-dependent 2′′-phosphorylation and acetyl-CoA-dependent 6′-acetylation of aminoglycosides. J. Am. Chem. Soc. 119:2317-2318. [Google Scholar]
- 7.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of Archaea from soil. Proc. Natl. Acad. Sci. U. S. A. 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G. Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucher, H. W., G. H. Talbot, J. S. Bradley, J. E. Edwards, D. Gilbert, L. B. Rice, M. Scheld, B. Spellberg, and J. Bartlett. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1-12. [DOI] [PubMed] [Google Scholar]
- 11.Centrón, D., and P. H. Roy. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 46:1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiou, C. S., and A. L. Jones. 1995. Expression and identification of the strA-strB gene pair from streptomycin-resistant Erwinia amylovora. Gene 152:47-51. [DOI] [PubMed] [Google Scholar]
- 13.Chiou, C. S., and A. L. Jones. 1993. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J. Bacteriol. 175:732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conway, S. P., K. G. Brownlee, M. Denton, and D. G. Peckham. 2003. Antibiotic treatment of multidrug-resistant organisms in cystic fibrosis. Am. J. Respir. Med. 2:321-332. [DOI] [PubMed] [Google Scholar]
- 15.D'Costa, V. M., K. M. McGrann, D. W. Hughes, and G. D. Wright. 2006. Sampling the antibiotic resistome. Science 311:374-377. [DOI] [PubMed] [Google Scholar]
- 16.Enright, M. C. 2003. The evolution of a resistant pathogen—the case of MRSA. Curr. Opin. Pharmacol. 3:474-479. [DOI] [PubMed] [Google Scholar]
- 17.Kim, C., A. Villegas-Estrada, D. Hesek, and S. Mobashery. 2007. Mechanistic characterization of the bifunctional aminoglycoside-modifying enzyme AAC(3)-Ib/AAC(6′)-Ib′ from Pseudomonas aeruginosa. Biochemistry 46:5270-5282. [DOI] [PubMed] [Google Scholar]
- 18.Kuske, C. R., L. O. Ticknor, M. E. Miller, J. M. Dunbar, J. A. Davis, S. M. Barns, and J. Belnap. 2002. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeCleir, G. R., A. Buchan, J. Maurer, M. A. Moran, and J. T. Hollibaugh. 2007. Comparison of chitinolytic enzymes from an alkaline, hypersaline lake and an estuary. Environ. Microbiol. 9:197-205. [DOI] [PubMed] [Google Scholar]
- 20.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liles, M. R., L. L. Williamson, J. Rodbumrer, V. Torsvik, R. M. Goodman, and J. Handelsman. 2008. Recovery, purification, and cloning of high-molecular-weight DNA from soil microorganisms. Appl. Environ. Microbiol. 74:3302-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llanes, C., C. Neuwirth, F. El Garch, D. Hocquet, and P. Plesiat. 2006. Genetic analysis of a multiresistant strain of Pseudomonas aeruginosa producing PER-1 beta-lactamase. Clin. Microbiol. Infect. 12:270-278. [DOI] [PubMed] [Google Scholar]
- 23.Mantengoli, E., and G. M. Rossolini. 2005. Tn5393d, a complex Tn5393 derivative carrying the PER-1 extended-spectrum beta-lactamase gene and other resistance determinants. Antimicrob. Agents Chemother. 49:3289-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCaig, A. E., S. J. Grayston, J. I. Prosser, and L. A. Glover. 2001. Impact of cultivation on characterisation of species composition of soil bacterial communities. FEMS Microbiol. Ecol. 35:37-48. [DOI] [PubMed] [Google Scholar]
- 25.Poirel, L., L. Cabanne, H. Vahaboglu, and P. Nordmann. 2005. Genetic environment and expression of the extended-spectrum beta-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob. Agents Chemother. 49:1708-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Power, P., J. Di Conza, M. M. Rodriguez, B. Ghiglione, J. A. Ayala, J. M. Casellas, M. Radice, and G. Gutkind. 2007. Biochemical characterization of PER-2 and genetic environment of blaPER-2. Antimicrob. Agents Chemother. 51:2359-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riesenfeld, C. S., R. M. Goodman, and J. Handelsman. 2004. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 6:981-989. [DOI] [PubMed] [Google Scholar]
- 28.Sundin, G. W. 2002. Distinct recent lineages of the strA-strB streptomycin-resistance genes in clinical and environmental bacteria. Curr. Microbiol. 45:63-69. [DOI] [PubMed] [Google Scholar]
- 29.Tranier, S., A. T. Bouthors, L. Maveyraud, V. Guillet, W. Sougakoff, and J. P. Samama. 2000. The high resolution crystal structure for class A beta-lactamase PER-1 reveals the bases for its increase in breadth of activity. J. Biol. Chem. 275:28075-28082. [DOI] [PubMed] [Google Scholar]
- 30.van Overbeek, L. S., E. M. H. Wellington, S. Egan, K. Smalla, H. Heuer, J.-M. Collard, G. Guillaume, A. D. Karagouni, T. L. Nikolakopoulou, and J. D. van Elsas. 2002. Prevalence of streptomycin-resistance genes in bacterial populations in European habitats. FEMS Microbiol. Ecol. 42:277-288. [DOI] [PubMed] [Google Scholar]
- 31.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]