Abstract
Protein secretion plays an eminent role in cell maintenance and adaptation to the extracellular environment of microorganisms. Although protein secretion is an extremely efficient process in filamentous fungi, the mechanisms underlying protein secretion have remained largely uncharacterized in these organisms. In this study, we analyzed the effects of the d-xylose induction of cellulase and hemicellulase enzyme secretion on the protein composition of secretory organelles in Aspergillus niger. We aimed to systematically identify the components involved in the secretion of these enzymes via mass spectrometry of enriched subcellular microsomal fractions. Under each condition, fractions enriched for secretory organelles were processed for tandem mass spectrometry, resulting in the identification of peptides that originate from 1,081 proteins, 254 of which—many of them hypothetical proteins—were predicted to play direct roles in the secretory pathway. d-Xylose induction led to an increase in specific small GTPases known to be associated with polarized growth, exocytosis, and endocytosis. Moreover, the endoplasmic-reticulum-associated degradation (ERAD) components Cdc48 and all 14 of the 20S proteasomal subunits were recruited to the secretory organelles. In conclusion, induction of extracellular enzymes results in specific changes in the secretory subproteome of A. niger, and the most prominent change found in this study was the recruitment of the 20S proteasomal subunits to the secretory organelles.
Aspergillus niger is a soil-dwelling filamentous fungus with a high capacity for decomposing plant materials. Many of the secreted depolymerizing enzymes have important biotechnological applications, e.g., to modify plant-derived food products. Homologous protein secretion in filamentous fungi may yield up to 20 g per liter of extracellular medium (14). For this reason, A. niger has been used intensively as a cell factory for enzyme production (3, 14, 32). In contrast to this, yields are much lower for heterologous protein secretion, with exceptions (11, 18, 35, 59).
The secretory potential of A. niger is not well understood, and only a limited number of functional studies have been performed to investigate the major components of the fungal secretion pathway. These studies have addressed overall transcriptional and translational responses in A. niger by studying the impact of secretion stress-inducing chemicals, temperature shifts, protein overproduction, or growth on carbon sources that induce changes in secretory states (20, 23, 26).
In recent years, high-throughput shotgun proteomics has been used to study cell organelle makeup and function. Through the combined use of liquid chromatography and tandem mass spectrometry (LC-MS/MS), various studies have identified many organelle-specific proteins, including proteins related to protein secretion (18, 24, 29, 51, 51). In the case of aspergilli, such studies have focused mainly on the secretome and not on the actual components of the secretory pathway (34, 36, 55).
In a previous study, we established defined culture conditions for the induced expression of the cellulase and hemicellulase enzyme system of A. niger (56). The expression of the corresponding genes is controlled by the dedicated transcriptional activator XlnR and its inducer, d-xylose (58). Using identical conditions with a different strain of A. niger, we applied shotgun proteomics for the analysis of enriched microsomal fractions containing endoplasmic reticulum (ER) membranes with associated ribosomes and Golgi membranes. The microsomal fractions were isolated from d-xylose-induced and non-d-sorbitol-induced mycelium. Following this approach, we were able to identify major protein components of the secretory organelles and to estimate their relative abundances in enriched microsomal samples.
MATERIALS AND METHODS
Strain and culture conditions.
The fungal strain used in this study was A. niger wild type N400 (CBS 120.49). For preculture, 1.0 × 106 spores per milliliter were inoculated into 2.5-liter fermentors (Applikon) containing 2.2 liters of minimal medium (44) with 0.05% yeast extract and 100 mM d-sorbitol at 30°C. Spore germination in bioreactors was as described previously (56), with headspace aeration and a stirring speed of 300 rpm; when dissolved-oxygen levels were below 60%, the stirring speed was increased to 750 rpm and aeration was switched to the sparger inlet. This moment was defined as the actual culture starting point (t = 0). At 14 h from the starting point, d-xylose or d-sorbitol (10 mM) was added to each culture, and at a t of 16 h, mycelia were harvested.
Subcellular fractionation and marker enzyme assays.
Subcellular fractionation was based on the work of Record and coworkers (46) with important modifications. Mycelia were harvested by Büchner filtration on nylon gauze and washed with sterile cold 0.25 M sucrose. From that moment until storage of the samples, all procedures were conducted at 4°C. Mycelia were press dried, and under each of the two conditions (addition of d-xylose or d-sorbitol), 6 g of mycelial sample was added to 80 ml of homogenization buffer (0.25 M sucrose, 1 mM EDTA, 20 mM HEPES, pH 7.6) containing 1% (vol/vol) protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Cells were disrupted by French press cell homogenization at 3,000 lb/in2 (20.6 MPa). The resulting homogeneous suspension was filtered through two layers of nylon gauze, and the homogenate suspensions (60 ml per condition) were centrifuged at 6,500 × g for 15 min. The supernatants were centrifuged at 29,000 × g for 18 min. The mitochondrial pellets were resuspended in 2 ml of homogenization buffer with protease inhibitors, and the supernatants were centrifuged at 100,000 × g for 1 h 12 min. The microsome-enriched pellets were resuspended in 2 ml of 0.4 M sucrose, 1 mM EDTA, 20 mM HEPES, pH 7.6, in a Dounce homogenizer (10 gentle strokes with a loose piston), and 200 μl of these suspensions (1 mg protein) was loaded onto each linear 9-ml buffered sucrose gradient (1 mM EDTA, 20 mM HEPES, pH 7.6; densities, 1.06 to 1.17 g·ml−1) and centrifuged at 130,000 × g for 13 h 42 min at 4°C. Fractions of 360 μl were collected, and samples were stored at −80°C until further processing was done. Protein concentrations were estimated using the Bradford method (7), with bovine serum albumin as a standard. The NADPH-cytochrome c reductase assay, as a marker enzyme for microsomes, was done using a commercially available kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions.
Sample preparation for LC-MS/MS.
Enriched microsomal fractions from ultracentrifugation were pooled in two groups according to density (one of higher density and one of lower density) and loaded onto Microcon YM-10 columns (cutoff, 10 kDa; Millipore, Eschborn, Germany). The concentrated enriched samples were run on 12% SDS-polyacrylamide gels, and the gels were stained according to the manufacturer's instructions using Colloidal Blue staining (Invitrogen, Carlsbad, CA). Each gel lane was cut into five slices, and the slices were cut into smaller pieces of about 1 mm3 and washed with ultrapure water and 50% acetonitrile in 50 mM NH4HCO3, pH 8.0. The gel samples were reduced with 50 mM dithiothreitol (DTT) in 50 mM NH4HCO3, pH 8.0, for 2 h at 60°C. Subsequently, the DTT solutions were removed and samples were alkylated with 100 mM iodoacetamide in NH4HCO3, pH 8.0, for 1 h at room temperature in the dark with occasional mixing. The iodoacetamide solutions were removed, and samples were washed with NH4HCO3, pH 8.0. Gel pieces were frozen at −20°C and thawed at least three times to increase the accessible area for trypsin digestion. All gel pieces were rehydrated in 25 ng·μl−1 trypsin (sequencing-grade modified trypsin; Promega, Madison, WI) and digested overnight at room temperature. In order to maximize peptide extraction, all solutions from trypsin digestion were transferred to new tubes, and the gel pieces were subjected to two rounds of 1 min of sonication, the first round with 10% trifluoroacetic acid (TFA) and the second round with 5% acetonitrile, 1% TFA. After each of these two rounds, the solutions were removed and added to the original trypsin digests. The final pH was adjusted to 2.5 by addition of 10% TFA.
Liquid chromatography-tandem mass spectrometric analysis.
LC-MS/MS was performed at Biqualys, as described previously (57). Briefly, samples were loaded on a preconcentration column, and peptides were eluted to an analytical column with an acetonitrile gradient and a fixed concentration of formic acid. The resulting eluent was subjected to an electrospray potential via a coupled platinum electrode. MS spectra were measured on an LTQ-Orbitrap (Thermo Electron, San Jose, CA), and MS scans of the four most abundant peaks were recorded in data-dependent mode.
Mass spectrometry database searching.
The resulting spectra from the MS analysis of the A. niger samples were submitted to a local implementation of the open mass spectrometry search algorithm (OMSSA) search engine (15). MS/MS spectra were independently searched against peptide databases derived from the predicted proteomes of A. niger strain CBS513.88 and strain ATCC 1015, and a decoy reverse database was constructed from the reverse CBS513.88 proteome.
All OMSSA searches used the following parameters: a precursor ion tolerance of 0.2 Da, a fragment ion tolerance of 0.3 Da, a missed-cleavage allowance of ≤2, fixed carbamide methylation, variable oxidation of methionine, and deamination of glutamine and asparagine.
The E value threshold was determined iteratively from the false-discovery rate (FDR) and was set to 0.01. With this setting, an FDR of <5% is expected.
FDR calculation was done as follows. Peptide spectrum matches with each individual peptide database were ranked by their E values for each identified spectrum with a threshold E value of <0.01, and the top hit identified peptide sequence was selected. For FDR calculation, top hit spectral matches to peptides in the reversed database rCBS were taken, and the number of false positives was divided by the number of total positives.
Functional annotation groups and sequence analysis.
Proteins predicted to play a role in secretion were grouped in functional annotation groups guided by previously published functional classification schemes (16), the Functional Catalogue (FunCat) annotation scheme, and the predicted molecular function as provided with the A. niger CBS531.88 genome annotation (42). Proteins considered to be involved in secretion were used for a BLAST search (NCBI) against Saccharomyces cerevisiae and Homo sapiens protein databases. Only the yeast and human reference sequence proteins that presented the lowest E values and highest percentages of positives were included in this study. Protein similarity analyses were done using the ClustalW and Jalview version 2 tools (54, 60). Transmembrane domains (TMD) were assessed using the TMHMM tool (50).
Relative protein abundance and statistics.
A normalized spectral abundance factor was used as a parameter to estimate relative protein abundance using the following equation (63):
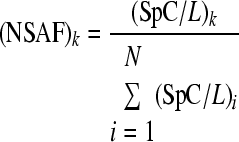 |
where (NSAF)k is the normalized spectral abundance factor for a protein k, SpC is the number of spectral counts (the total number of MS/MS spectra) for a given protein plus a pseudocount factor of 0.5, and L is the protein's length. The pseudocount was necessary to analyze the differential distribution of secretion-related proteins between the two conditions (addition of xylose or sorbitol). For the proteins predicted to be involved in secretion, significant differences in NSAF values were evaluated by applying a likelihood ratio test (G test) for independence using a null hypothesis of equal protein distribution between the two conditions.
QPCR.
RNA isolation, cDNA synthesis, quantitative real-time PCR (QPCR), and data analysis were performed as described previously (37). In brief, total RNA was extracted, quantified, and mixed with an exogenous RNA reference transcript. After enzymatic DNA degradation, oligo(dT)-mediated cDNA synthesis was performed. QPCR was performed using QPCR SYBR green mix (ABgene, Epsom, United Kingdom) and specific oligonucleotide primers (see Table S1 in the supplemental material). Two independent PCR runs were performed per cDNA sample from each biological sample (xylose or control sorbitol addition). The Pfaffl method (43) was used to calculate relative gene expression levels double normalized to the added exogenous RNA transcript and to the standard condition, i.e., the average normalized expression of the two biological samples from the sorbitol control condition.
RESULTS
Global protein analysis of the secretory machinery of A. niger.
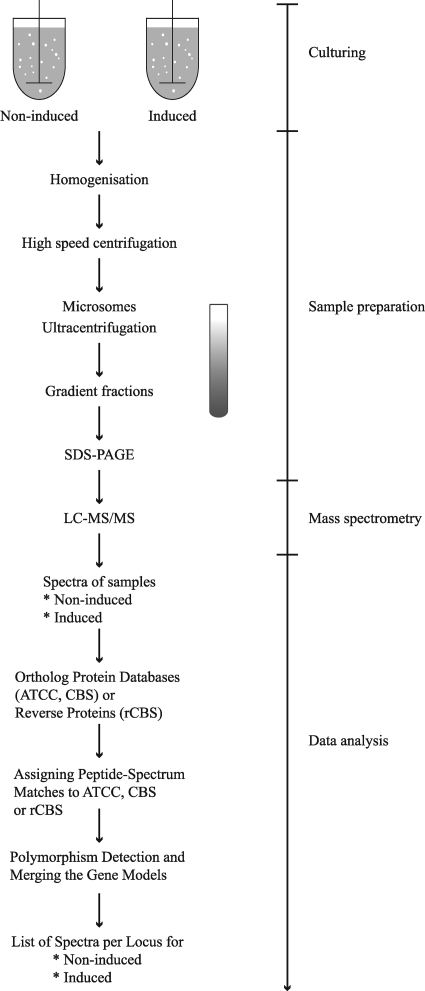
Shotgun proteomics was applied to analyze the secretory machinery of A. niger under induction of secretion, i.e., induction of cellulases and hemicellulases by d-xylose. For each of the two conditions, addition of d-xylose (induced) or d-sorbitol (noninduced), biological duplicates were taken and proteins from different pooled gradient fractions were analyzed (Fig. 1) to increase the resolution, similarly to previous studies (17, 29). Overall, between 32,000 and 50,000 MS/MS spectra were obtained per condition, and 25 to 30% of these spectra could be assigned to proteins. In total, 1,081 proteins were identified: 638 proteins were present under both conditions, 282 proteins were found only in the d-xylose-induced samples, and 161 proteins were found exclusively in the samples from sorbitol addition. A protein database is available for A. niger CBS120.49, the strain used in this work (52). However, despite the improved annotation, many open reading frames (∼5,672) lack a translation start, resulting from errors in the genome sequence. Although not fully identical to the strain used in our laboratory, the A. niger CBS513.88 genome-derived database was used, since from this genome, an additional 3,000 proteins are predicted compared to the other high-coverage genome-sequenced strains, such as A. niger ATCC 1015 (5). For a limited number of spectra, no proteins could be matched to the CBS513.88 proteome, and in these cases, the A. niger ATCC 1015 database was used. The proteins identified in this way were included in this study (see Table S2 in the supplemental material). Upon initial curation, the total number of peptides per condition could be primarily divided in the following manner: related to membrane traffic and protein secretion, 26%; mitochondrial, 15%; ribosomal and translation, 13%; metabolism, 13%; lipid biosynthesis and cytochrome P450 enzymes, 10%; cargo proteins, 5%; nuclear, 3%; and other functions or unknown, 15%. Proteins from nonsecretory organelles, i.e., the nucleus and the mitochondrion, are listed in Table S3 in the supplemental material.
FIG. 1.
Workflow of the subcellular organelle sample preparation and data analysis.
Many secretion components in A. niger are highly similar to mammalian proteins.
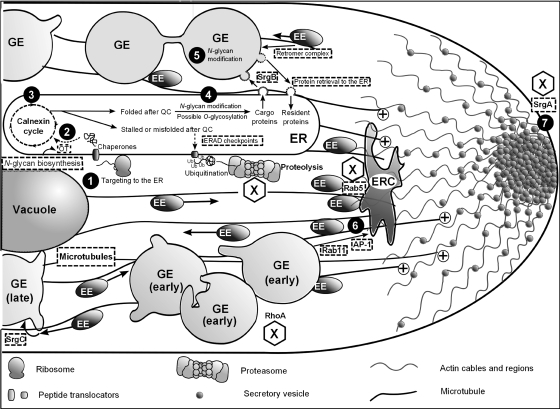
Many of the membrane traffic and secretion components identified were found to be more similar to mammalian than to yeast proteins (see Tables S4 to S7, boldface, in the supplemental material). These proteins (Fig. 2) were predicted to be involved in (i) N-glycan biosynthesis and transfer to asparagine in normally glycosylated target proteins, (ii) ER-to-Golgi network anterograde and retrograde transport, (iii) processes mediated by Rab GTPases and interacting factors, (iv) microtubule-related processes, and (v) the early checkpoints of the ER-associated degradation (ERAD) pathway. Furthermore, proteins, such as UDP-glucose-glycoprotein glucosyltransferase, were found to be much more similar to human than to yeast counterparts, particularly at the catalytic core (see Fig. S1 in the supplemental material). In addition, we were able to detect five secretory pathway candidate proteins from A. niger that lacked a corresponding yeast homologue: a protein for utilization of Dol-P-mannose (An04g03130), a UDP-galactose transporter (An08g10400), a tetratricopeptide repeat domain 35 protein (54765; JGI), annexin AnxC3.1 (An02g05210), and a dynamin-related Ras GTPase (An06g02180).
FIG. 2.
Secretory processes controlled by mammalian-like proteins in A. niger. The dashed arrows and text boxes indicate processes that are carried out by a majority of proteins more similar to human than to yeast proteins. 1, peptide biosynthesis and translocation to the ER; 2, addition of N-glycan; 3, glycoprotein folding; 4, N and O glycosylation; 5, N glycosylation after transport to Golgi equivalents (GEs); 6, protein sorting and formation of secretory vesicles; 7, vesicle fusion with the plasma membrane; EE, early endosomes; ERC, endosomal recycling compartment; OT, oligosaccharyltransferase complex; QC, quality control; X, proteins abundant after d-xylose induction.
Secretory pathway proteins of A. niger.
From all proteins identified in this study, 254 proteins were predicted to play a direct role in membrane traffic and protein secretion (see Tables S4 to S7 in the supplemental material). The secretion-related proteins with the highest abundances identified in this study were molecular chaperones (e.g., PdiA and BipA), the structural molecules actin and tubulin, oligosaccharyltransferase subunits, and small GTPases (Table 1 ). Of the 34 high-abundance proteins identified, 11 have been previously characterized (Table 1, boldface). What is more, 21 secretory pathway proteins were differentially expressed in microsomal samples after the addition of d-xylose or d-sorbitol (Table 2; see Table S2 in the supplemental material). The secretion-related proteins overexpressed after d-xylose induction comprised the subunits of the 20S core particle of the proteasome, the ER retrotranslocation ATPase Cdc48, Gpi12 for glycosylphosphatidylinositol (GPI) anchor biosynthesis, the GTPases RhoA and SrgA, and a homologue of the human Rab5C protein. For the d-sorbitol condition, the cyclophilin CypA and proteins involved in targeting to the ER were found to be more abundant in the microsomal fractions.
TABLE 1.
Most abundant secretory pathway proteins
| A. niger locus taga | Yeast homologue | Specific process and function | NSAF (104)b |
|
|---|---|---|---|---|
| S | X | |||
| An02g14800 | PDI1 | Protein disulfide isomerase PdiA: formation and unscrambling of S-S bonds (essential) | 75.6 | 59 |
| An07g09990 | SSA4 | Hsp70 protein: involved in SRP-dependent protein targeting to the ER | 72.5 | 40 |
| An11g04180 | KAR2 | ER ATPase BipA: protein import, protein folding; regulates UPRd via Ire1; role in karyogamy | 71.8 | 52.7 |
| An07g08300 | CPR1 | Peptidyl-proline cis-trans isomerase CypA: cyclophilin | 65.1 | 32.7 |
| An17g01420 | YOP1 | Membrane protein: Yip1 interaction; ER tubular morphology | 56.2 | 40 |
| An15g00560 | ACT1 | Actin structural protein ActA: cell polarity, endocytosis | 53.8 | 42.1 |
| An07g04570 | HYP2 | HexA protein: assembly of Woronin body (septal pore sealing) | 50 | 28 |
| An09g06590 | HSC82 | Hsp90 chaperone SspB: nearly identical to Hsp82 | 46.9 | 38.7 |
| An01g08420 | CNE1 | Integral membrane ER chaperone ClxA: folding/QCc of glycoproteins (calnexin cycle) | 42.4 | 46.4 |
| An07g04190 | WBP1 | OTe complex β subunit (essential) | 42.3 | 38 |
| An02g14560 | OST1 | OT complex α subunit OstA: ribophorin I (essential) | 41.1 | 35.7 |
| An01g04040 | SAR1 | ARFf GTPase SarA: ER-to-Golgi vesicle formation | 39.2 | 26.2 |
| An04g02020 | CPR1 | ER peptidyl-prolyl cis-trans isomerase CypB: cyclophilin | 37.4 | 41.3 |
| An01g12810 | YET3 | Mutant that decreases level of secreted invertase | 34.6 | 20.7 |
| An08g03190 | TUB2 | Structural protein TubB: β-tubulin forms dimers with α-tubulin, and these form microtubules | 34.1 | 29.3 |
| An18g05980 | RHO1 | Ras GTPase RhoA: Rho subfamily; involved in cell polarity | 33 | 61.2 |
| An18g02020 | PDI1 | Protein disulfide isomerase TigA: S-S bonds | 33 | 25.9 |
| An04g02070 | CHC1 | Structural molecule: clathrin heavy chain | 31.4 | 15 |
| An07g07760 | BMH1 | 14-3-3 protein similar to ArtA: DNA/protein binding, exocytosis, Ras/MAPK signaling | 30.5 | 29.5 |
| An18g06270 | BMH2 | 1 14-3-3 protein ArtA: DNA/protein binding, exocytosis, Ras/MAPK signaling | 30.1 | 28.8 |
| An09g05880 | ROT2 | ER glucosidase II α subunit: sequential removal of 2 Glc residues (calnexin cycle) | 27.6 | 28.9 |
| An01g04600 | MPD1 | Protein disulfide isomerase PrpA: N terminus similar to Mpd1, C terminus unknown | 26.6 | 26.2 |
| An04g07440 | SHR3 | ER packaging chaperone: incorporation of amino acid permeases into COPII-coated vesicles | 26.4 | 15.5 |
| An16g04330 | DPM1 | ER dolichyl-phosphate β-d-mannosyltransferase, also required for O mannosylation | 26 | 23.9 |
| An05g00140 | SRP102 | Signal recognition particle (SRP) receptor β subunit: anchors Srp101 to the ER membrane | 25.6 | 11.6 |
| An01g03820 | SBH2 | GEFg for ARFs: protein translocation to the ER | 25.6 | 7.8 |
| An09g06790 | YPT1 | Rab GTPase SrgB: ER-to-Golgi traffic | 24 | 21.1 |
| An12g00820 | UGP1 | UGPase: Glc-1-P + UTP ↔ UDP-Glc | 23.2 | 26.5 |
| An01g02500 | TRX1 | Thioredoxin: ER-Golgi transport and vacuole inheritance | 22.9 | 9.2 |
| An03g04340 | SEC61 | ER SRP-dependent and -independent protein targeting, P-P bond hydrolysis-driven transporter | 22.1 | 12.3 |
| An01g06060 | YPT31 | Rab GTPase: intra-Golgi network traffic, budding of post-Golgi vesicles | 21.6 | 24.4 |
| An15g05740 | YPT6 | Rab GTPase SrgC: fusion endosomal vesicles with the late Golgi network | 20.5 | 15.1 |
| An08g04480 | SEY1 | Dynamin GTPase: ER morphology | 20.2 | 19.9 |
| An01g13220 | LHS1 | ER nucleotide exchange factor for Kar2/BipA (LhsA): UPR-regulated translocation/folding | 20.2 | 22.1 |
Boldface indicates characterized genes.
S, sorbitol addition; X, xylose addition.
QC, quality control.
UPR, unfolded protein response.
OT, oligosaccharyltransferase.
ARF, ADP ribosylation factor.
GEF, guanine nucleotide exchange factor.
TABLE 2.
Relative abundances of secretion-related proteins in microsomes after addition of d-xylose or d-sorbitol
| Overrepresentation in microsomes | Locus tag | Description | G scorea |
|---|---|---|---|
| d-Xylose addition | An18g06700 | Pre7 20S CP β subunit of the proteasome | 26.20 |
| An04g09170 | Cdc48 retrotranslocation ATPase | 20.47 | |
| An15g00510 | Pre5 20S CP α subunit of the proteasome | 16.83 | |
| An02g10790 | Pre6 20S CP α subunit of the proteasome | 11.95 | |
| An11g06720 | Pre9 20S CP α subunit of the proteasome | 9.91 | |
| An05g02300 | Gpi12 ER protein: GPI anchor assembly | 9.47 | |
| An02g07040 | Scl1 20S CP α subunit of the proteasome | 9.23 | |
| An18g05980 | RhoA Ras GTPase: cell polarity | 8.56 | |
| An18g06800 | Pre10 20S CP α subunit of the proteasome | 8.52 | |
| An07g02010 | Pre8 20S CP α subunit of the proteasome | 8.36 | |
| An02g03400 | Pup2 20S CP α subunit of the proteasome | 7.48 | |
| An04g02470 | Rab5-like GTPase: endocytosis, protein sorting | 6.39 | |
| An14g00010 | SrgA Rab GTPase: exocytosis | 5.70 | |
| d-Sorbitol addition | An07g08300 | PPIase CypA: cyclophilin | 10.95 |
| An01g03820 | Shb2/Sbh1: protein translocation to the ER | 9.99 | |
| An07g09990 | Ssa4/3/1/2: SRP-dependent targeting to the ER | 9.49 | |
| An07g04570 | HexA protein: assembly of Woronin body | 6.31 | |
| An01g02500 | Thioredoxin: ER-Golgi network transport | 6.05 | |
| An04g02070 | Clathrin heavy chain: structural | 5.91 | |
| An05g00140 | SRP receptor β subunit: binds Srp101 to the ER | 5.45 | |
| 202202 (JGI) | Sac6 actin-bundling protein | 4.13 |
Normalized spectral abundance factors were compared by G test (P < 0.05).
d-Xylose induction leads to 20S proteasome recruitment to the microsomal fraction.
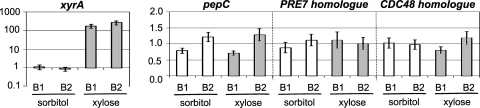
In a previous study, we analyzed transcript levels resulting from d-xylose induction by microarray analysis (56). In this data set, no significant change in the transcript levels of the gene encoding the 20S core particle of the proteasome was found. Since the strain used in our previous study was derived from A. niger CBS 120.49, we determined whether the significant increase in abundance of 20S proteasome complex proteins under the secretion-induced conditions was a result of increased transcription levels. For this, we compared the transcription levels of three distinct types of genes by quantitative PCR: the d-xylose reductase gene, which is strongly induced by d-xylose (21); the vacuolar serine proteinase C gene, which is stably transcribed under a wide range of conditions (25); and the homologues of the yeast genes PRE7 and CDC48. The corresponding proteins Pre7 and Cdc48 showed the largest difference in relative protein abundance in microsomes under the xylose-induced condition compared to the sorbitol control condition (Table 2). However, transcript levels for the A. niger homologues of yeast PRE7 and CDC48 were identical under xylose-induced and sorbitol control conditions (Fig. 3). In contrast to this and as expected, the transcript levels of the xylose reductase gene were strongly increased in the presence of d-xylose, while there was no significant change in the transcript levels of pepC.
FIG. 3.
Relative gene expression of the A. niger homologues of the yeast genes PRE7 and CDC48. B1, biological replicate 1; B2, biological replicate 2. Relative levels of xyrA transcripts are given in logarithmic scale. The bars represent the mean relative gene expression ± standard error.
DISCUSSION
Protein secretion is a fundamental feature of filamentous fungi that is strongly related to the fungal saprophytic lifestyle. Hyphal growth requires coordination of protein secretion with cell wall synthesis, polarized growth, recycling by endocytosis, and transport of extracellular hydrolytic enzymes. Ironically, protein secretion in the kingdom Fungi has been best described for typically modest secretors, such as the yeast S. cerevisiae. Thus, compared to yeast, more sophisticated secretory mechanisms exist in the protein secretor A. niger. Here, we aimed to identify candidate microsomal proteins involved in intracellular membrane traffic and protein secretion upon cellulase and hemicellulase induction by d-xylose.
In order to decrease false-positive identifications of peptides, experiments were done in duplicate, and a conservative criterion was applied for peptide identification (at least 2 unique peptides in one of the biological samples and another unique peptide in the replicate), resulting in the identification of 30% of ∼40,000 MS peptide spectra per condition. Organelle isolation is prone to contamination by cell components other than the target organelle, as has become clear after the use of sensitive methods such as mass spectrometry (22, 31). Apart from technical challenges, other factors may obscure shotgun proteomic analyses, such as the multiple localization of a given protein or the direct physical interaction of target organelles with other organelles or intracellular structures (33, 47, 48).
Despite these limitations, we were able to identify key components of the secretory pathway of A. niger and to estimate their relative abundances. With regard to the relative abundance in microsomal samples, only 11 of the 34 most abundant protein components found have been previously characterized or described for A. niger (Table 1, boldface). Besides ranking high-abundance proteins, NSAF calculation also allows the estimation of the stoichiometry of protein subunits in protein complexes. Many protein complexes in A. niger were found present with the same stoichiometry as the ones observed in yeast protein complexes in vivo (Table 3). These protein complexes are predicted to be involved in a diversity of biological processes, namely (i) protein targeting to the ER by the Sec63-Sec66-Sec62 complex; (ii) addition of N-glycan to the protein asparagine by the oligosaccharyltransferase complex; (iii) ER-to-Golgi network transport by the complexes formed by Sec23-Sec24, Emp24-Erv25, and Erv41-Erv46; (iii) Golgi network protein sorting by the Apl2-Apm1-Apl4 complex; and (iv) endosome-to-Golgi network transport by the Pep8-Vps29-Vps35 complex. On the other hand, one protein complex was not completely identified in this study. This complex, termed the chaperonin-containing T complex (CCT), is required in yeast for the cytosolic assembly of actin and tubulins in vivo. This discrepancy could be the result of selective peptide picking during MS, incorrect gene models to identify peptides, different stoichiometry in A. niger, or undersampling, as the NSAF values of the identified CCT subunits were low compared to the NSAF values of the aforementioned protein complexes found with the expected stoichiometry. Moreover, we were unable to detect a few other protein complexes predicted to play a role in membrane traffic or protein secretion in A. niger, such as the HOPS complex for vacuole fusion and vacuole protein sorting, the GARP complex for traffic from the early endosome to the late Golgi, the ESCRT endosomal sorting complexes, and the Exocyst complex for targeting of vesicles to active sites of exocytosis. It is possible either that these complexes rarely occur in secretory organelles in A. niger or that they are more present in vesicle subpopulations that were incompletely covered during organelle enrichment procedures.
TABLE 3.
Secretory pathway and membrane-trafficking protein complexes identified in this study and protein subunit relative abundances
| Complexa | Reference | S. cerevisiae protein | A. niger locus tag | NSAF (10,000)b |
|
|---|---|---|---|---|---|
| Sorbitol | Xylose | ||||
| Sec62/Sec63 | 13 | Sec63 | An01g13070 | 16.9 | 14.8 |
| Sec66 | An16g08830 | 19.8 | 16.4 | ||
| Sec62 | An02g01510 | 14.3 | 9.1 | ||
| OT subcomplexes | 28 | Stt3 | An16g08570 | 11.4 | 15.7 |
| Ost3 | An02g14930 | 8.2 | 15.3 | ||
| Ost4c | An08g07485 (N terminus) | ND | ND | ||
| Ost1 | An02g14560 | 41.1 | 35.7 | ||
| Wbp1 | An07g04190 | 42.3 | 38.0 | ||
| Sec23-Sec24 heterodimer | 12 | Sec23 | An01g04730 | 9.0 | 4.1 |
| Sec24 | An08g10650 | 6.4 | 6.6 | ||
| p24 | 6 | Emp24 | An08g03590 | 12.2 | 17.3 |
| Erv25 | An01g08870 | 16.7 | 22.7 | ||
| Erv41/Erv46 | 39 | Erv41 | An03g04940 | 7.5 | 11.1 |
| Erv46 | An01g04320 | 12.5 | 14.9 | ||
| AP-1 adaptor | 62 | Apl2 | An16g02490 | 2.6 | 0.8 |
| Apm1 | An07g03200 | 2.8 | 1.5 | ||
| Apl4 | An01g02600 | 2.6 | 1.4 | ||
| Retromer complex inner shell | 49 | Pep8 | An01g07320 | 9.2 | 4.4 |
| Vps29 | An08g01030 | 10.0 | 5.1 | ||
| Vps35 | An16g04270 | 11.3 | 9.5 | ||
| Chaperonin-containing T complex | 27 | Tcp1 | An15g06370 | ND | ND |
| Cct2 | An01g06480 | 1.2 | 1.2 | ||
| Cct3 | An02g08920 | ND | ND | ||
| Cct4 | An02g12750 | 2.2 | 1.2 | ||
| Cct5 | An11g02360 | ND | ND | ||
| Cct6 | An16g03100 | ND | ND | ||
| Cct7 | An18g05770 | 0.2 | 1.0 | ||
| Cct8 | An04g07340 | ND | ND | ||
OT, oligosaccharyltransferase.
ND, not detected.
S. cerevisiae Ost4 is 36 amino acids long, too small for the 10-kDa cutoff used in this study.
In A. niger, specific secretory processes are carried out by proteins more similar to mammalian proteins than to yeast proteins, a phenomenon that has been reported for filamentous fungi in general (8, 30). For example, most A. niger Rab GTPases identified here show higher similarity to mammalian than to yeast proteins. Since these proteins are involved in vesicle transport and fusion, it is very likely that this similarity to mammalian proteins would explain some fundamental differences in protein secretion of yeast and A. niger. Moreover, the controlled interplay between exo- and endocytosis is thought to be necessary for adequate polarized growth in filamentous fungi, and this interplay must in part be determined by the primary sequence of secretory pathway proteins.
In addition to the differences in terms of protein sequence in comparison to yeast, this study also reveals that important changes in the composition of the secretory pathway may be explained by different relative protein abundances in secretory organelles. During induction of cellulase and hemicellulase secretion by d-xylose, hydrolytic enzymes are efficiently synthesized and exported from the cell. These enzymes include proteins previously reported to be significantly upregulated by d-xylose induction, namely, β-xylosidase, a protein similar to bacterial xylosidase S (AxlA; An09g003300), and β-glucosidase (56). From the current study, it becomes clear that in addition to bringing changes in the amount of cargo hydrolytic enzymes, the presence of d-xylose also promotes changes in the relative protein abundance of specific components within the secretory organelles. Unexpectedly, proteins involved in targeting to the ER or quality control of protein glycosylation and folding were not overrepresented after d-xylose induction. On the contrary, these proteins were in some cases even underrepresented in comparison to the noninduced state of d-sorbitol addition. In this study, it was found that d-xylose induction is associated with an increase of three GTPases that are predicted to play roles in endocytosis, exocytosis, and polarized growth and with an increase of specific components of protein degradation pathways. With regard to the three overexpressed GTPases, the RhoA and SrgA GTPases have been described as necessary for polar growth and proper hyphal branching (4, 19, 38, 45), whereas Rab5 is known to play a role in early endocytosis in eukaryotes (7, 8). This important role in endocytosis has also been investigated recently in Aspergillus (1). From this study, it has become clear that Rab5-rich endosomes cycle rapidly along the hyphal cell in a microtubule-dependent manner. It is plausible that an increased state of protein secretion, such as induction by d-xylose in A. niger, increases the number of Rab5-rich early endosomes, thereby facilitating exchange of material with the Golgi apparatus equivalents and the hyphal tip. One observation that confirms this hypothesis is that Golgi apparatus equivalents in Aspergillus are relatively slow in motion and are usually found in a gradient across the hyphal cell, more concentrated near the hyphal tip but excluded from the apical body (41, 53).
Previous microarray analyses investigated the effects of d-xylose on the transcriptome of Aspergillus species and linked it to metabolic pathways or cellular processes (2, 26, 40, 56). In our study, and in agreement with the above-mentioned transcriptome studies, at the proteome level, d-xylose induction did not affect the microsomal abundance of most proteins belonging to the ERAD pathway. However, the presence of d-xylose did increase the protein microsomal abundance of Cdc48 homologue and the proteasomal 20S CP. Since there was no increase in the transcription levels of the proteasomal 20S CP, both previously by microarray analysis of a daughter fungal strain (56) and in this study by quantitative PCR, this strongly suggests that these proteins are recruited and that they can attach and detach from microsomes according to the cell's needs.
This study shows different strategies that A. niger undertakes when forced into a secretory state. Some functional routes of the cell's metabolism show differences between induced and noninduced states via different gene expression profiles, like the cargo proteins, followed by changes in the protein abundances. In other cases, no change in the transcriptional status of the organism was found, and changes in the protein abundance in the secretory organelles can be attributed to specific recruitment of functional complexes (e.g., the proteasome). Together, this shows that secretion is a dynamic and interacting process, several aspects of which can be studied using proteomic analyses.
Supplementary Material
Acknowledgments
M.W.J.V.P. is funded by the Netherlands Organization for Scientific Research (NWO) via a VENI grant.
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abenza, J. F., A. Pantazopoulou, J. M. Rodriguez, A. Galindo, and M. A. Peñalva. 2009. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic 10:57-75. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, M. R., W. Vongsangnak, G. Panagiotou, M. P. Salazar, L. Lehmann, and J. Nielsen. 2008. A trispecies Aspergillus microarray: comparative transcriptomics of three Aspergillus species. Proc. Natl. Acad. Sci. U. S. A. 105:4387-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, D. B., I. F. Connerton, and D. A. MacKenzie. 2008. Filamentous fungi for production of food additives and processing aids. Adv. Biochem. Eng. Biotechnol. 111:99-147. [DOI] [PubMed] [Google Scholar]
- 4.Asanuma, K., E. Yanagida-Asanuma, C. Faul, Y. Tomino, K. Kim, and P. Mundel. 2006. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat. Cell Biol. 8:485-491. [DOI] [PubMed] [Google Scholar]
- 5.Baker, S. E. 2006. Aspergillus niger genomics: past, present and into the future. Med. Mycol. 44(Suppl. 1):S17-S21. [DOI] [PubMed] [Google Scholar]
- 6.Belden, W. J., and C. Barlowe. 1996. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J. Biol. Chem. 271:26939-26946. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Braun, E. L., S. Kang, M. A. Nelson, and D. O. Natvig. 1998. Identification of the first fungal annexin: analysis of annexin gene duplications and implications for eukaryotic evolution. J. Mol. Evol. 47:531-543. [DOI] [PubMed] [Google Scholar]
- 9.Chen, P. I., C. Kong, X. Su, and P. D. Stahl. 2009. Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J. Biol. Chem. 284:30328-30338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compagnon, J., L. Gervais, M. S. Roman, S. Chamot-Boeuf, and A. Guichet. 2009. Interplay between Rab5 and PtdIns(4,5)P2 controls early endocytosis in the Drosophila germline. J. Cell Sci. 122:25-35. [DOI] [PubMed] [Google Scholar]
- 11.Dunn-Coleman, N. S., P. Bloebaum, R. M. Berka, E. Bodie, N. Robinson, G. Armstrong, M. Ward, M. Przetak, G. L. Carter, R. LaCost, L. J. Wilson, K. H. Kodama, E. F. Baliu, B. Bower, M. Lamsa, and H. Heinsohn. 1991. Commercial levels of chymosin production by Aspergillus. Nat. Biotechnol. 9:976-981. [DOI] [PubMed] [Google Scholar]
- 12.Fath, S., J. D. Mancias, X. Bi, and J. Goldberg. 2007. Structure and organization of coat proteins in the COPII cage. Cell 129:1325-1336. [DOI] [PubMed] [Google Scholar]
- 13.Feldheim, D., K. Yoshimura, A. Admon, and R. Schekman. 1993. Structural and functional characterization of Sec66p, a new subunit of the polypeptide translocation apparatus in the yeast endoplasmic reticulum. Mol. Biol. Cell 4:931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkelstein, D. B. 1987. Improvement of enzyme production in Aspergillus. Antonie Van Leeuwenhoek 53:349-352. [DOI] [PubMed] [Google Scholar]
- 15.Geer, L. Y., S. P. Markey, J. A. Kowalak, L. Wagner, M. Xu, D. M. Maynard, X. Yang, W. Shi, and S. H. Bryant. 2004. Open mass spectrometry search algorithm. J. Proteome Res. 3:958-964. [DOI] [PubMed] [Google Scholar]
- 16.Geysens, S., G. Whyteside, and D. B. Archer. 2009. Genomics of protein folding in the endoplasmic reticulum, secretion stress and glycosylation in the aspergilli. Fungal Genet. Biol. 46:S121-S140. [Google Scholar]
- 17.Gilchrist, A., C. E. Au, J. Hiding, A. W. Bell, J. Fernandez-Rodriguez, S. Lesimple, H. Nagaya, L. Roy, S. J. Gosline, M. Hallett, J. Paiement, R. E. Kearney, T. Nilsson, and J. J. Bergeron. 2006. Quantitative proteomics analysis of the secretory pathway. Cell 127:1265-1281. [DOI] [PubMed] [Google Scholar]
- 18.Gouka, R. J., P. J. Punt, and C. A. van den Hondel. 1997. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl. Microbiol. Biotechnol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 19.Guest, G. M., X. Lin, and M. Momany. 2004. Aspergillus nidulans RhoA is involved in polar growth, branching, and cell wall synthesis. Fungal. Genet. Biol. 41:13-22. [DOI] [PubMed] [Google Scholar]
- 20.Guillemette, T., N. N. van Peij, T. Goosen, K. Lanthaler, G. D. Robson, C. A. van den Hondel, H. Stam, and D. B. Archer. 2007. Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics 8:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasper, A. A., L. M. Trindade, D. van der Veen, A. J. van Ooyen, and L. H. de Graaff. 2004. Functional analysis of the transcriptional activator XlnR from Aspergillus niger. Microbiology 150:1367-1375. [DOI] [PubMed] [Google Scholar]
- 22.Huber, L. A., K. Pfaller, and I. Vietor. 2003. Organelle proteomics: implications for subcellular fractionation in proteomics. Circ. Res. 92:962-968. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs, D. I., M. M. Olsthoorn, I. Maillet, M. Akeroyd, S. Breestraat, S. Donkers, R. A. van der Hoeven, C. A. van den Hondel, R. Kooistra, T. Lapointe, H. Menke, R. Meulenberg, M. Misset, W. H. Muller, N. N. van Peij, A. Ram, S. Rodriguez, M. S. Roelofs, J. A. Roubos, M. W. van Tilborg, A. J. Verkleij, H. J. Pel, H. Stam, and C. M. Sagt. 2009. Effective lead selection for improved protein production in Aspergillus niger based on integrated genomics. Fungal Genet. Biol. 46(Suppl. 1):S141-S152. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs, M. E., L. V. DeSouza, H. Samaranayake, R. E. Pearlman, K. W. Siu, and L. A. Klobutcher. 2006. The Tetrahymena thermophila phagosome proteome. Eukaryot. Cell 5:1990-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarai, G., H. van den Hombergh, and F. P. Buxton. 1994. Cloning and characterization of the pepE gene of Aspergillus niger encoding a new aspartic protease and regulation of pepE and pepC. Gene 145:171-178. [DOI] [PubMed] [Google Scholar]
- 26.Jørgensen, T. R., T. Goosen, C. A. van den Hondel, A. F. Ram, and J. J. Iversen. 2009. Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway. BMC Genomics 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabir, M. A., J. Kaminska, G. B. Segel, G. Bethlendy, P. Lin, F. Della Seta, C. Blegen, K. M. Swiderek, T. Zoladek, K. T. Arndt, and F. Sherman. 2005. Physiological effects of unassembled chaperonin Cct subunits in the yeast Saccharomyces cerevisiae. Yeast 22:219-239. [DOI] [PubMed] [Google Scholar]
- 28.Karaoglu, D., D. J. Kelleher, and R. Gilmore. 1997. The highly conserved Stt3 protein is a subunit of the yeast oligosaccharyltransferase and forms a subcomplex with Ost3p and Ost4p. J. Biol. Chem. 272:32513-32520. [DOI] [PubMed] [Google Scholar]
- 29.Kislinger, T., B. Cox, A. Kannan, C. Chung, P. Hu, A. Ignatchenko, M. S. Scott, A. O. Gramolini, Q. Morris, M. T. Hallett, J. Rossant, T. R. Hughes, B. Frey, and A. Emili. 2006. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125:173-186. [DOI] [PubMed] [Google Scholar]
- 30.Kruszewska, J. S., M. Saloheimo, A. Migdalski, P. Orlean, M. Penttilä, and G. Palamarczyk. 2000. Dolichol phosphate mannose synthase from the filamentous fungus Trichoderma reesei belongs to the human and Schizosaccharomyces pombe class of the enzyme. Glycobiology 10:983-991. [DOI] [PubMed] [Google Scholar]
- 31.Lilley, K. S., and P. Dupree. 2007. Plant organelle proteomics. Curr. Opin. Plant Biol. 10:594-599. [DOI] [PubMed] [Google Scholar]
- 32.Lubertozzi, D., and J. D. Keasling. 2009. Developing Aspergillus as a host for heterologous expression. Biotechnol. Adv. 27:53-75. [DOI] [PubMed] [Google Scholar]
- 33.Mannella, C. A. 2006. The relevance of mitochondrial membrane topology to mitochondrial function. Biochim. Biophys. Acta 1762:140-147. [DOI] [PubMed] [Google Scholar]
- 34.Medina, M. L., P. A. Haynes, L. Breci, and W. A. Francisco. 2005. Analysis of secreted proteins from Aspergillus flavus. Proteomics 5:3153-3161. [DOI] [PubMed] [Google Scholar]
- 35.Moralejo, F. J., A. J. Watson, D. J. Jeenes, D. B. Archer, and J. F. Martin. 2001. A defined level of protein disulfide isomerase expression is required for optimal secretion of thaumatin by Aspegillus awamori. Mol. Genet. Genomics 266:246-253. [DOI] [PubMed] [Google Scholar]
- 36.Oda, K., D. Kakizono, O. Yamada, H. Iefuji, O. Akita, and K. Iwashita. 2006. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 72:3448-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira, J. M., D. van der Veen, L. H. de Graaff, and L. Qin. 2008. Efficient cloning system for construction of gene silencing vectors in Aspergillus niger. Appl. Microbiol. Biotechnol. 80:917-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olivo, C., C. Vanni, P. Mancini, L. Silengo, M. R. Torrisi, G. Tarone, P. Defilippi, and A. Eva. 2000. Distinct involvement of cdc42 and RhoA GTPases in actin organization and cell shape in untransformed and Dbl oncogene transformed NIH3T3 cells. Oncogene 19:1428-1436. [DOI] [PubMed] [Google Scholar]
- 39.Otte, S., W. J. Belden, M. Heidtman, J. Liu, O. N. Jensen, and C. Barlowe. 2001. Erv41p and Erv46p: new components of COPII vesicles involved in transport between the ER and Golgi complex. J. Cell Biol. 152:503-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panagiotou, G., M. R. Andersen, T. Grotkjaer, T. B. Regueira, J. Nielsen, and L. Olsson. 2009. Studies of the production of fungal polyketides in Aspergillus nidulans by using systems biology tools. Appl. Environ. Microbiol. 75:2212-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pantazopoulou, A., and M. A. Peñalva. 2009. Organization and dynamics of the Aspergillus nidulans Golgi during apical extension and mitosis. Mol. Biol. Cell 20:4335-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pel, H. J., J. H. de Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. de Vries, R. Albang, K. Albermann, M. R. Andersen, J. D. Bendtsen, J. A. E. Benen, M. van den Berg, S. Breestraat, M. X. Caddick, R. Contreras, M. Cornell, P. M. Coutinho, E. G. J. Danchin, A. J. M. Debets, P. Dekker, P. W. M. van Dijck, A. van Dijk, L. Dijkhuizen, A. J. M. Driessen, C. d'Enfert, S. Geysens, C. Goosen, G. S. P. Groot, P. W. J. de Groot, T. Guillemette, B. Henrissat, M. Herweijer, J. P. T. W. van den Hombergh, C. A. M. J. J. van den Hondel, R. T. J. M. van der Heijden, R. M. van der Kaaij, F. M. Klis, H. J. Kools, C. P. Kubicek, P. A. van Kuyk, J. Lauber, X. Lu, M. J. E. C. van der Maarel, R. Meulenberg, H. Menke, M. A. Mortimer, J. Nielsen, S. G. Oliver, M. Olsthoorn, K. Pal, N. N. M. E. van Peij, A. F. J. Ram, U. Rinas, J. A. Roubos, C. M. J. Sagt, M. Schmoll, J. B. Sun, D. Ussery, J. Varga, W. Vervecken, P. J. J. V. de Vondervoort, H. Wedler, H. A. B. Wösten, A. P. Zeng, A. J. J. van Ooyen, J. Visser, and H. Stam. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221-231. [DOI] [PubMed] [Google Scholar]
- 43.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. Macdonald, and A. W. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 45.Punt, P. J., B. Seiboth, X. O. Weenink, C. van Zeijl, M. Lenders, C. Konetschny, A. F. Ram, R. Montijn, C. P. Kubicek, and C. A. van den Hondel. 2001. Identification and characterization of a family of secretion-related small GTPase-encoding genes from the filamentous fungus Aspergillus niger: a putative SEC4 homologue is not essential for growth. Mol. Microbiol. 41:513-525. [DOI] [PubMed] [Google Scholar]
- 46.Record, E., M. Asther, S. Moukha, D. Marion, V. Burlat, K. Ruel, and M. Asther. 1998. Localization of a phosphatidylglycerol/phosphatidylinositol transfer protein in Aspergillus oryzae. Can. J. Microbiol. 44:945-953. [PubMed] [Google Scholar]
- 47.Rizzuto, R., P. Pinton, W. Carrington, F. S. Fay, K. E. Fogarty, L. M. Lifshitz, R. A. Tuft, and T. Pozzan. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280:1763-1766. [DOI] [PubMed] [Google Scholar]
- 48.Rusiñol, A. E., Z. Cui, M. H. Chen, and J. E. Vance. 1994. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 269:27494-27502. [PubMed] [Google Scholar]
- 49.Seaman, M. N., J. M. McCaffery, and S. D. Emr. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 51.Štefanić, S., D. Palm, S. G. Svärd, and A. B. Hehl. 2006. Organelle proteomics reveals cargo maturation mechanisms associated with Golgi-like encystation vesicles in the early-diverged protozoan Giardia lamblia. J. Biol. Chem. 281:7595-7604. [DOI] [PubMed] [Google Scholar]
- 52.Sun, J., X. Lu, U. Rinas, and A. P. Zeng. 2007. Metabolic peculiarities of Aspergillus niger disclosed by comparative metabolic genomics. Genome Biol. 8:R182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taheri-Talesh, N., T. Horio, L. Araújo-Bazán, X. Dou, E. A. Espeso, M. A. Peñalva, S. A. Osmani, and B. R. Oakley. 2008. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell 19:1439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsang, A., G. Butler, J. Powlowski, E. A. Panisko, and S. E. Baker. 2009. Analytical and computational approaches to define the Aspergillus niger secretome. Fungal. Genet. Biol. 46(Suppl. 1):S153-S160. [DOI] [PubMed] [Google Scholar]
- 56.van der Veen, D., J. M. Oliveira, W. A. van den Berg, and L. H. de Graaff. 2009. Analysis of variance components reveals the contribution of sample processing to transcript variation. Appl. Environ. Microbiol. 75:2414-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Esse, H. P., J. W. van't Klooster, M. D. Bolton, K. A. Yadeta, P. van Baarlen, S. Boeren, J. Vervoort, P. J. de Wit, and B. P. Thomma. 2008. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20:1948-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Peij, N. N., J. Visser, and L. H. de Graaff. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27:131-142. [DOI] [PubMed] [Google Scholar]
- 59.Ward, P. P., H. Chu, X. Zhou, and O. M. Conneely. 1997. Expression and characterization of recombinant murine lactoferrin. Gene 204:171-176. [DOI] [PubMed] [Google Scholar]
- 60.Waterhouse, A. M., J. B. Procter, D. M. Martin, M. Clamp, and G. J. Barton. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, C. C., M. J. MacCoss, G. Mardones, C. Finnigan, S. Mogelsvang, J. R. Yates III, and K. E. Howell. 2004. Organellar proteomics reveals Golgi arginine dimethylation. Mol. Biol. Cell 15:2907-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeung, B. G., H. L. Phan, and G. S. Payne. 1999. Adaptor complex-independent clathrin function in yeast. Mol. Biol. Cell 10:3643-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zybailov, B., A. L. Mosley, M. E. Sardiu, M. K. Coleman, L. Florens, and M. P. Washburn. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5:2339-2347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.