Abstract
Verotoxin (VT) has been implicated in the promotion of adherence to and colonization of intestinal epithelial cells by enterohemorrhagic Escherichia coli (EHEC) O157:H7. The present study investigated the effect of VT2 on the adherence of EHEC O157:H7 strain 86-24 to porcine jejunal (IPEC-J2), human colon (CaCo-2), and human laryngeal carcinoma (HEp-2) cell lines and on the expression in IPEC-J2 cells of synthases for β1-integrin and nucleolin, both of which are implicated in bacterial adherence. The effect on expression of globotriaosylceramide (Gb3) synthase, the receptor for VT, was also examined. Data were obtained by adherence assays and quantitative reverse transcriptase PCR, using EHEC O157 strain 86-24, a vt2 deletion mutant, a vt2 phage-negative strain, and complemented mutants in which the vt2 gene was restored. Compared with the adherence of the parent and complemented mutant strains, the vt2-negative strains adhered significantly less to all three types of cells. Adherence of the wild-type EHEC strain to IPEC-J2 cells was accompanied by increased expression of β1-integrin, nucleolin, and Gb3 synthase. IPEC-J2 cells in association with wild-type EHEC O157:H7 or the complemented mutants expressed higher levels of β1-integrin than did cells in association with the vt2-negative strains or with no bacteria. Expression of nucleolin was decreased by association with the vt2-negative mutant, but complementation failed to restore wild-type expression. The data indicate that VT2 plays a role in the adherence of EHEC O157:H7 to intestinal epithelial cells, possibly by increasing the expression of the host receptor β1-integrin.
Enterohemorrhagic Escherichia coli (EHEC) is a subgroup of verotoxin-producing E. coli (VTEC), also referred to as Shiga-toxin-producing E. coli (STEC). Serotype O157:H7 is the prototypic EHEC strain, associated with large outbreaks of disease in North America due to the ingestion of food contaminated with feces from cattle, which are the primary reservoir for EHEC O157:H7 (11). The disease syndromes caused by EHEC infection include mild diarrhea to hemorrhagic colitis (HC) and, in the most severe cases, hemolytic-uremic syndrome (HUS) (28).
The key virulence factors for EHEC are a phage-encoded verotoxin (VT) (12) and adherence mediated by the locus for enterocyte effacement (LEE) pathogenicity island (PAI). LEE encodes proteins responsible for the attaching-and-effacing lesion (AE) (5), which is characterized by intimate adhesion of the pathogen to the enterocyte surface, formation of an actin-rich pedestal on the host cell surface beneath the bacterium, and destruction of brush border microvilli (14). The LEE PAI consists of five major operons (LEE1, LEE2, LEE3, tir/LEE5, and LEE4) (3) that encode a type III secretion system involved in secretion of translocon and effector proteins and the outer membrane surface adhesin intimin and its receptor, the translocated intimin receptor (Tir). In addition to binding to its primary receptor, Tir, intimin also interacts with host cell intimin receptors (HIRs), such as β1-integrin and nucleolin, on the surface of enterocytes (4, 27). It is hypothesized that prior to the formation of a stable intimin-Tir interaction, intimin binds to HIRs and brings the bacteria close enough for injection of Tir and other effectors into the host epithelial cells (6).
Nucleolin is a multifunctional nuclear protein that can be expressed at the surface of many cell types and that serves as a receptor for some viruses and intimin (27). Integrins are a large family of heterodimeric receptors that are associated with a wide range of cell-to-cell interactions (9). The intimin of enteropathogenic E. coli (EPEC) specifically binds to β1-integrin (4). Immunostained β1-integrin clusters at the sites of bacterial adherence to porcine and bovine tissues, suggesting that β1-integrin potentially serves as a receptor for intimin during EHEC O157:H7 infection (4, 26).
VT, an AB5 toxin that inhibits protein synthesis in eukaryotic cells, is responsible for the severe clinical manifestations of EHEC infection, such as HC and HUS (10, 28). The toxins are internalized by receptor-mediated endocytosis after binding of the B pentamer to the target cell glycolipid receptor globotriaosylceramide (Gb3). Recent studies have indicated that VT2 also promotes EHEC colonization of the intestine by stimulating expression of nucleolin (22, 30). However, this role is still controversial.
In a previous study, we showed that a nalidixic acid-resistant (Nalr) vt2 insertion mutant caused a reduction in the level of adherence of EHEC O157:H7 strain 86-24 to IPEC-J2 and HEp-2 cells (30). In the present study, new vt2 deletion and vt2 phage-cured mutants were generated in Nal-sensitive strain 86-24, and the effects of the mutations on the adherence of EHEC O157:H7 to IPEC-J2 cells were assessed and compared to their effects on adherence to HEp-2 and CaCo-2 cells. To further characterize the new in vitro model of porcine jejunal IPEC-J2 cells for use in combination with pig gut loops for the study of EHEC O157:H7 pathogenesis, we analyzed the effect of EHEC O157:H7 strain 86-24 and its isogenic vt2-negative mutant on the expression of nucleolin, β1-integrin, and Gb3 synthase in IPEC-J2 cells.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Mutant strains were constructed in EHEC O157:H7 strain 86-24.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype and description | Source or reference |
|---|---|---|
| E. coli strains | ||
| EHEC 86-24 | EHEC O157:H7 strain 86-24 | 29 |
| 86-24Δ933W | EHEC O157:H7 strain 86-24 with cured BP933W phage | This study |
| 86-24Δ933W(pVT2) | 86-24Δ933W containing pVT2 | This study |
| 86-24Δvt2S | 86-24Δvt2::Gm | This study |
| 86-24Δvt2S(pVT2) | 86-24Δvt2::Gm containing pVT2 | This study |
| Plasmids | ||
| pUCGm | Template plasmid for gentamicin resistance | 24 |
| pBAD-TOPO | Cloning and expression vector PBAD, Kanr | Invitrogen |
| pKM208 | pMAK700 derivative with red and gam expressed from Ptac, AprlacI IPTGa inducible | 18 |
| pCR2.1-TOPO | TA cloning vector, Apr | Invitrogen |
| pVT2A | pCR2.1-TOPO containing vt2-A | This study |
| pVT2B | pCR2.1-TOPO containing vt2-B | This study |
| pVT2B-Gm | pCR2.1-TOPO containing vt2-B and gentamicin resistance gene | This study |
| pVT2-Gm | pVT2B-Gm containing vt2-A | This study |
| pVT2 | pBAD-TOPO containing vt2 genes | 30 |
IPTG, isopropyl-β-d-thiogalactopyranoside.
Generation of a vt2 deletion mutant.
Long sequences homologous to the vt2 gene were used to make a deletion mutant of the vt2 gene using the bacteriophage λ red recombination system. The upstream and downstream regions of the vt2 gene containing sequences homologous to vt2 fragment A (vt2-A) and vt2-B, respectively, were amplified from wild-type EHEC O157:H7 strain 86-24 by the Expand High Fidelity PCR system (Roche Diagnostics, Montreal, Quebec, Canada) using primer pairs vt2-F1/vt2-R1 and vt2-F2/vt2-R2 (Table 2 and Fig. 1). The two fragments (vt2-A and vt2-B) were cloned into the pCR2.1-TOPO vector, generating pVT2A and pVT2B, respectively. A gentamicin resistance gene (Gm; 855 bp) was released with SmaI from plasmid pUGm and was cloned into the EcoRV site before the vt2-B fragment in pVT2B, resulting in pVT2B-Gm. Fragment vt2-A was released from pVT2A by SalI and XhoI and was subcloned into the corresponding sites before Gm-VT2B in pVT2B-Gm, resulting in pVT2-Gm. This construct contains 1,634-bp and 1,654-bp DNA sequences homologous to regions upstream and downstream, respectively, of the vt2 gene. A PCR amplicon was obtained with the primer pair vt2-F1 and vt2-R2 using pVT2-Gm as the template and was electroporated into competent EHEC O157:H7 strain 86-24 cells containing pKM208. After electroporation, the cells were incubated at 37°C overnight with shaking and plated on medium containing 20 μg/ml gentamicin. The resulting colonies were screened for the absence of the vt2 gene by colony PCR (performed directly from bacterial colonies lysed in sterile water at 95°C for 5 min) using primers vt2-F03 and vt2-R03 flanking the deleted region (data not shown). Positive colonies were examined by colony PCR using primers Gm-F and vt2-R77 targeting the inserted gentamicin resistance gene and flanking the deleted region, respectively (Fig. 1). Clones that produced an amplicon of 3,512 bp were positive in the PCR and had the vt2 gene deleted, while the wild type produced no amplicon (Fig. 1). The resultant vt2 deletion mutant strain was confirmed by sequencing and was named 86-24Δvt2S. To complement the mutant, the wild-type vt2 gene was amplified by PCR with primers vt2-F06B and vt2-R06 and cloned into the expression vector pBAD-TOPO (behaving as a low-copy-number plasmid under uninduced conditions), resulting in pVT2. The latter was then electroporated into 86-24Δvt2S, yielding 86-24Δvt2S(pVT2) (Tables 1 and 2).
TABLE 2.
Primers used for PCRa
| Primer | Nucleotide sequence (5′-3′) | Target gene (or gene type) | GenBank accession no. |
|---|---|---|---|
| vt2-F1(SalI) | gtcgacGCTGGACTGTCGAAGAATGb | Z1458 | NC_002655 |
| vt2-R1 (XbaI) | tctagaGGGGTCGATATCTCTGc | vt2-A | NC_002655 |
| vt2-F2 (XbaI) | tctagaGGTTGACGGGAAAGAATACc | vt2-B | NC_002655 |
| vt2-R2 (SacI) | gagctcTTCGTCGGCACATTCACCd | Z1466 | NC_002655 |
| vt2-F03 | TATTTAAATGGGTACTGTGCCT | vt2-A | NC_002655 |
| vt2-R32 | GTATACACAGGAGCAGTTTCAG | vt2-A | NC_002655 |
| vt2-R03 | AAACTGCACTTCAGCAAATCCG | vt2-B | NC_002655 |
| 1459R | CTCAACATACCGATCCATCTC | Z1459/Q | NC_002655 |
| 1475F | AAGCTGGACTGGAAAAAGCTG | Z1475/terminase | NC_002655 |
| 1475R | TGCAGCGATGACACGATGTC | Z1475/terminase | NC_002655 |
| vt2-F06B | CACCCAGAATGTAGTCAGTCAGAACe | Z1460 | NC_002655 |
| vt2-R06 | CCCTGACAACATCATAGTGT | Z1466 | NC_002655 |
| vt2-R77 | CAGAGTACGAAAGTATCGTTC | Z1466 adjacent | NC_002655 |
| Gm-F | CGAGCTCGAATTGACATAAG | Gentamicin resistance | U04610.1 |
| N1 | AGGATGACGATGACGATGAGG | Nucleolin | M60858.1 |
| N2 | CTCCAGGTCTTCAGCAGATTC | Nucleolin | M60858.1 |
| N3f | AAAGCGTTGGAACTCACTGG | Nucleolin | M60858.1 |
| N4f | TCCTTGCTGACTAATCTGATC | Nucleolin | M60858.1 |
| G1 | GATCTACTGGCACGTTGTGG | Gb3 synthase | AB037883 |
| G2 | TGGTCCGGTCTGAAGTCTCC | Gb3 synthase | AB037883 |
| G3f | AAGTTCGGCGGCATCTACC | Gb3 synthase | AB037883 |
| G4f | GATCCAGCCGTTGTAGTGG | Gb3 synthase | AB037883 |
| IntgF | CCTTATGGACCTGTCTTACTC | β1-integrin | NM_213968 |
| IntgR | AGATAATGTTCCTACTGCTGAC | β1-integrin | NM_213968 |
| IntgF3f | ACTTGTGAGATGTGTCAGACC | β1-integrin | NM_213968 |
| IntgR3f | CAATCATCAACGTCCTTCTCC | β1-integrin | NM_213968 |
| GAPDH-Ff | TGGGAAGCTTGTCATCAATGG | GAPDH | AF017079 |
| GAPDH-R2f | CTGTTGTCATACTTCTCATGG | GAPDH | AF017079 |
All the primers were designed in the present study.
Lowercase and boldface letters show the SalI site.
Lowercase and boldface letters show the XbaI site.
Lowercase and boldface letters show SacI site.
Underlined letters show the 5′ overhang required for directional cloning into pBAD-TOPO.
Used for quantitative PCR.
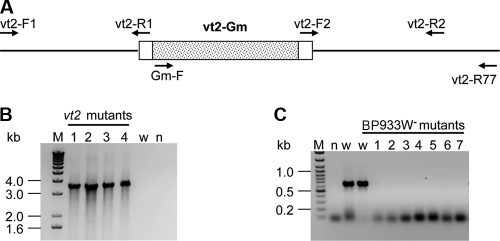
FIG. 1.
(A) Schematic diagram showing the vt2 deletion mutation with an insertion of Gm (shaded area within the vt2 gene). (B) Confirmation of the mutation generated by the bacteriophage λ red recombination system by PCR using primers Gm-F/vt2-R77. With DNA from the mutants (lanes 1 to 4), an expected 3,512-bp amplicon was produced, while with DNA from the wild type, there was no amplicon. (C) Confirmation of the absence of phage DNA by PCR using primers vt2-F1 and 1459R. With the wild-type DNA, there was an amplicon of 643 bp; with the phage-cured (BP933W−) strain DNA, there was no PCR product (lanes 1 to 7). (B and C) Lanes M, DNA mass ladder (kb); lanes w, wild-type strain; lanes n, no-template control.
Selection of a strain lacking the vt2 phage.
VT2 is produced by the gene carried by bacteriophage BP933W (19). To select a vt2 phage-cured mutant, wild-type EHEC O157:H7 strain 86-24 was grown in brain heart infusion (BHI) broth plus 44 mM NaHCO3 (BHIN) overnight at 37°C without shaking, streaked on MacConkey agar, and incubated at 37°C overnight. Single colonies were tested for the spontaneous loss of the vt2 phage by colony PCR using primer pairs vt2-F03/vt2-R32 and vt2-F1/vt2-R1, which target the vt2 gene and its upstream region, respectively (data not shown). The phage-cured colonies yielded no PCR product; phage loss was confirmed by PCR with primer pair 1475F/1475R, which detects the phage terminase small subunit (data not shown), and primer pair vt2-F1/1459R, which detects the protein Q encoded by the bacteriophage. The phage-cured colonies produced no PCR amplicons (Fig. 1C and Table 2).
In vitro adherence assay.
IPEC-J2 and CaCo-2 cells were maintained in Dulbecco's minimal essential medium (DMEM; Invitrogen, Carlsbad, CA). HEp-2 cells were maintained in Eagle's minimal essential medium (EMEM) (Invitrogen). Both media were supplemented with 10% fetal bovine serum (FBS), penicillin (100 IU/ml), and streptomycin (100 μg/ml). The adherence assays and the quantification of adherence were conducted as previously detailed, with approximately 107 bacteria of an overnight culture of each strain cultured in 3 ml BHIN for 16 to 18 h without shaking (30). Briefly, the cells were infected for 6 h at 37°C under 5% CO2 with a medium change at 3 h, washed with phosphate-buffered saline (PBS) to remove unbound bacteria, fixed with 70% methanol for 10 min, and stained with 1:40 Giemsa (Sigma) for 30 min. Adherence was examined by light microscopy and was quantified by counting 100 consecutive cells per well and recording the percentage of cultured cells with clusters of 5 to 9, 10 to 19, and ≥20 bacterial clusters. The percentage of cells with at least five adherent bacteria per cell was calculated as a measure of total adherence. Data are expressed as the mean of at least three separate experiments ± standard deviation (SD). Growth rates for the wild-type, mutant, and complemented strains in EMEM for 6 h were examined to determine whether there was a growth defect for the mutant.
In preparation for RNA isolation, IPEC-J2 cells were incubated with mutant strain 86-24Δvt2S, complemented strain 86-24Δvt2S(pVT2), wild-type strain 86-24, or BHIN medium alone for 6 h with a medium change at 3 h. The IPEC-J2 cells were washed vigorously, collected in RNAlater solution (Invitrogen), and held at 4°C overnight before RNA isolation.
VT detection by VCA.
The Vero cell cytotoxicity assay (VCA) was performed as described previously (30).
RNA isolation.
IPEC-J2, CaCo-2, and HEp-2 cells were collected in RNAlater and incubated at 4°C overnight before RNA isolation. Total RNA was extracted by an RNeasy minikit (Qiagen, Mississauga, Ontario, Canada), according to the manufacturer's instruction. The extracted RNA was treated several times with DNase I until it was confirmed to be free of genomic DNA contamination, as determined by PCR using RNA as the template. RNA integrity was verified by visualization on 1.0% agarose gel.
RT-PCR and qPCR.
First-strand cDNA was synthesized from a 0.5 μg quantity of the DNase I-treated total RNA using SuperScript II-RT with oligo(dT)12, as recommended by the supplier (Invitrogen). Reverse transcription (RT)-PCR was carried out as described previously (30). Quantitative PCR (qPCR) was performed using a Stratagene (La Jolla, CA) MX4000 thermal cycler with the brilliant SYBR green qPCR master mixture. cDNA (1 μl) was added in a 25-μl reaction mixture which contained 1× master mixture, 150 nM each primer, and 30 nM 6-carboxy-X-rhodamine (ROX). The program was 3 min at 95°C and then 40 cycles of 95°C for 30 s, 55° to 57°C (depending on the primers used) for 30 s, and 72°C for 30 s. On the completion of amplification, a dissociation curve was produced by holding the reaction mixture at 95°C for 1 min and at 55°C for 30 s and ending at 95°C for 30 s. The housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The relative levels of the mRNA of the genes of interest were normalized to that of the GAPDH mRNA and were determined using a modified 2−ΔΔCT method (where CT is the threshold cycle) (15).
Statistical analysis.
All analyses were performed with SAS for Windows, version 8.02 (SAS Institute Inc., Cary, NC). The levels of adherence of the various strains to cultured cells were compared by analysis of variance of the percent adherence of clusters with 5 to 9, 10 to 19, and ≥20 bacteria per cell, as well as the total percent adherence (≥5 adherent bacteria per cell), using the PROC GLM program. Differences with P values of ≤0.05 were considered significant.
RESULTS
VT2 production by vt2 mutant and parental strains of EHEC O157:H7.
Mutant strain 86-24Δvt2S produced no cytotoxicity, whereas the wild-type EHEC O157:H7 strain 86-24 produced 5.1 × 104 50% cytotoxic doses (CD50s)/ml in the VCA (data not shown). Complementation of the vt2-negative mutant restored the level of VT2 production to 4 × 104 CD50s/ml, approximately 80% of the wild-type level. VT2 phage-cured strain 86-24Δ933W showed no cytotoxicity, and complementation of the strain with a plasmid carrying the vt2 gene partially restored VT2 production with a VCA value of 1.3 × 104 CD50s/ml, approximately 25% of the wild-type level (data not shown).
Effect of vt2 deletion on adherence of EHEC O157:H7 to IPEC-J2, CaCo-2, and HEp-2 cells.
There was no difference in the growth rates among the wild-type, mutant, and complemented strains. The patterns of adherence by the wild-type strain and its isogenic mutant, 86-24Δvt2S, to IPEC-J2 and CaCo-2 cells are illustrated in Fig. 2. With all three cell types, loss of the vt2 gene alone was associated with a reduction in total adherence (P < 0.0261) and in the formation of large clusters (P < 0.0048; Fig. 3). For the phage-cured strain there was also a reduction in both total adherence and large clusters on CaCo-2 cells (P = 0.0216 and 0.019, respectively) but only a reduction in large clusters on IPEC-J2 and HEp-2 cells (P < 0.0475) (Fig. 3). Complementation of the vt2-negative and phage-cured mutants by a plasmid carrying the vt2 gene (pVT2) partially restored the adherence phenotypes relative to the wild-type phenotype (Fig. 3).
FIG. 2.
Qualitative comparison of adherence of wild-type strain 86-24 and its derivative vt2-negative mutants and complemented strains to IPEC-J2 and CaCo-2 cells. Adherence assays were performed for 6 h, as described in Materials and Methods. The vt2-negative mutants showed decreased adherence compared to that of wild-type strain 86-24; complementation restored the adherence phenotype.
FIG. 3.
Quantitative comparison of bacterial adherence of vt2-negative mutants 86-24Δvt2S and 86-24Δ933W and complemented strains 86-24Δvt2S(pVT2) and 86-24Δ933W(pVT2) to IPEC-J2, CaCo-2, and HEp-2 cells. The bacterial strains were incubated with the cultured cells for 6 h, and adherence was quantified by examining 100 cells for each assay and determining the mean percentage of cells (+SDs) with clusters. 5 to 9a, percentage of cells with a cluster of 5 to 9 adherent bacteria; 10 to 20b, percentage of cells with a cluster of 10 to 20 adherent bacteria; >20c, percentage of cells with a cluster of >20 adherent bacteria; Totald, percentage of cells with a cluster of ≥5 adherent bacteria. *, P < 0.05.
Detection of gene transcripts for nucleolin, β1-integrin, and Gb3 synthase in IPEC-J2 cells.
It was not known whether the three cell lines expressed nucleolin, β1-integrin, and Gb3 synthase. Therefore, RT-PCR was conducted to detect these genes in the cells. Gb3 synthase is the key enzyme required for the synthesis of Gb3, which serves as the receptor for VTs (10, 13). With initial RT-PCR experiments, primer pairs N1/N2 and G1/G2 were designed on the basis of the sequences of the genes for human nucleolin (hNCL; GenBank accession number M60858.1) and Gb3 synthase (A4GALT or Gb3-CD77 synthase) (Fig. 4 A). Transcripts of both genes were detected in CaCo-2 and HEp-2 cells but not in IPEC-J2 cells. Homologous genes were not found in the pig (Sus scrofa) genome; however, searches of porcine expressed sequence tags (EST) using the hNCL and Gb3 synthase sequences identified clones from porcine intestinal cDNA libraries that showed homology to genes for hNCL and Gb3 synthase. Clone ITT010089D05 (GenBank accession number BW983899.1) has 874 bp which has 92.2% nucleotide identity and 94.8% deduced amino acid identity with hNCL from positions 865 to 1738 of 2,124 bp (Fig. 4A). Clone c11b08 (GenBank accession number Z81218.1) has 339 bp which has 87.6% nucleotide identity and 87% deduced amino acid identity with Gb3 synthase from positions 431 to 770 of 1,062 bp (Fig. 4A). Primer pairs N3/N4, and G3/G4 were then made to target these regions that were homologous with hNCL and Gb3 synthase, respectively, and RT-PCR with these primers produced amplicons of the expected sizes in all three cell lines (Fig. 4B). Therefore, primer pairs N3/N4 and G3/G4 were used to target porcine NCL (pNCL) and Gb3 synthase (pGb3 syn), respectively, in the qPCR analysis for IPEC-J2 cells. β1-integrin was also detected in the three cell lines (Fig. 4B).
FIG. 4.
(A) Comparison of human nucleolin and Gb3 synthase genes with selected pig intestinal cDNA library clones. The percent nucleotide identity of each clone with its corresponding human gene is shown below. (B) RT-PCR analysis with primer pairs IntgF/IntgR, N3/N4, and G3/G4 targeting β1-integrin, nucleolin, and Gb3 synthase transcripts, respectively, in IPEC-J2 cells (lanes I), CaCo-2 cells (lanes C), and HEp-2 cells (lanes H). Lanes N, no-template control.
Effects of vt2 deletion on expression of nucleolin, β1-integrin, and Gb3 synthase in IPEC-J2 cells.
RNA was isolated from IPEC-J2 cells infected with wild-type strain 86-24, mutant strain 86-24Δvt2S, and its complemented strain, 86-24Δvt2S(pVT2). Uninfected cells served as a control for background levels of expression. Transcripts of the genes for nucleolin, β1-integrin, and Gb3 synthase were quantified by qPCR analysis. Association of the IPEC-J2 cells with wild-type strain 86-24 or complemented strains 86-24Δvt2S(pVT2) and 86-24Δ933W(pVT2) increased the mRNA level of β1-integrin compared to the level in the control IPEC-J2 cells incubated without bacteria (Fig. 5 A). The vt2-negative mutant strain had no effect on the background expression of β1-integrin. The level of transcription for nucleolin was also significantly increased by the association of IPEC-J2 cells with wild-type strain 86-24, and a decreased transcript level was observed in association with the vt2-negative mutant compared to that for the control, but complementation did not restore the level of nucleolin mRNA to that of the wild type (Fig. 5B). Transcription of Gb3 synthase was induced by wild-type strain 86-24, but the difference did not reach significance; the vt2-negative mutant had a significant reduction in transcript levels, but in the complemented mutant the transcript level was not restored to the wild-type level (Fig. 5C).
FIG. 5.
Transcriptional profiles of β1-integrin (A), nucleolin (B), and Gb3 synthase (C) in IPEC-J2 cells infected with wild-type strain 86-24, 86-24Δvt2S, or its complement, 86-24Δvt2S(pVT2) or cells cultured in BHIN medium (control). Data are expressed as the means + SDs for RNA extracted in triplicate experiments. Relative fold expression represents the changes in transcription compared to that for the control (black bars; value of 1.0). The levels of GAPDH transcript were used to normalize the CT values. * and †, P < 0.05 compared to the control and wild-type 86-24, respectively.
DISCUSSION
In the present study, we examined the effect of a vt2 deletion mutation on adherence of an EHEC O157:H7 strain to epithelial cells and the effect of the wild-type strain and its isogenic vt2-negative mutant on the expression of β1-integrin, nucleolin, and Gb3 synthase in IPEC-J2 cells. Loss of the ability to produce VT2 was associated with reduced adherence to the three epithelial cell lines and with reduced expression of β1-integrin in IPEC-J2 cells. These findings support the notion that VT2 increases the expression of host cell-encoded intimin receptors, thereby enhancing EHEC attachment. Intimin has been shown to bind to β1-integrin, and immunostained β1-integrin was demonstrated to be recruited beneath the site of adherent O157:H7 bacteria in porcine and bovine tissues (4, 26). These results are consistent with the proposal that, prior to the formation of a stable intimin-Tir interaction, intimin binds to HIRs and brings the bacteria close enough for injection of Tir and other effectors into the host epithelial cell (4, 6, 27).
Surprisingly, it was β1-integrin rather than nucleolin whose expression was clearly shown to be related to the effect of VT2 in the present study. There were similar increases in the expression of β1-integrin, nucleolin, and Gb3 synthase in IPEC-J2 cells associated with the wild-type EHEC O157:H7 strain and decreases associated with the vt2-negative mutant. In the cases of nucleolin and Gb3 synthase, the levels of expression accompanying loss of the vt2 gene were significantly lower than the background levels, but complementation failed to restore the levels to those associated with the wild-type bacteria. One possibility is that the levels of VT2 produced in the cultures by the complemented mutant were adequate for restoration of the expression levels of β1-integrin but not of nucleolin and Gb3 synthase. If this is the case, then VT2 production influences the expression of both HIRs and the VT2 receptor. This hypothesis may be tested by adding exogenous VT2 to the cultures of IPEC-J2 cells with the complemented strains. Nucleolin has been associated with enhanced adherence by VT-positive EHEC O157:H7 to HEp-2 cells; increased expression of nucleolin at the surface of the HEp-2 cells occurred at 2 h and peaked at 24 h after treatment with 100 ng/ml of exogenous VT2 (22). With respect to expression of nucleolin and β1-integrin, it appears that HEp-2 and IPEC-J2 cells are affected differently by VT2, but the difference may be quantitative.
Gb3 expression is regulated by the key enzyme Gb3 synthase (13). Gb3 synthesis is also induced by cytokines, including tumor necrosis factor (TNF), that are induced by VT2 and lipopolysaccharide (LPS) during infection (8, 17). Binding of VT to Gb3 in endothelial cells further increases the expression of Gb3 and exacerbates the disease process by EHEC O157:H7 (16, 20). Binding of VT to Gb3 may indirectly promote EHEC O157:H7 adherence, because it represses antibacterial peptide secretion and host cellular immune response (7, 23). It is possible that VT2-Gb3 binding may play a role in signal transduction for the recruitment of β1-integrin, as mentioned above. Therefore, this study examined the effect of adherence of a wild-type EHEC O157 strain and its vt2-negative mutant on Gb3 expression in IPEC-J2 cells by investigating the expression of Gb3 synthase. Our data show that adherence of the wild-type bacteria resulted in an elevation of the level of expression of Gb3 synthase, but the role of VT2 in this phenomenon was not demonstrated.
Complementation of the vt2 mutants restored the adherence phenotype to that of the wild type in IPEC-J2 and CaCo-2 cells but only partially in HEp-2 cells. This shows that different results may be obtained with different cell lines and that there is value in using more than one cell type for studies of adherence by EHEC O157:H7. IPEC-J2 cells have only recently been used for the study of EHEC O157:H7 pathogenesis and adherence. However, it was not clear whether significant differences might exist between this cell type and HEp-2 or CaCo2 cells with respect to adherence of EHEC O157:H7. Therefore, we examined the effect of VT2 on adherence of O157:H7 to IPEC-J2 cells and compared the findings with those from human intestinal epithelial CaCo-2 cells and laryngeal carcinoma HEp-2 cells. The similarities in the adherence of EHEC O157:H7 to IPEC-J2, CaCo-2, and HEp-2 cells suggest that porcine IPEC-J2 cells may be a good in vitro model. This model of adherence may be particularly relevant in combination with a porcine gut loop model for studies of interactions between EHEC O157:H7 and intestinal epithelia. In vivo studies have resulted in conflicting results on VT's role in colonization in a variety of animal models (1, 2, 21, 22, 25). The reasons for the conflicting results are not known, but they could be due to the different animal species, bacterial strains, and routes of infection.
In an earlier study (30), two vt2 mutants were created. One (86-24NalrΔvt2) was produced by insertion in a nalidixic acid-resistant 86-24 strain. This mutant showed a decreased adherence, and the effect was reversed by complementation. The second mutant (86-24NalsΔvt2-1) was generated by the bacteriophage lambda red method in a nalidixic acid-sensitive 86-24 strain and showed decreased adherence, but the mutation was not complemented. It was noted that other mutations might account for the failure of complementation because the lambda red methodology is known to cause secondary mutations (18). For the present study, we therefore made another vt2 mutant (86-24Δvt2S) by deleting the vt2 gene from the nalidixic acid-sensitive 86-24 strain. Both 86-24NalrΔvt2 and 86-24Δvt2S had reduced adherence, and this effect was complemented. Data from the present study further consolidate the results from the mutant 86-24NalrΔvt2. Our finding of a role for VT2 in adherence is consistent with the findings described in a 2006 publication in which deletion of the vt2 gene resulted in decreased colonization of the mouse intestine (22). It is well established that the type III secretion system is a major factor in the adherence phenotype, but VT2 also appears to affect adherence, possibly due to an effect on expression of host receptors.
The phage-cured mutant had a less pronounced effect on adherence of EHEC than the vt2 deletion mutant, and the phage-cured mutant was associated with a reduction in large cluster formation but not in the total adherence to HEp-2 and IPEC-J2 cells. While the reason for this difference is not clear, there are two possible interpretations of the data. One is that the loss of vt2 had nothing to do with the decreased adherence and that there are other mutations that caused the reduction in adherence. This seems unlikely in this spontaneous phage-loss mutant and in the face of other evidence that vt2 does play a role in adherence (22). Another interpretation is that phage carrying genes other than vt2 played a role in adherence or in the production of VT2. Our finding that the complemented phage mutant produced much less VT2 than the complemented vt2 mutant is consistent with the latter interpretation and suggests that some trans-acting phage-encoded factor influenced the plasmid-encoded VT2 production.
In summary, this study showed that deletion of the vt2 gene was associated with the reduced adherence of EHEC O157:H7 strain 86-24 to one porcine and two human cultured epithelial cell lines. This effect was correlated with a decrease in the gene expression of β1-integrin in IPEC-J2 cells. Adherence of the wild-type EHEC strain was associated with a significant increase in the gene expression of both β1-integrin and nucleolin in IPEC-J2 cells. Reduction in the gene expression of Gb3 synthase was also associated with a loss of VT production by EHEC O157:H7.
Acknowledgments
We thank Musafiri Karama for critical review of the manuscript.
This research was financially supported by Agriculture and Agri-Food Canada. B.L., Y.F., and J.Z. were visiting graduates to the Guelph Food Research Centre, Agriculture and Agri-Food Canada, financially supported by the China Scholar Council.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Best, A., R. M. La Ragione, W. A. Cooley, C. D. O'Connor, P. Velge, and M. J. Woodward. 2003. Interaction with avian cells and colonisation of specific pathogen free chicks by Shiga-toxin negative Escherichia coli O157:H7 (NCTC 12900). Vet. Microbiol. 93:207-222. [DOI] [PubMed] [Google Scholar]
- 2.Cornick, N. A., A. F. Helgerson, and V. Sharma. 2007. Shiga toxin and Shiga toxin-encoding phage do not facilitate Escherichia coli O157:H7 colonization in sheep. Appl. Environ. Microbiol. 73:344-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elliott, S. J., S. W. Hutcheson, M. S. Dubois, J. L. Mellies, L. A. Wainwright, M. Batchelor, G. Frankel, S. Knutton, and J. B. Kaper. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176-1189. [DOI] [PubMed] [Google Scholar]
- 4.Frankel, G., O. Lider, R. Hershkoviz, A. P. Mould, S. G. Kachalsky, D. C. Candy, L. Cahalon, M. J. Humphries, and G. Dougan. 1996. The cell-binding domain of intimin from enteropathogenic Escherichia coli binds to beta1 integrins. J. Biol. Chem. 271:20359-20364. [DOI] [PubMed] [Google Scholar]
- 5.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 6.Frankel, G., A. D. Phillips, L. R. Trabulsi, S. Knutton, G. Dougan, and S. Matthews. 2001. Intimin and the host cell—is it bound to end in Tir(s)? Trends Microbiol. 9:214-218. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman, M. A., C. Menge, T. A. Casey, W. Laegreid, B. T. Bosworth, and E. A. Dean-Nystrom. 2006. Bovine immune response to Shiga-toxigenic Escherichia coli O157:H7. Clin. Vaccine Immunol. 13:1322-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes, A. K., Z. Ergonul, P. K. Stricklett, and D. E. Kohan. 2002. Molecular basis for high renal cell sensitivity to the cytotoxic effects of shigatoxin-1: upregulation of globotriaosylceramide expression. J. Am. Soc. Nephrol. 13:2239-2245. [DOI] [PubMed] [Google Scholar]
- 9.Hynes, R. O. 1992. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11-25. [DOI] [PubMed] [Google Scholar]
- 10.Johannes, L., and W. Romer. 2009. Shiga toxins—from cell biology to biomedical applications. Nat. Rev. Microbiol. 8:105-116. [DOI] [PubMed] [Google Scholar]
- 11.Kaper, J. B., and M. A. Karmali. 2008. The continuing evolution of a bacterial pathogen. Proc. Natl. Acad. Sci. U. S. A. 105:4535-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 13.Kojima, Y., S. Fukumoto, K. Furukawa, T. Okajima, J. Wiels, K. Yokoyama, Y. Suzuki, T. Urano, and M. Ohta. 2000. Molecular cloning of globotriaosylceramide/CD77 synthase, a glycosyltransferase that initiates the synthesis of globo series glycosphingolipids. J. Biol. Chem. 275:15152-15156. [DOI] [PubMed] [Google Scholar]
- 14.LeBlanc, J. J. 2003. Implication of virulence factors in Escherichia coli O157:H7 pathogenesis. Crit. Rev. Microbiol. 29:277-296. [DOI] [PubMed] [Google Scholar]
- 15.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 16.Louise, C. B., and T. G. Obrig. 1991. Shiga toxin-associated hemolytic-uremic syndrome: combined cytotoxic effects of Shiga toxin, interleukin-1 beta, and tumor necrosis factor alpha on human vascular endothelial cells in vitro. Infect. Immun. 59:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyers, K. E., and B. S. Kaplan. 2000. Many cell types are Shiga toxin targets. Kidney Int. 57:2650-2651. [DOI] [PubMed] [Google Scholar]
- 18.Murphy, K. C., and K. G. Campellone. 2003. Lambda red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 20.Poirier, K., S. P. Faucher, M. Beland, R. Brousseau, V. Gannon, C. Martin, J. Harel, and F. Daigle. 2008. Escherichia coli O157:H7 survives within human macrophages: global gene expression profile and involvement of the Shiga toxins. Infect. Immun. 76:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie, J. M., C. M. Thorpe, A. B. Rogers, and M. K. Waldor. 2003. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect. Immun. 71:7129-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson, C. M., J. F. Sinclair, M. J. Smith, and A. D. O'Brien. 2006. Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. U. S. A. 103:9667-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuller, S., R. Heuschkel, F. Torrente, J. B. Kaper, and A. D. Phillips. 2007. Shiga toxin binding in normal and inflamed human intestinal mucosa. Microbes Infect. 9:35-39. [DOI] [PubMed] [Google Scholar]
- 24.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831-834. [PubMed] [Google Scholar]
- 25.Sheng, H., J. Y. Lim, H. J. Knecht, J. Li, and C. J. Hovde. 2006. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 74:4685-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinclair, J. F., E. A. Dean-Nystrom, and A. D. O'Brien. 2006. The established intimin receptor Tir and the putative eucaryotic intimin receptors nucleolin and beta1 integrin localize at or near the site of enterohemorrhagic Escherichia coli O157:H7 adherence to enterocytes in vivo. Infect. Immun. 74:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinclair, J. F., and A. D. O'Brien. 2004. Intimin types alpha, beta, and gamma bind to nucleolin with equivalent affinity but lower avidity than to the translocated intimin receptor. J. Biol. Chem. 279:33751-33758. [DOI] [PubMed] [Google Scholar]
- 28.Tarr, P. I., C. A. Gordon, and W. L. Chandler. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073-1086. [DOI] [PubMed] [Google Scholar]
- 29.Tarr, P. I., M. A. Neill, C. R. Clausen, J. W. Newland, R. J. Neill, and S. L. Moseley. 1989. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984-1987. J. Infect. Dis. 159:344-347. [DOI] [PubMed] [Google Scholar]
- 30.Yin, X., J. R. Chambers, R. Wheatcroft, R. P. Johnson, J. Zhu, B. Liu, and C. L. Gyles. 2009. Adherence of Escherichia coli O157:H7 mutants in vitro and in ligated pig intestines. Appl. Environ. Microbiol. 75:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]