Abstract
Given its availability and low price, glycerol has become an ideal feedstock for the production of fuels and chemicals. We recently reported the pathways mediating the metabolism of glycerol in Escherichia coli under anaerobic and microaerobic conditions. In this work, we engineer E. coli for the efficient conversion of glycerol to d-lactic acid (d-lactate), a negligible product of glycerol metabolism in wild-type strains. A homofermentative route for d-lactate production was engineered by overexpressing pathways involved in the conversion of glycerol to this product and blocking those leading to the synthesis of competing by-products. The former included the overexpression of the enzymes involved in the conversion of glycerol to glycolytic intermediates (GlpK-GlpD and GldA-DHAK pathways) and the synthesis of d-lactate from pyruvate (d-lactate dehydrogenase). On the other hand, the synthesis of succinate, acetate, and ethanol was minimized through two strategies: (i) inactivation of pyruvate-formate lyase (ΔpflB) and fumarate reductase (ΔfrdA) (strain LA01) and (ii) inactivation of fumarate reductase (ΔfrdA), phosphate acetyltransferase (Δpta), and alcohol/acetaldehyde dehydrogenase (ΔadhE) (strain LA02). A mutation that blocked the aerobic d-lactate dehydrogenase (Δdld) also was introduced in both LA01 and LA02 to prevent the utilization of d-lactate. The most efficient strain (LA02Δdld, with GlpK-GlpD overexpressed) produced 32 g/liter of d-lactate from 40 g/liter of glycerol at a yield of 85% of the theoretical maximum and with a chiral purity higher than 99.9%. This strain exhibited maximum volumetric and specific productivities for d-lactate production of 1.5 g/liter/h and 1.25 g/g cell mass/h, respectively. The engineered homolactic route generates 1 to 2 mol of ATP per mol of d-lactate and is redox balanced, thus representing a viable metabolic pathway.
Lactic acid (lactate) and its derivatives have many applications in the food, pharmaceutical, and polymer industries (13, 30). An example is polylactic acid, a renewable, biodegradable, and environmentally friendly polymer produced from d- and l-lactate (19). In this context, biological processes have the advantage of being able to produce chirally pure lactate from inexpensive media containing only the carbon source and mineral salts (43). While lactic acid bacteria traditionally have been used in the production of d-lactate from carbohydrate-rich feedstocks, several laboratories recently have reported alternative biocatalysts (13, 30), many of which are engineered Escherichia coli strains that produce d- or l-lactate (4, 8, 50, 51, 52).
Unlike the aforementioned reports, which have dealt with the use of carbohydrates, our work focuses on the use of glycerol as a carbon source for the production of d-lactate. Glycerol has become an inexpensive and abundant substrate due to its generation in large amounts as a by-product of biodiesel and bioethanol production (18, 32, 47). The conversion of glycerol to higher-value products has been proposed as a path to economic viability for the biofuels industry (47). One such product is lactate, whose production could be readily integrated into existing biodiesel and bioethanol facilities, thus establishing true biorefineries.
Although many microorganisms are able to metabolize glycerol (25), the use of industrial microbes such as E. coli could greatly accelerate the development of platforms to produce fuels and chemicals from this carbon source. We recently reported on the ability of E. coli to metabolize glycerol under either anaerobic or microaerobic conditions and identified the environmental and metabolic determinants of these processes (9, 11, 28). In one of the studies, the pathways involved in the microaerobic utilization of glycerol were elucidated, and they are shown in Fig. 1 (9). A common characteristic of glycerol metabolism under either anaerobic or microaerobic conditions is the generation of ethanol as the primary product and the negligible production of lactate (6, 9, 11, 28). In the work reported here, the knowledge base created by the aforementioned studies was used to engineer E. coli for the efficient conversion of glycerol to d-lactate in minimal medium. The engineered strains hold great promise as potential biocatalysts for the conversion of low-value glycerol streams to a higher-value product like d-lactate.
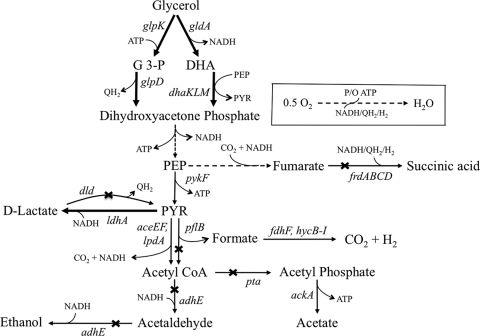
FIG. 1.
Pathways involved in the microaerobic utilization of glycerol in E. coli (9). Genetic modifications supporting the metabolic engineering strategies employed in this work are illustrated by thicker lines (overexpression of gldA-dhaKLM, glpK-glpD, and ldhA) or cross bars (disruption of pflB, pta, adhE, frdA, and dld). Broken lines illustrate multiple steps. Relevant reactions are represented by the names of the gene(s) coding for the enzymes: aceEF-lpdA, pyruvate dehydrogenase complex; adhE, acetaldehyde/alcohol dehydrogenase; ackA, acetate kinase; dhaKLM, dihydroxyacetone kinase; dld, respiratory d-lactate dehydrogenase; fdhF, formate dehydrogenase, part of the formate hydrogenlyase complex; frdABCD, fumarate reductase; gldA, glycerol dehydrogenase; glpD, aerobic glycerol-3-phosphate dehydrogenase; glpK, glycerol kinase; hycB-I, hydrogenase 3, part of the formate hydrogenlyase complex; ldhA, fermentative d-lactate dehydrogenase; pflB, pyruvate formate-lyase; pta, phosphate acetyltransferase; pykF, pyruvate kinase. Abbreviations: DHA, dihydroxyacetone; DHAP, DHA phosphate; G-3-P, glycerol-3-phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; P/O, amount of ATP produced in the oxidative phosphorylation per pair of electrons transferred through the electron transport system; QH2, reduced quinones.
MATERIALS AND METHODS
Strains, plasmids, and genetic methods.
Wild-type E. coli K12 strain MG1655 (F− λ− ilvG-negative rfb50 rph1) was obtained from the University of Wisconsin E. coli Genome Project (16) and used as the host to engineer the production of d-lactate. Specific details about the procedures used in gene knockout and overexpression are provided below. All resulting strains, along with primers and plasmids used in this study, are listed in Table 1.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain/plasmid/primer | Description | Source or reference |
|---|---|---|
| Strainsa | ||
| MG1655 | F− λ−ilvG-negative rfb50 rph1 | 16 |
| LA01 | MG1655 ΔpflB::FRT ΔfrdA::FRT-Kan-FRT; sequential deletion of pflB and frdA in MG1655 | This study |
| LA02 | MG1655 Δpta::FRT ΔadhE::FRT ΔfrdA::FRT-Kan-FRT; sequential deletion of pta, adhE, and frdA in MG1655 | This study |
| LA01Δdld | MG1655 ΔpflB::FRT ΔfrdA::FRT Δdld::FRT-Kan-FRT; dld deletion in LA01 | This study |
| LA02Δdld | MG1655 Δpta::FRT ΔadhE::FRT ΔfrdA::FRT Δdld::FRT-Kan-FRT; dld deletion in LA02 | This study |
| Plasmids | ||
| pCP20 | reppSC101ts Apr CmrcI857 l PRflp+ | 5 |
| pZSblank | Blank plasmid created by removing Citrobacter freundii dhaKL from pZSKLcf and self ligating the plasmid (tetR, oriR SC101*, cat) | 47a |
| pZSKLMgldA | E. coli dhaKLM and gldA under the control of PLtetO-1 (tetR, oriR SC101*, cat) | 47a |
| pZSKLcf | C. freundii dhaKL under the control of PLtetO-1 (tetR, oriR SC101*, cat) | 2 |
| pZSglpK | E. coli glpK gene control of PLtetO-1 (tetR, oriR SC101*, cat) | This study |
| pZSglpKglpD | E. coli glpK and glpD under the control of PLtetO-1 (tetR, oriR SC101*, cat) | This study |
| pZSldhA | E. coli ldhA under the control of PLtetO-1 (tetR, oriR SC101*, cat) | This study |
| Primersb | ||
| v-pflB | AAATCCACTTAAGAAGGTAGGTG | This study |
| TCGTGGAGCCTTTATTGTAC | ||
| v-frdA | ACCCTGAAGTACGTGGCTG | This study |
| GCACCACCTCAATTTTCAGG | ||
| v-pta | GCTGTTTTGTAACCCGCC | This study |
| GCAGCGCAAAGCTGCGG | ||
| v-adhE | CAAATCATCACCGCACTGAC | This study |
| CCTTAACTGATCGGCATTG | ||
| v-dld | TGATATTTTTTCGCCACCACAAG | This study |
| AAACAAAAAGCCGCCCAAATG | ||
| c-glpK | ACTGCAGGTACCATGACTGAAAAAAAATATATCGTTG | This study |
| TGCTACCTGCAGTTATTCGTCGTGTTCTTCCCAC | ||
| c-glpD | TCGATCCTGCAGGAAAGTGAATGAGGGCAGCATG | This study |
| ACTTGTGGATCCTTACGACGCCAGCGATAACC | ||
| c-ldhA | CATTAAAGAGGAGAAAGGTACCATGAAACTCGCCG | This study |
| GATGCCTCTAGCACGCGTTTAAACCAGTTCGTT |
Deletions were moved into each strain in the order they appear in the description column.
v and c indicate the primer sequences (5′ to 3′) that were used for verification purposes during the creation of disruption mutants and cloning purposes, respectively. The forward sequence follows the reverse sequence in each case. Genes or operons deleted or cloned are apparent from primer names.
Gene knockouts were introduced in MG1655 and its derivatives by P1 phage transduction as described elsewhere (27). Single-gene knockout mutants from the National BioResource Project (NIG; Japan) (1) were used as donors of specific mutations. Details of the protocol used have been described elsewhere (47a). All mutations created in the host cells were confirmed by PCR using the verification primers listed in Table 1. The disruption of multiple genes in a common host was achieved by sequentially implementing the procedure described above and rechecking strains in all disruption sites by PCR at every step where a new mutation was introduced.
Gene overexpression was achieved by cloning the desired gene(s) in a low-copy vector as described in what follows. Plasmids pZSblank (control plasmid, empty vector) and pZSKLMgldA were constructed as previously reported (47a). To construct plasmid pZSldhA, the ldhA gene was PCR amplified using genomic DNA from strain MG1655 and c-ldhA primers (Table 1). The resulting PCR product was cloned within the KpnI and MluI sites of pZSKLM (2) using In-Fusion PCR cloning (Clontech Laboratories, Inc., Mountain View, CA). Plasmid pZSglpKglpD was constructed in two steps. First, glpK was PCR amplified using MG1655 genomic DNA and c-glpK primers (Table 1) and cloned into the KpnI and PstI sites of pZSKLcf, thus obtaining pZSglpK. glpD then was PCR amplified using MG1655 genomic DNA and c-glpD primers (Table 1) and cloned into the PstI and BamHI sites of pZSglpK to obtain pZSglpKglpD. PCR was performed using Pfu turbo DNA polymerase (Stratagene, CA) under standard conditions described by the supplier. The ligated products described above were used to transform E. coli DH5αT1 (Invitrogen, Carlsbad, CA). Positive clones were screened by plasmid isolation and restriction digestion.
Standard recombinant DNA procedures were used for gene cloning, plasmid isolation, and electroporation. Manufacturer protocols and standard methods (27, 33) were followed for DNA purification (Qiagen, Valencia, CA), restriction endonuclease digestion (New England Biolabs, Ipswich, MA), and DNA amplification (Stratagene, La Jolla, CA, and Invitrogen, Carlsbad, CA). The strains were kept in 32.5% glycerol stocks at −80°C. Plates were prepared using LB medium containing 1.5% agar, and appropriate antibiotics were included at the following concentrations: ampicillin (50 μg/ml), kanamycin (50 μg/ml), chloramphenicol (12.5 μg/ml), and tetracycline (3.33 μg/ml).
Culture medium and cultivation conditions.
Unless otherwise stated, all fermentations were conducted using the minimal medium designed by Neidhardt et al. (29) with Na2HPO4 in place of K2HPO4 and supplemented with 20 g/liter glycerol, 5 μM sodium selenite, 3.96 mM Na2HPO4, 5 mM (NH4)2SO4, and 30 mM NH4Cl. Chemicals were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma-Aldrich Co. (St. Louis, MO), except crude glycerol (85%, wt/wt), which was provided by Renewable Energy Group, Inc. (Ames, IA).
Fermentations in shake flasks were performed in 25-ml Pyrex Erlenmeyer flasks (narrow mouth/heavy-duty rim; Corning Inc., Corning, NY) filled with 15 ml of 1× morpholinepropanesulfonic acid (MOPS) minimal medium supplemented with appropriate drugs or inducers when needed at the following concentrations: ampicillin (50 μg/ml), kanamycin (50 μg/ml), chloramphenicol (12.5 μg/ml), tetracycline (3.33 μg/ml), and anhydrotetracycline (100 ng/ml). Unless otherwise stated, calcium carbonate (5% wt/wt) was used in all of the fermentation flasks to help buffer the pH. The flasks (with foam plugs filling the necks) were incubated at 37°C and 200 rpm in an NBS C24 benchtop incubator shaker (New Brunswick Scientific Co., Inc., Edison, NJ). The fermentations were run for 36 h (unless otherwise stated), at which time the supernatant was collected, the pH measured (UB-10; Denver Instruments Co., Arvada, CO), the optical density taken (Genesys 20, 4001/4; Thermo Spectronic, MA), and when necessary, cell pellets collected for enzyme activities. To determine the optical densities of the cultures in the presence of calcium carbonate, the cultures were allowed to briefly sit, in which time the calcium carbonate quickly settled to the bottom.
Prior to use, the cultures (stored as glycerol stocks at −80°C) were streaked onto LB plates and incubated overnight at 37°C. Three colonies were used to inoculate 25-ml flasks containing 5 ml of minimal medium supplemented with 10 g/liter of glycerol, 10 g/liter tryptone, and 5 g/liter yeast extract. The flasks were incubated at 37°C and 150 rpm in an NBS C24 benchtop incubator shaker until an optical density at 550 nm (OD550) of ∼0.7 was reached. An appropriate volume of this actively growing preculture was centrifuged, and the pellet was washed and used to inoculate 15 ml of medium in shake flasks with a target initial optical density at 550 nm of 0.05.
Analytical methods.
Optical density was measured at 550 nm (Thermo Spectronic Genesys 20; MA) and used as an estimate of cell mass (1 OD550 = 0.34 g dry weight/liter) (6). After centrifugation, the supernatant was stored at −20°C for HPLC (high-performance liquid chromatography) and NMR (nuclear magnetic resonance) analysis. Glycerol, organic acids, and ethanol were measured via HPLC (6, 7), and the identity of fermentation products was verified via one-dimension 1H NMR (6, 28).
The enantiomeric purity of lactate was determined enzymatically as previously described (37). The reaction mixture (3 ml) for l-lactate determination contained 0.92 ml hydrazine/glycine buffer (0.6 M glycine and 0.5 M hydrazine; pH 9.2), 55 U l-lactate dehydrogenase, 5 mg NAD, and 200 μl of the fermentation sample of interest. d-Lactate was measured in a similar mixture by replacing l-lactate dehydrogenase with 15 U of d-lactate dehydrogenase. After the addition of the sample, the reaction mixture was incubated at 25°C for 3 h, after which the absorbance at 340 nm was used as a measure of the concentration of d- or l-lactate present.
The oxygen transfer rate was characterized by estimating the volumetric oxygen transfer coefficient (kLa, in h−1) from previous reports on gas-liquid mass transfer in shake flask systems (26, 44) and further confirmed by in-vessel measurements conducted by a conventional dynamic gassing-out technique as previously described (9).
Enzyme activities.
Cells from microaerobic cultures were harvested by centrifugation (2 min, 10,000 × g), washed twice with 9 g/liter NaCl, and stored as cell pellets at −20°C. Cell pellets were resuspended in 0.1 M potassium phosphate buffer to obtain ∼3.4 mg dry cell weight/ml and were permeabilized with chloroform (28).
Glycerol kinase activity was assayed in a manner similar to that described by Hayashi and Lin (12) by measuring the change in absorbance at 340 nm and 25°C in a 1-ml reaction mixture containing 0.15 M glycine (pH 9), 11 mM MgCl2, 0.27 M hydrazine, 1.2 mM NAD+, 5 mM ATP, 2 mM glycerol, 20 U of α-glycerolphosphate dehydrogenase, and 50 μl crude cell extract prepared as described above. Aerobic-glycerol-3-phosphate dehydrogenase was assayed by the phenazine methosulfate (PMS)-mediated reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), which was monitored spectrophotometrically at 570 nm (20). The 1-ml reaction mixture contained 75 μM MTT, 600 μM PMS, 10 mM dl-glycerol 3-phosphate, 0.2% Triton X-100, 10 μM sodium cyanide, and 50 μl crude cell extract. An extinction coefficient of 17 mM−1 cm−1 for reduced MTT was used for the calculation of activities. The activity of glycerol dehydrogenase in the oxidation of glycerol was measured as previously described (11, 41) with potassium carbonate at pH 9.5 as the buffer. Phosphoenolpyruvate (PEP)-dependent dihydroxyacetone kinase activity was assayed using the method reported by Kornberg and Reeves (22), with minor modifications as previously described (47a). d-Lactate dehydrogenase activity was determined by monitored the NADH-dependent reduction of pyruvate at 340 nm and 25°C in a 1-ml reaction mixture containing 0.1 M potassium phosphate buffer (pH 7.5), 30 mM sodium pyruvate, 0.33 mM NADH, and 50 μl crude cell extract (39).
The linearity of the reactions (protein concentration and time) was established for all preparations. All spectrophotometric measurements were conducted in a BioMate 5 spectrophotometer (Thermo Scientific, MA). The nonenzymatic rates were subtracted from the observed initial reaction rates. Enzyme activities are reported as μmol of substrate/min/mg of cell protein and represent averages for at least three cell preparations. A protein content of 50% (wt/wt) was assumed in these calculations.
Calculation of fermentation parameters.
Data from cell growth, glycerol consumption, and product synthesis were used to calculate volumetric (mmol/liter/h) and specific rates (mmol/g cell mass/h) and product yields as previously described (9, 47a).
RESULTS
Rational design of a homolactic pathway for the production of d-lactate from glycerol.
In previous studies of glycerol metabolism under anaerobic and microaerobic conditions, wild-type E. coli exhibited a heterofermentative behavior that resulted in the synthesis of significant amounts of ethanol, acetate, succinate, and formate but only negligible production of d-lactate (9, 11, 28). Although ATP generating, the synthesis of d-lactate from glycerol also represents a redox-generating pathway (Fig. 1) and therefore is not favored under conditions of the limited availability of external electron acceptors (e.g., anaerobic or microaerobic conditions). Despite the use of a higher kLa than that in our previous reports (a kLa of 14.5 h−1 compared to 2.5 h−1 in Durnin et al. [9]), wild-type strain MG1655 produced only very small amounts of d-lactate (Fig. 2A). Therefore, several metabolic engineering strategies were used to increase the production of d-lactate from glycerol, as briefly described below.
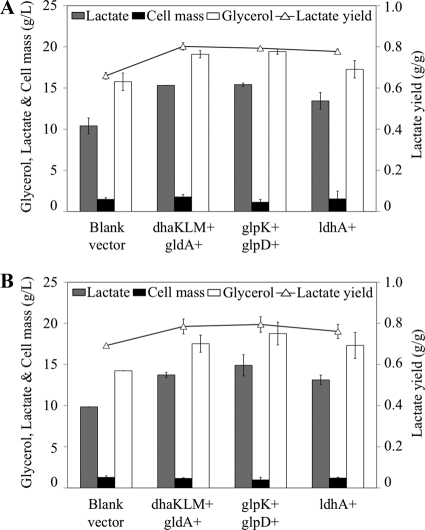
FIG. 2.
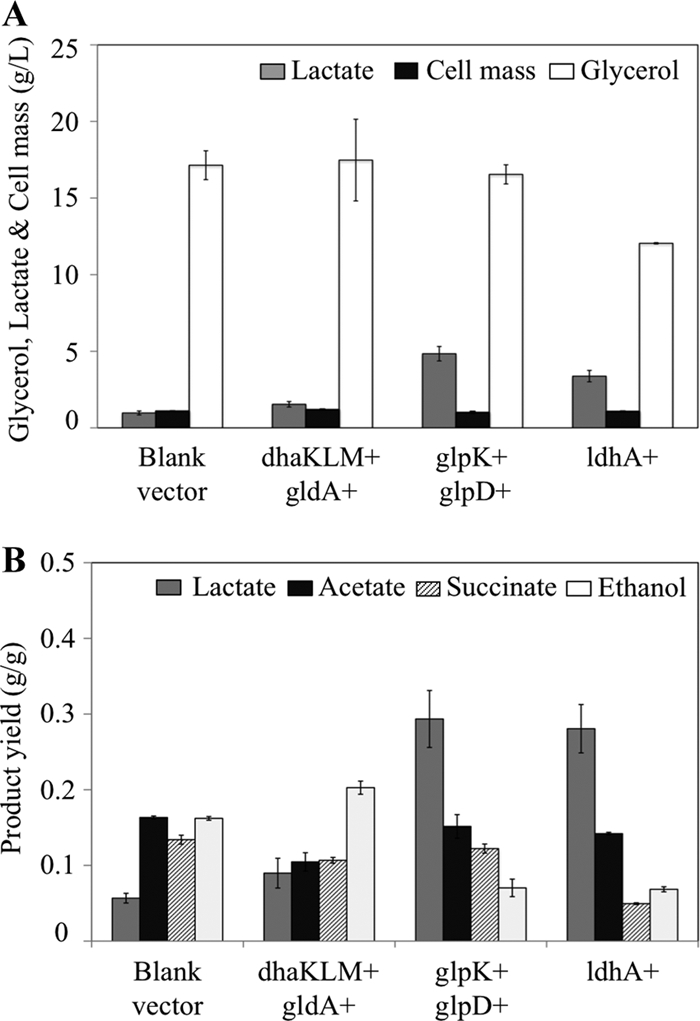
Cell growth, glycerol utilization, and product synthesis in MG1655 with amplified glycerol utilization and d-lactate synthesis pathways. Gene overexpressions are indicated by a plus sign next to the corresponding gene(s) or operon(s). (A) d-Lactate produced, cell growth, and glycerol consumed in shake flask cultures grown for 36 h. (B) Product yield for the cultures represented in panel A. Error bars represent standard deviations from triplicate measurements.
Metabolic targets to improve the synthesis of d-lactate from glycerol are apparent from an inspection of the three metabolic blocks composing the overall pathway involved in this conversion (Fig. 1): (i) the generation of glycolytic intermediate dihydroxyacetone phosphate (DHAP) from glycerol, (ii) the conversion of DHAP to pyruvate via the Embden-Meyerhof-Parnas (EMP) pathway, and (iii) the reduction of pyruvate to d-lactate by d-lactate dehydrogenase (LDH). Since the enzymes of the EMP are known to support high fluxes and are not uniquely involved in glycerol utilization or lactate synthesis, it was postulated that efficient d-lactate production from glycerol can be realized by improving the conversion of glycerol to glycolytic intermediate DHAP and/or the conversion of pyruvate to d-lactate. Since two different pathways, one fermentative and one respiratory, are involved in glycerol utilization (Fig. 1), the impact of their amplification on d-lactate synthesis might differ. On the other hand, the low levels of d-lactate produced by wild-type MG1655 may result from the inability of LDH to compete with enzymes involved in pyruvate dissimilation, such as pyruvate-formate lyase and pyruvate dehydrogenase (Fig. 1). Moreover, LDH is fully activated allosterically only in the presence of ∼5 mM pyruvate (35), and thus d-lactate synthesis may require a relatively large pyruvate pool.
In addition to the approach described above, channeling carbon toward the synthesis of d-lactate could be realized by blocking the synthesis of ethanol, acetate, and succinate. Since the disruption of pyruvate-formate lyase (PFL; pflB gene) or phosphoacetyltransferase (PTA; pta gene) (Fig. 1) has been reported to lead to d-lactate accumulation (9), two platforms for d-lactate production are proposed by using base strains that contain deletions in either the pflB or the pta gene. In the former case, the simultaneous inactivation of pyruvate-formate lyase (ΔpflB) and fumarate reductase (ΔfrdA) is proposed (Fig. 1). For the platform based on the pta deletion, the inactivation of fumarate reductase (ΔfrdA) and alcohol/acetaldehyde dehydrogenase (ΔadhE) also is proposed (Fig. 1). These manipulations are anticipated to impact ATP synthesis (acetate pathway) and the consumption of reducing equivalents (succinate and ethanol pathways) (Fig. 1).
Effect of amplifying the pathways involved in the conversion of glycerol to DHA phosphate.
Two primary routes mediate the conversion of glycerol to DHAP under microaerobic conditions (9) (Fig. 1). A fermentative pathway converts glycerol to dihydroxyacteone (DHA) (glycerol dehydrogenase, GDH; encoded by gldA) and then DHA to DHAP (DHA kinase, DHAK; encoded by dhaKLM). On the other hand, a respiratory pathway composed of the enzymes glycerol kinase (GK; encoded by glpK) and aerobic glycerol-3-phosphate (G3P) dehydrogenase (G3PDH; encoded by glpD) mediates the conversion of glycerol to G3P and G3P to DHAP, respectively. Plasmids encoding each of these pathways were constructed (Table 1) and functionally characterized (Table 2). While the overexpression of neither one of these pathways affected glycerol utilization or cell growth, significant changes in product synthesis, including d-lactate production, were observed (Fig. 2 and Table 3).
TABLE 2.
Functional characterization of constructs used in the overexpression of glycerol utilization and lactate synthesis enzymes
| Enzyme tested | Activitya (nmol/mg protein/min) |
|
|---|---|---|
| Wild type | Overexpressedb | |
| Glycerol kinase | 118 ± 12 | 420 ± 10 |
| Aerobic glycerol-3-phosphate dehydrogenase | 3 ± 1 | 21 ± 1 |
| Glycerol dehydrogenase | 77 ± 5 | 475 ± 30 |
| Dihydroxyacetone kinase | 11 ± 1 | 41 ± 10 |
| d-Lactate dehydrogenase | 82 ± 5 | 960 ± 60 |
All activities were measured as described in Materials and Methods, and values are reported as averages ± standard deviations from triplicate assays. Reported values are from shake flask cultures grown for 36 h.
Activities measured in wild-type strain MG1655 containing a plasmid overexpressing the specified enzyme; i.e., pZSKLMgldA for glycerol dehydrogenase and dihydroxyacetone kinase, pZSglpKglpD for glycerol kinase and aerobic glycerol-3-phosphate dehydrogenase, and pZSldhA for lactate dehydrogenase.
TABLE 3.
Carbon recovery in cell mass and fermentation products during the microaerobic utilization of glycerol in minimal medium by wild-type and engineered strains
| Strain | Carbon recoverya |
|||||||
|---|---|---|---|---|---|---|---|---|
| Acetate | Succinate | Ethanol | Lactate | Formate | Biomass | Products | Overall | |
| Wild type | ||||||||
| MG1655 | 23.4 (0.5) | 10.0 (0.1) | 22.7 (1.3) | 8.5 (1.6) | 2.9 (1.1) | 8.0 (0.2) | 67.4 (1.4) | 75.4 (1.6) |
| Wild type overexpressing genes in glycerol utilization and lactate synthesis pathway | ||||||||
| MG1655(pZSKLMgldA) | 16.1 (1.9) | 8.3 (0.3) | 40.5 (1.7) | 9.2 (2.0) | 4.1 (0.6) | 8.6 (1.3) | 78.3 (2.4) | 86.9 (1.1) |
| MG1655(pZSglpKglpD) | 23.2 (2.4) | 9.5 (0.5) | 14.1 (2.3) | 30.0 (3.8) | 1.1 (1.4) | 7.6 (0.8) | 78.4 (2.8) | 86.0 (2.3) |
| MG1655(pZSldhA) | 21.8 (0.3) | 3.9 (0.1) | 13.7 (0.7) | 28.7 (3.3) | 0.0 (0.0) | 11.1 (0.3) | 68.0 (4.0) | 79.1 (4.2) |
| Engineered with deletions in enzymes involved in by-product synthesis | ||||||||
| LA01 | 5.4 (0.9) | 0.2 (0.1) | 0.0 (0.0) | 69.3 (3.8) | 0.0 (0.0) | 9.3 (0.6) | 74.9 (3.4) | 84.3 (3.7) |
| LA02 | 5.6 (0.8) | 0.3 (0.2) | 0.0 (0.0) | 71.8 (2.6) | 0.0 (0.0) | 9.7 (0.7) | 77.6 (2.8) | 87.4 (3.4) |
| Engineered with amplification of desired pathways and elimination of competing by-products | ||||||||
| LA01(pZSKLMgldA) | 2.1 (0.0) | 0.0 (0.0) | 2.4 (0.3) | 82.0 (1.8) | 0.0 (0.0) | 11.5 (0.4) | 86.6 (1.5) | 98.0 (1.1) |
| LA01(pZSglpKglpD) | 5.8 (0.8) | 0.1 (0.0) | 0.0 (0.0) | 77.8 (5.7) | 0.0 (0.0) | 7.0 (0.6) | 84.0 (5.1) | 91.0 (5.5) |
| LA01(pZSldhA) | 4.9 (1.0) | 0.2 (0.1) | 0.0 (0.0) | 79.5 (1.3) | 0.0 (0.0) | 10.9 (1.8) | 84.5 (0.3) | 95.4 (2.0) |
| LA02(pZSKLMgldA) | 3.8 (0.3) | 0.5 (0.1) | 0.0 (0.0) | 80.3 (3.7) | 0.0 (0.0) | 8.2 (0.3) | 84.5 (4.1) | 92.8 (4.4) |
| LA02(pZSglpKglpD) | 4.4 (0.9) | 0.8 (0.2) | 0.0 (0.0) | 81.2 (3.8) | 0.0 (0.0) | 6.5 (0.8) | 86.6 (3.8) | 93.1 (4.3) |
| LA02(pZSldhA) | 3.6 (0.4) | 0.3 (0.1) | 0.0 (0.0) | 76.3 (3.7) | 0.0 (0.0) | 9.6 (1.6) | 80.2 (3.4) | 89.8 (3.2) |
| LA01Δdld(pZSglpKglpD)b | 8.3 (2.0) | 0.3 (0.1) | 3.2 (0.2) | 80.2 (2.5) | 0.0 (0.0) | 4.3 (0.1) | 92.1 (0.2) | 96.4 (0.2) |
| LA02Δdld(pZSglpKglpD)b | 4.8 (0.6) | 0.76 (0.1) | 0.0 (0.0) | 83.3 (0.1) | 0.0 (0.0) | 3.4 (0.4) | 89.1 (0.7) | 92.4 (1.1) |
| LA02Δdld(pZSglpKglpD)c | 4.5 (0.1) | 0.8 (0.1) | 0.0 (0.0) | 85.9 (0.1) | 0.6 (0.1) | 4.5 (0.1) | 92.4 (0.2) | 96.9 (0.2) |
| LA02Δdld(pZSglpKglpD)d | 4.6 (0.1) | 0.95 (0.2) | 0.0 (0.0) | 93.4 (1.3) | 0.0 (0.0) | 5.0 (0.1) | 99.2 (1.1) | 104.2 (1.2) |
Data represent the averages from three samples (standard deviations are shown in parentheses) taken from shake flask cultures grown for 36 h on minimal medium supplemented with 20 g/liter of glycerol, unless otherwise specified. Carbon recovery is expressed as the percent moles of carbon in the product, including biomass, per mole of carbon in glycerol consumed. The column labeled product shows the total recovery of carbon in products, assuming that moles of acetate plus moles of ethanol equals moles of 1-C compounds (formate plus CO2) generated by the dissimilation of pyruvate. The column overall shows the overall carbon recovery, including products and biomass.
Cultures in which 40 g/liter of glycerol was used; samples were taken at 72 h (all glycerol was consumed).
A culture in which 40 g/liter of crude glycerol derived from biodiesel production was used; samples were taken at 72 h (all glycerol was consumed).
A culture in which 60 g/liter of crude glycerol (40 g/liter initially present and 20 g/liter added at 48 h) was used; samples were taken at 84 h (∼54 g/liter of glycerol was consumed).
The amplification of the fermentative pathway caused a modest increase in d-lactate concentration and yield but also increased ethanol yield and decreased acetate yield. These changes reflect the fact that glycerol utilization through the gldA-dhaKLM pathway generates reducing equivalents in the form of NADH (Fig. 1). The accumulation of NADH has been shown to increase the levels of alcohol/acetaldehyde dehydrogenase (23, 24), thus explaining the observed increase in ethanol synthesis. Since both ethanol and acetate are synthesized from acetyl-coenzyme A (AcCoA), the increase in ethanol synthesis would, in turn, lead to lower acetate production. Moreover, the potential decrease in the PEP pool and increase of the pyruvate pool upon the overexpression of the enzyme DHAK (DHAK uses PEP as a phosphate donor in the phosphorylation of DHA [2] and therefore converts PEP to pyruvate) could be contributing to the observed decrease in succinate yield and increase in d-lactate yield (Fig. 2B and Table 3).
Unlike the fermentative pathway, the overexpression of the respiratory pathway for glycerol utilization led to a major increase in d-lactate production (an approximately 5-fold increase in concentration and yield) and a significant reduction in ethanol synthesis (Fig. 2 and Table 3). This pathway involves a respiratory glycerol-3-phosphate dehydrogenase (GlpD) that donates electrons directly to quinones (36, 45), which ultimately are transferred to oxygen. Due to this coupling between glycerol-3-phosphate oxidation and oxygen reduction, the overall conversion of glycerol to d-lactate becomes a redox-balanced pathway that also generates ATP via substrate-level phosphorylation (Fig. 1). This explains why d-lactate production is preferred when the GlpD-mediated respiratory pathway is the primary route for glycerol utilization.
Overall, the overexpression of the respiratory pathway seems to be a better alternative for d-lactate production, while the fermentative pathway leads to an increase in the production of highly reduced product ethanol.
Effect of overexpressing d-lactate dehydrogenase.
d-Lactate dehydrogenase overexpression (LDH; encoded by ldhA) could lead to an increase in the fraction of carbon diverted toward the synthesis of d-lactate (increasing d-lactate yield) and/or the flux of the glycerol-to-d-lactate pathway (increasing the rate of d-lactate production). However, given the allosteric properties of the enzyme (i.e., allosterically activated by pyruvate), this is likely to happen only if the pyruvate levels are high enough to support LDH activity (10, 35, 51). Figure 2 and Table 3 show the impact of amplifying the LDH pathway on cell growth, product synthesis, and glycerol utilization. Both the concentration and yield of d-lactate significantly increased (3.5- and 5-fold, respectively), with the concomitant reduction in the synthesis of competing by-products (Fig. 2 and Table 3). Glycerol consumption, however, decreased significantly (Fig. 2A), and the yield of competing by-products, especially acetate, still was high (Fig. 2B and Table 3). The observed increases in d-lactate yield and concentration arguably were caused by the 12-fold increase in the activity of LDH (Table 2). The pyruvate node thus appears to respond similarly during the microaerobic utilization of glycerol and the anaerobic fermentation of glucose, as a 10-fold overproduction of LDH doubled d-lactate synthesis during glucose fermentation (46).
Effect of eliminating by-product pathways on d-lactate production from glycerol.
An alternative approach to divert carbon toward the synthesis of d-lactate is eliminating the pathways to the undesired products ethanol, acetate, and succinate. In our previous studies of glycerol metabolism in E. coli under microaerobic conditions, two mutations that led to d-lactate accumulation were reported (9) (Fig. 1): (i) the disruption of pyruvate-formate lyase (PFL; pflB gene), which catalyzes the conversion of pyruvate to acetyl-CoA and formate, and (ii) the disruption of phosphoacetyltransferase (PTA; pta gene), which converts acetyl-CoA to acetyl-phosphate (first step in acetate production from AcCoA). The increase in lactate accumulation upon the introduction of these mutations appears to be mediated by an increase in the pyruvate pool, as the introduction of defects in pta and pflB do not affect ldhA expression (15).
Based on the evidence described above, two platforms for d-lactate production were engineered by using base strains that contain deletions in either the pflB or the pta gene. Since PFL is the primary route for pyruvate conversion to AcCoA during the microaerobic utilization of glycerol (9), the pflB mutation should effectively minimize both acetate and ethanol production from AcCoA (Fig. 1). A double mutant, ΔpflB ΔfrdA (named LA01), then was constructed in which succinate production was minimized by blocking the enzyme fumarate reductase (ΔfrdA) (Fig. 1). Strain LA01 produced d-lactate as the primary product of glycerol metabolism at high concentration (12.4 g/liter) and yield (0.69 g lactate/g glycerol) and with minimum by-product formation (Fig. 3 and Table 3).
FIG. 3.
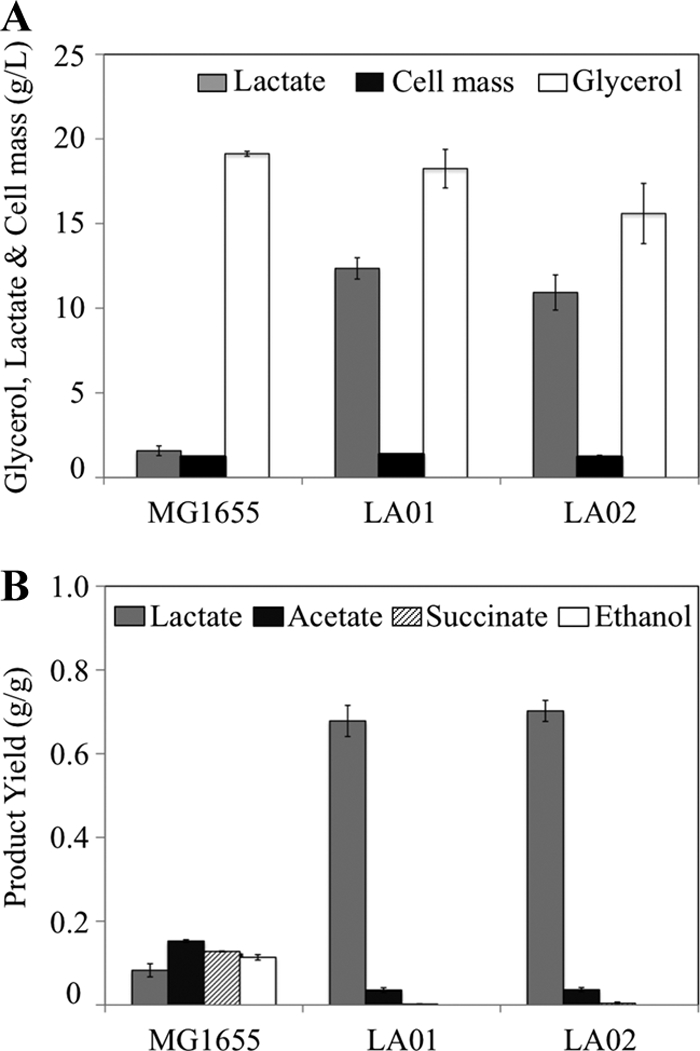
Cell growth, glycerol utilization, and product synthesis in strains LA01 and LA02 containing mutations in the pathways that lead to the synthesis of competing by-products. LA01 is a double mutant (ΔpflB ΔfrdA) and LA02 a triple mutant (ΔadhE Δpta ΔfrdA). (A) d-Lactate production, cell growth, and glycerol consumption in shake flask cultures grown for 36 h. (B) Product yield for the cultures represented in panel A. Error bars represent standard deviations from triplicate measurements.
A second platform, based on the pta mutant, was constructed by adding mutations that minimized the synthesis of ethanol (ΔadhE, blocking alcohol/acetaldehyde dehydrogenase) and succinate (ΔfrdA). The resulting triple mutant (Δpta ΔadhE ΔfrdA) was named LA02 and exhibited a performance very similar to that of LA01 (Fig. 3 and Table 3), although with slightly lower concentrations of d-lactate produced (10.9 g/liter). The production of d-lactate in both strains presumably is triggered by an increase in the pyruvate pool, which is known to allosterically activate LDH (15, 35).
Combined effect of amplification of desired pathways and elimination of competing by-products.
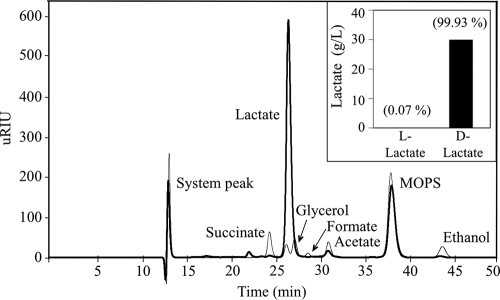
In previous sections, the beneficial effect of separately overexpressing LDH or the glycerol utilization pathways as well as the reduction in the synthesis of by-products acetate, ethanol, and succinate has been demonstrated. In this section, the combined effect of those modifications is evaluated. As shown in Fig. 4 and Table 3, the overexpression of glycerol utilization and d-lactate synthesis pathways in mutants LA01 and LA02 had a beneficial effect on d-lactate titer and yield. Overall, the overexpression of the respiratory glycerol utilization pathway (GlpK-GlpD) in either the LA01 or LA02 background led to the highest production of d-lactate (Fig. 4 and Table 3). A sample chromatogram from the fermentation conducted using strain LA02(pZSglpKglpD) is shown in Fig. 5, clearly demonstrating the chemical purity of the product. To establish its chiral purity, the fermentation broth was assayed with l- and d-lactate dehydrogenase, and the results are shown in the inset of Fig. 5. Based on these analyses, the lactate produced by strain LA02(pZSglpKglpD) was found to be 99.9% d-lactate. Similar results were found for strain LA01(pZSglpKglpD) (data not shown).
FIG. 4.
Effect of amplifying the glycerol utilization and d-lactate synthesis pathways in engineered strains LA01 (A) and LA02 (B). Bars indicate d-lactate produced, cell growth, and glycerol consumed, and lines indicate d-lactate yield for shake flask cultures grown for 36 h. Overexpressed genes are indicated by a plus sign next to the corresponding gene(s) or operon(s). Error bars represent standard deviations from triplicate measurements.
FIG. 5.
Chromatogram of 36-h fermentation samples taken from cultures of wild-type MG1655 (gray line) and LA02(pZSglpKglpD) (black line) demonstrating the chemical purity of lactate produced in the engineered strain. The inset shows the quantification of d- and l-lactate in the LA02(pZSglpKglpD) sample to assess chiral purity: the concentration (bars; g/liter) and percentage (number in parenthesis; wt/wt) of each enantiomer are shown.
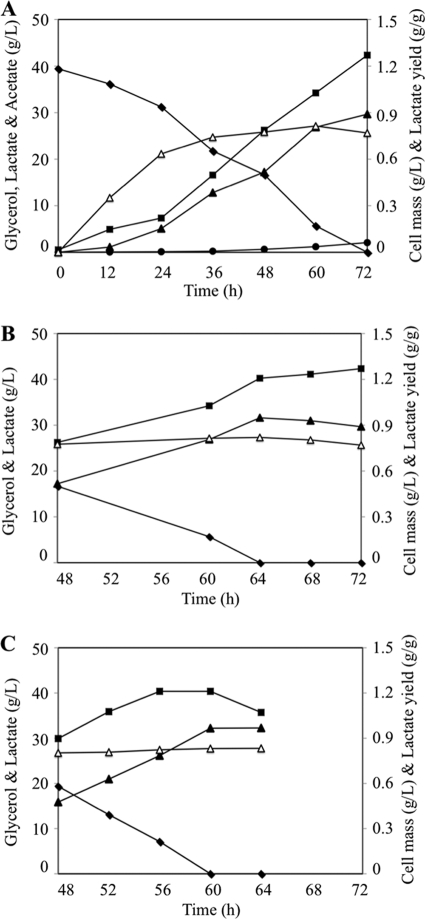
A more detailed characterization of strain LA02 carrying the amplified GlpK-GlpD pathway was conducted by investigating the dynamics of cell growth, glycerol consumption, and product synthesis in the presence of higher concentrations of glycerol (Fig. 6 A and Table 2). This strain consumed about 40 g/liter of glycerol and produced about 30 g/liter of d-lactate in less than 72 h at a yield of 0.80 g d-lactate/g glycerol. Besides d-lactate, only small amounts of acetate were found in the culture medium, demonstrating the homolactic nature of the fermentation. However, a closer examination of the data revealed that the cultures never reached stationary phase (Fig. 6A and B), which indicates the consumption of a second carbon source after glycerol has been exhausted. More importantly, in the final hours of the fermentation, a clear decrease in lactate concentration and yield was observed (Fig. 6B). Based on these observations, it was hypothesized that the accumulation of large amounts of lactate in the medium triggers its consumption by the respiratory d-lactate dehydrogenase, which catalyzes the oxidation of d-lactate to pyruvate (31). Since this enzyme is encoded by dld (34), that gene was knocked out in strain LA02 (new strain named LA02Δdld). When the GlpK-GlpD pathway was amplified in strain LA02Δdld, a clear stationary phase followed glycerol depletion from the medium, and no decrease in lactate yield or concentration was observed (Fig. 6C). This strain produced 32.3 g/liter from 39.5 g/liter of glycerol in about 60 h. The overall product yield was 0.83 g d-lactate/g of glycerol. In the final 12 h of the culture (Fig. 6C), d-lactate was produced at maximum volumetric and specific rates of 1.5 g/liter/h and 1.25 g/g cell mass/h, respectively.
FIG. 6.
Kinetics of d-lactate production by strains LA02(pZSglpKglpD) (A and B) and LA02Δdld(pZSglpKglpD) (C) in shake flasks containing minimal medium with 40 g/liter of glycerol. (A) Fermentation profile for strain LA02(pZSglpKglpD): data are shown for the concentration of cells (▪), glycerol (⧫), d-lactate (▴), and acetate (•). The d-lactate yield also is shown (▵). (B) High-resolution fermentation profile for strain LA02(pZSglpKglpD) at late stages of cultivation. Symbols are the same as those described for panel A. (C) Fermentation profile for strain LA02Δdld(pZSglpKglpD): symbols are as described for panel B. Coefficients of variation (i.e., standard deviations/average × 100) were below 5% in all cases.
Production of d-lactate at high concentrations from crude glycerol.
The use of an industrial medium containing crude glycerol generated as a by-product in the biodiesel industry is of great relevance for the biocatalysts developed in this work. Engineered strains performed very well when crude glycerol was used as a carbon source. Glycerol consumption and d-lactate synthesis by strain LA02Δdld(pZSglpKglpD) using 40 g/liter crude glycerol were similar to those reported for the consumption of pure glycerol (Table 3). In fact, d-lactate concentration (∼34 g/liter) and yield (0.845 g/g glycerol) were slightly higher when the crude glycerol was used. Maximum specific and volumetric rates for d-lactate production also were similar to those from the experiments with pure glycerol. In another experiment using higher concentrations of crude glycerol, strain LA02Δdld(pZSglpKglpD) produced 45 g/liter of d-lactate in 84 h at a yield of 0.83 g/g glycerol (Table 3).
DISCUSSION
Current processes for d-lactate production based on native lactic acid bacteria use sugars as the carbon source and suffer from requirements of complex nutrients for cell growth and limited product selectivity and enantiomeric purity (13, 30). To address these issues, E. coli and other bacteria have been engineered to produce d-lactate from sugars as the primary fermentation product (4, 8, 50, 51, 52). Unlike sugars, when glycerol is used as a carbon source under anaerobic or microaerobic conditions by wild-type E. coli, only very small amounts of d-lactate are found in the fermentation broth (9, 28). d-Lactate also has been reported to be a by-product of 1,3-propanediol (1,3-PDO) production in an E. coli strain engineered to produce 1,3-PDO from glycerol in rich medium (40). This strain was constructed by importing the glycerol utilization and 1,3-PDO-producing pathways from Klebsiella pneumoniae, an organism that synthesizes lactate as a by-product of glycerol fermentation. However, E. coli has not been engineered for the homofermentative production of d-lactate from glycerol. The work reported here focused on the metabolic engineering of E. coli for the production of chemically (>96%) and optically (>99.9%) pure d-lactic acid from glycerol in minimal salts medium. The engineered biocatalysts offer several advantages, such as the use of minimal medium without rich supplements, high yields and productivities, and the use of crude glycerol as the carbon source, which is an abundant and inexpensive feedstock (47).
The three metabolic engineering strategies designed and implemented in this work led to significant changes in the partition of carbon at the pyruvate node and favored d-lactate synthesis to different extents. First, the amplification of the respiratory pathway for glycerol utilization brought about a significant increase in d-lactate yield and titer. The direct coupling of glycerol-3-phosphate oxidation and oxygen reduction made the glycerol-to-d-lactate pathway redox balanced, thus favoring its functioning. A second strategy based on the overexpression of LDH, the enzyme responsible for the synthesis of d-lactate from pyruvate, also led to significant increases in d-lactate yield and titer. However, this strategy is limited by the higher Km of LDH for pyruvate (Km = 7.0 to 7.2 mM [39]) compared to those of pyruvate-dissimilating enzymes PFL (Km = 2 mM [21]) and PDH (Km = 0.2 to 0.4 mM [3]). Since none of the strategies described above was effective in minimizing the accumulation of ethanol, acetate, and succinate, a third approach based on blocking the pathways leading to the synthesis of these competing by-products was implemented. Two platforms were explored based on the previous finding that pta and pflB knockouts lead to the accumulation of lactate (9). Of the two, the strain based on the pta mutation, LA02 (also containing frdA and adhE mutations), yielded better results. The amplification of the respiratory GlpK-GlpD pathway in LA02 resulted in the production of 30 g/liter of d-lactate in less than 72 h at a yield of 0.80 g d-lactate/g glycerol and with a chiral purity higher than 99.9%. This strain was further improved by introducing a dld mutation (strain LA02Δdld), which prevented the utilization of d-lactate as a carbon source. When the GlpK-GlpD pathway was amplified in LA02Δdld, 32 g/liter of d-lactate was produced in about 60 h at a yield of 0.83 g d-lactate/g glycerol. Maximum volumetric and specific rates of d-lactate production in this strain reached 1.5 g/liter/h and 1.25 g/g cell/h, respectively.
In the strain describe above, i.e., LA02Δdld(pZSglpKglpD), the oxidation of glycerol-3-phosphate by GlpD is coupled to oxygen reduction, and therefore only one molecule of NADH is generated in the glycolytic pathway (Fig. 1). The synthesis of d-lactate from pyruvate also consumes an NADH, and thus the overall conversion of glycerol to d-lactate in this strain is redox balanced. In terms of energy, the synthesis of one molecule of d-lactate from glycerol is accompanied by the net generation of one molecule of ATP via substrate-level phosphorylation (Fig. 1). Additional ATP is generated by the coupling between the transfer of electrons from glycerol-3-phosphate to oxygen and oxidative phosphorylation. Although GlpD does not conserve the redox energy in a proton potential, the overall transfer of electrons from glycerol-3-phosphate to oxygen (a combination of GlpD and CyoABCD) results in a proton translocation ratio of four H+/2e− (42). With an average H+/ATP ratio of 3.5 for E. coli F0F1 ATP synthetase, it follows that the oxidation of one molecule of glycerol-3-phosphate theoretically can generate 1.14 molecules of ATP. However, given the lower experimental values typically observed (17), the synthesis of 1 ATP molecule per glycerol-3-phosphate molecule oxidized is probably a more reasonable assumption. The overall stoichiometry for the conversion of glycerol to d-lactate in strain LA02Δdld(pZSglpKglpD) then can be represented by the equation
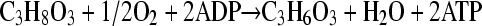 |
From this equation it also is apparent that the maximum theoretical yield for the conversion of glycerol to d-lactate is 100% on a mole or carbon basis and 98% on a weight basis. The engineered strain LA02Δdld(pZSglpKglpD) exhibited a d-lactate yield of 0.83 g/g glycerol, which is 85% of the theoretical maximum. It also is noteworthy that d-lactate yields and specific productivities reported here are similar to those achieved with the use of glucose. Moreover, the production of d-lactate from glycerol generates up to twice the amount of ATP synthesized in the glucose-based fermentation (see the analysis above). This higher ATP yield will be advantageous in maintaining high cell viability and productivity at high concentrations of d-lactate or during prolonged cultivations (e.g., fed-batch or continuous operation). Finally, the engineered strains performed very well in an industrial medium based on inexpensive glycerol generated as the by-product in the production of biodiesel supplemented only with mineral salts.
Additional improvements of the engineered strains are envisioned through the use of metabolic evolution techniques like those successfully implemented in E. coli for the efficient production of biofuels and other products (14, 48, 49). Our engineered strains are likely to evolve more productive phenotypes given the link between cell growth and their ability to produce d-lactate due to the redox-balanced and ATP-generating nature of the overall conversion of glycerol to d-lactate. Process-based modifications, including fed-batch cultivations and high-density cultures, also are envisioned to further improve the volumetric rates of d-lactate production.
Acknowledgments
This work was supported by grants from the U.S. National Science Foundation (CBET-0645188) and the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (2005-35504-16698).
We thank H. Mori and B. Erni for providing research materials, C. Rivera, S. Doneske, and S. S. Yazdani for assistance with genetic methods, and Paul Campbell for fruitful discussions.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bächler, C., P. Schneider, P. Bahler, A. Lustig, and B. Erni. 2005. Escherichia coli dihydroxyacetone kinase controls gene expression by binding to transcription factor DhaR. EMBO J. 24:283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswanger, H., and U. Henning. 1971. Regulatory properties of the pyruvate-dehydrogenase complex from Escherichia coli: positive and negative cooperativity. Eur. J. Biochem. 24:376-384. [DOI] [PubMed] [Google Scholar]
- 4.Chang, D. E., H. C. Jung, J. S. Rhee, and J. G. Pan. 1999. Homofermentative production of d- or l-lactate in metabolically engineered Escherichia coli RR1. Appl. Environ. Microbiol. 65:1384-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dharmadi, Y., A. Murarka, and R. Gonzalez. 2006. Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnol. Bioeng. 94:821-829. [DOI] [PubMed] [Google Scholar]
- 7.Dharmadi, Y., and R. Gonzalez. 2005. A better global resolution function and a novel iterative stochastic search method for optimization of high-performance liquid chromatographic separation. J. Chromatogr. A 1070:89-101. [DOI] [PubMed] [Google Scholar]
- 8.Dien, B. S., N. N. Nichols, and R. J. Bothast. 2001. Recombinant Escherichia coli engineered for production of l-lactic acid from hexose and pentose sugars. J. Ind. Microbiol. Biotechnol. 27:259-264. [DOI] [PubMed] [Google Scholar]
- 9.Durnin, G., J. Clomburg, Z. Yeates, P. J. J. Alvarez, K. Zygourakis, P. Campbell, and R. Gonzalez. 2009. Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol. Bioeng. 103:148-161. [DOI] [PubMed] [Google Scholar]
- 10.Gokarn, R. R., M. A. Eiteman, and E. Altman. 2000. Metabolic analysis of Escherichia coli in the presence and absence of carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase. Appl. Environ. Microbiol. 66:1844-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez, R., A. Murarka, Y. Dharmadi, and S. S. Yazdani. 2008. A new model for the anaerobic fermentation of glycerol in enteric bacteria: trunk and auxiliary pathways in Escherichia coli. Metab. Eng. 10:234-245. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi, S., and E. C. C. Lin. 1967. Purification and properties of glycerol kinase from Escherichia coli. J. Biol. Chem. 242:1030-1035. [PubMed] [Google Scholar]
- 13.Hofvendahl, K., and B. Hahn-Hagerdal. 2000. Factors affecting the fermentative lactic acid production from renewable resources. Enzyme Microb. Technol. 26:87-107. [DOI] [PubMed] [Google Scholar]
- 14.Jantama, K., M. J. Haupt, S. A. Svoronos, X. L. Zhang, J. C. Moore, K. T. Shanmugam, and L. O. Ingram. 2008. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 99:1140-1153. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, G. R., S. Nikolova, and D. P. Clark. 2001. Regulation of the ldhA gene, encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 147:2437-2446. [DOI] [PubMed] [Google Scholar]
- 16.Kang, Y., T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch, K. M. Winterberg, and F. R. Blattner. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kashket, E. R. 1983. Stoichiometry of the H+-ATPase of Escherichia coli cells during anaerobic growth. FEBS Lett. 154:343-346. [DOI] [PubMed] [Google Scholar]
- 18.Khanal, S. K., M. Rasmussen, P. Shrestha, H. Van Leeuwen, C. Visvanathan, and H. Liu. 2008. Bioenergy and biofuel production from wastes/residues of emerging biofuel industries. Water Environ. Res. 80:1625-1647. [Google Scholar]
- 19.Kharas, G. B., F. Sanchez-Riera, and D. K. Severson. 1994. Polymers of lactic acid, p. 93-137. In D. P. Mobley (ed.), Plastics from microbes: microbial synthesis of polymers and polymer precursors. Hanser Publishers, Munich, Germany.
- 20.Kistler, W. A., and E. C. C. Lin. 1971. Anaerobic l-α-glycerophosphate dehydrogenase of Escherichia coli: its genetic locus and its physiological role. J. Bacteriol. 108:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knappe, J., J. Schacht, W. Mochel, T. Hopner, J. Vetter, and R. Edenharder. 1969. Pyruvate formate lyase reaction in Escherichia coli. Eur. J. Biochem. 11:316-327. [DOI] [PubMed] [Google Scholar]
- 22.Kornberg, H. L., and R. E. Reeves. 1972. Inducible phosphoenolpyruvate-dependent hexose phosphotransferase activities in Escherichia coli. Biochem. J. 128:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonardo, M. R., P. R. Cunningham, and D. P. Clark. 1993. Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. J. Bacteriol. 175:870-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonardo, M. R., Y. P. Dailly, and D. P. Clark. 1996. Role of NAD in regulating the adhE gene of Escherichia coli. J. Bacteriol. 178:6013-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, E. C. 1976. Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 30:535-578. [DOI] [PubMed] [Google Scholar]
- 26.Maier, U., M. Losen, and J. Buchs. 2004. Advances in understanding and modeling the gas-liquid mass transfer in shake flasks. Biochem. Eng. J. 17:155-167. [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Murarka, A., Y. Dharmadi, S. S. Yazdani, and R. Gonzalez. 2008. Fermentative utilization of glycerol in Escherichia coli and its implications for the production of fuels and chemicals. Appl. Environ. Microbiol. 74:1124-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okano, K., T. Tanaka, C. Ogino, H. Fukuda, and A. Kondo. 2010. Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl. Microbiol. Biotechnol. 85:413-423. [DOI] [PubMed] [Google Scholar]
- 31.Pratt, E. A., L. W. Fung, J. A. Flowers, and C. Ho. 1979. Membrane-bound d-lactate dehydrogenase from Escherichia coli: purification and properties. Biochemistry 18:312-316. [DOI] [PubMed] [Google Scholar]
- 32.Rausch, K. D., and R. L. Belyea. 2006. The future of co-products from corn processing. Appl. Biochem. Biotechnol. 128:47-86. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Santos, E., H. Kung, I. G. Young, and H. R. Kaback. 1982. In vitro synthesis of the membrane-bound d-lactate dehydrogenase of Escherichia coli. Biochemistry 21:2085-2091. [DOI] [PubMed] [Google Scholar]
- 35.Sawers, R. G., and D. P. Clark. July 2004, posting date. Chapter 3.5.3, Fermentative pyruvate and acetyl-coenzyme A metabolism. In A. Böck, R. Curtiss III, J. B. Kaper, P. D. Karp, F. C. Neidhardt, T. Nyström, J. M. Slauch, C. L. Squires, and D. Ussery (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. doi: 10.1128/ecosal.3.5.3. [DOI]
- 36.Schweizer, H., and T. J. Larson. 1987. Cloning and characterization of the aerobic sn-glycerol-3-phosphate dehydrogenase structural gene glpD of Escherichia coli K-12. J. Bacteriol. 169:507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Severn, D. J., M. E. Johnson, and N. F. Olson. 1986. Determination of lactic acid in Cheddar cheese and calcium lactate crystals. J. Dairy Sci. 69:2027-2030. [Google Scholar]
- 38.Reference deleted.
- 39.Tarmy, E. M., and N. O. Kaplan. 1968. Kinetics of Escherichia coli B d-lactate dehydrogenase and evidence for pyruvate controlled change in conformation. J. Biol. Chem. 243:2587-2596. [PubMed] [Google Scholar]
- 40.Tong, I. T., H. H. Liao, and D. C. Cameron. 1991. 1,3-Propanediol production by Escherichia coli expressing genes from the Klebsiella pneumoniae dha regulon. Appl. Environ. Microbiol. 57:3541-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Truniger, V., and W. Boos. 1994. Mapping and cloning of gldA, the structural gene of the Escherichia coli glycerol dehydrogenase. J. Bacteriol. 176:1796-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unden, G., and P. Dünnwald. March 2008, posting date. Chapter 3.2.2, The aerobic and anaerobic respiratory chain of Escherichia coli and Salmonella enterica: enzymes and energetics. In A. Böck, R. Curtiss III, J. B. Kaper, P. D. Karp, F. C. Neidhardt, T. Nyström, J. M. Slauch, C. L. Squires, and D. Ussery (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. doi: 10.1128/ecosal.3.2.2. [DOI] [PubMed]
- 43.Vaidya, A. N., R. A. Pandey, S. Mudliar, M. S. Kumar, T. Chakrabarti, and S. Devotta. 2005. Production and recovery of lactic acid for polylactide—an overview. Crit. Rev. Environ. Sci. Technol. 35:429-467. [Google Scholar]
- 44.Van Suijdam, J. C., N. W. F. Kossen, and A. C. Joha. 1978. Model for oxygen transfer in a shake flask. Biotechnol. Bioeng. 20:1695-1710. [Google Scholar]
- 45.Walz, A. C., R. A. Demel, B. de Kruijff, and R. Mutzel. 2002. Aerobic sn-glycerol-3-phosphate dehydrogenase from Escherichia coli binds to the cytoplasmic membrane through an amphipathic alpha-helix. Biochem. J. 365:471-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, Y. T., K. Y. San, and G. N. Bennett. 1999. Redistribution of metabolic fluxes in Escherichia coli with fermentative lactate dehydrogenase overexpression and deletion. Metab. Eng. 1:141-152. [DOI] [PubMed] [Google Scholar]
- 47.Yazdani, S. S., and R. Gonzalez. 2007. Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr. Opin. Biotechnol. 18:213-219. [DOI] [PubMed] [Google Scholar]
- 47a.Yazdani, S. S., and R. Gonzalez. 2008. Engineering Escherichia coli for the efficient conversion of glycerol to ethanol and co-products. Metab. Eng. 10:340-351. [DOI] [PubMed] [Google Scholar]
- 48.Yomano, L. P., S. W. York, S. Zhou, K. T. Shanmugam, and L. O. Ingram. 2008. Re-engineering Escherichia coli for ethanol production. Biotechnol. Lett. 30:2097-2103. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, X., K. Jantama, J. C. Moore, K. T. Shanmugam, and L. O. Ingram. 2007. Production of l-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77:355-366. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, S., K. T. Shanmugam, and L. O. Ingram. 2003. Functional replacement of the Escherichia coli d(−)-lactate dehydrogenase gene (ldhA) with the l(+)-lactate dehydrogenase gene (ldhL) from Pediococcus acidilactici. Appl. Environ. Microbiol. 69:2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, S., T. B. Causey, A. Hasona, K. T. Shanmugam, and L. O. Ingram. 2003. Production of optically pure d-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl. Environ. Microbiol. 69:399-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, Y., M. A. Eiteman, K. DeWitt, and E. Altman. 2007. Homolactate fermentation by metabolically engineered Escherichia coli strains. Appl. Environ. Microbiol. 73:456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]