Abstract
We designed a new genetic tool to detect plasmid transfer under anaerobic and aerobic conditions. The system is based on the T7 RNA polymerase gene and a T7 promoter-driven oxygen-independent green fluorescent protein, evoglow, alone or in combination with red fluorescent protein DsRed. Constructs are available as plasmids and mini-mariner transposons.
Conjugative plasmid transfer allows exchange of genetic material within and between bacterial populations through direct cell-to-cell contact and plays an important role in bacterial adaptation. This role is exemplified by the rapid spread of multidrug resistance (8) and the abundance of diverse transferable pollutant degradation plasmids (14). While many environmental factors are known to affect plasmid transfer efficiency (5), few studies have addressed the effect of oxygen levels on plasmid transferability, in spite of growing concerns about antibiotic resistance and virulence among facultatively and strictly anaerobic pathogens (2, 3, 10, 12, 13). There is thus an obvious need for new methods that allow quantitative analysis of plasmid transfer under both aerobic and anaerobic conditions.
The traditional method of plasmid transfer detection is based on plating and enumeration of donor, recipient, and transconjugant cells on selective media. However, this approach does not allow in situ observation of conjugation in spatially structured populations like bacterial microcolonies and biofilms. Therefore, it cannot address questions about the effects of spatial cell organization on plasmid spread. In order to detect plasmid transfer in situ, confocal laser scanning microscopy (CLSM) is often used to detect either fluorescently labeled molecular probes or fluorescent proteins encoded by chromosome- or plasmid-located reporter genes (11). Here, the challenge is the level of fluorescence, which may depend on many factors. For the frequently used green fluorescent protein (GFP) family, an important limiting factor is the concentration of oxygen necessary for protein maturation (9). The fluorescent proteins in this family are therefore not useful under anaerobic or low-oxygen conditions, such as those in sediments, anaerobic digesters, the human gut, and thick biofilms.
Recently, a new flavin mononucleotide (FMN)-based oxygen-independent fluorescent protein, Escherichia coli FMN-based fluorescent protein (EcFbFP; evoglow), was described (4). We propose application of this fluorescent protein to provide a true representation of plasmid transfer in situ that is not confounded by the effects of O2 concentration on fluorescence levels.
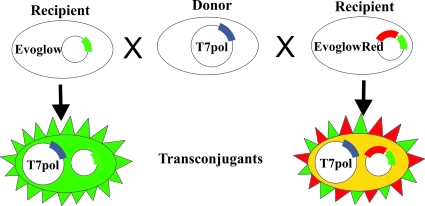
To allow in situ monitoring of plasmid transfer in both aerobic and anaerobic environments, we constructed a dual reporter system comprising the fluorescent reporter proteins DsRed and EcFbFP. The red fluorescent protein (RFP) DsRed provides very strong and stable fluorescence but requires oxygen in the maturation process (1). It can thus serve as an internal control to confirm conditions of limited oxygen. The system, presented in Fig. 1, consists of two independent elements: the phage T7 RNA polymerase gene and a reporter gene (or genes) driven by the T7 promoter (pT7). Thus, when T7 RNA polymerase is not present, the cells show no fluorescence at all. However, when both the polymerase and the reporter genes are present in the same cell, the latter are expressed and fluorescence is detected. Both components are available on plasmid vectors or as a part of the mariner minitransposon (see Fig. S1 in the supplemental material).
FIG. 1.
Schematic action of the reporter system. The donor strain contains plasmid F′ marked with the T7 RNA polymerase gene, and recipient strains contain the vector pGLOW-Txn-Bs2 (left) or pEvoGlowRed (right). Transconjugant cells with both the marked F′ plasmid and a reporter gene are green fluorescent (left) or green and red (yellow; right), respectively.
The T7 RNA polymerase gene was amplified from plasmid pBT20-Δbla-T7pol (6) and cloned into pGem3Zf+ under the control of two promoters, the lactose operon promoter plac and pT7 (resulting in plasmids pGemT7pol and pGemT7cat [see Fig. S1a in the supplemental material]), and subsequently into the mariner minitransposon delivery vector pBT20-Δbla (6), resulting in the pMaT7cat plasmid (see Fig. S1b in the supplemental material). The presence of two strong promoters should ensure expression of the T7 RNA polymerase gene under most growth conditions. Detailed descriptions of the cloning strategy and methods used in this paper are provided in the supplemental material.
The second part of the system consists of a broad-host-range plasmid or mariner transposon with one or two reporter genes driven by the pT7 promoter. Plasmid pGLOW-Txn-Bs2 (see Fig. S1c in the supplemental material), which contains the EcFbFp gene driven by the pT7 promoter, was obtained from evocatal GmbH (Düsseldorf, Germany). Vector pEvoGlowRed encodes both EcFbFP and DsRed, which are expressed using a single pT7 promoter (see Fig. S1d in the supplemental material). This construct can be mobilized into a broad range of strains that can be used as recipients in plasmid transfer studies. The vector can also be used as a promoter probe vector, since part of the pGem3Zf+ multiple-cloning site (MCS) is located between pT7 and the DsRed-EcFbFP genes. Single PstI, SalI, XbaI, and BamHI sites can thus be used to clone foreign DNA fragments and detect promoter activity under both aerobic and anaerobic conditions in bacterial strains not containing the T7 RNA polymerase gene.
Since the reporter plasmids described above are based on the broad-host-range vector pBBR1MCS (7), which can replicate and persist in many Gram-negative bacteria, problems due to occasional plasmid loss and mobilization could be encountered. To avoid this and to further extend the range of organisms in which our system can be used, the gene cassettes containing either the EcFbFP gene or both DsRed and EcFbFP genes driven by the pT7 promoter were also cloned into the mariner transposon delivery vector pBT20-Δbla (6), resulting in constructs carrying different antibiotic resistance genes (see Fig. S1e to k in the supplemental material). The mariner transposon can be used for in vivo as well as in vitro mutagenesis and has been successfully applied in Gram-negative and Gram-positive bacteria (15).
To quantify the efficiency of plasmid transfer in relation to the parental strains, one typically also counts donor and/or recipient cells. To simultaneously detect recipients and transconjugants under aerobic conditions with one vector, we first constructed the vector pGlowRed. In this vector, the plac-DsRed gene cassette was cloned into the EcoRV site of the pGLOW-Txn-Bs2 vector (see Fig. S1e in the supplemental material). In this case, expression of the DsRed gene is constitutive, driven by the lac promoter as well as the distantly located pKm promoter (yielding red fluorescence), but EcFbFP is expressed only in T7 polymerase-carrying strains (data not shown). Thus, when pGlowRed is present in the recipient, the cells show only red fluorescence, but transconjugants are green and red (yellow). In parallel, a mariner transposon containing a constitutively expressed DsRed gene and a pT7-driven EcFbFP gene with gentamicin (Gm) and kanamycin (Km) resistance genes was constructed (data not shown).
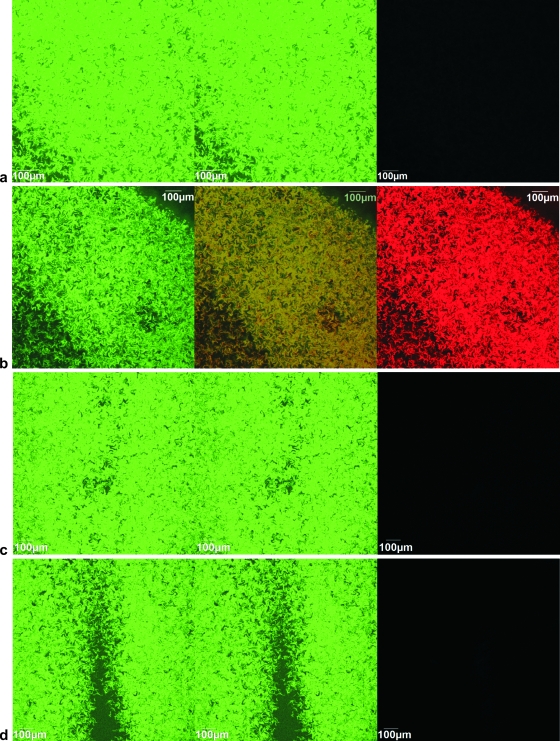
To evaluate the feasibility of using EcFbFP and DsRed in plasmid transfer studies under both aerobic and anaerobic conditions, we monitored the transfer of plasmid F′, as it is known to transfer efficiently under both aerobic and anaerobic conditions (12). The plasmid was first marked using pMaT7cat. In two marked plasmids (F′::T7cat1 and F′::T7cat2), insertions were located within the lacI gene of the F′ plasmid (see Materials and Methods in the supplemental material). Thus, the locations of the mariner transposon carrying T7cat (MaT7cat) in this region do not disturb any of the core genes involved in replication, maintenance/control, or transfer functions. Transfer of the marked plasmids to three E. coli K-12 recipient strains was observed in simple cross-streak experiments (see Fig. S2 in the supplemental material). The first two recipient strains carried reporter plasmids pGLOW-Txn-Bs2 and pEvoGlowRed; the third strain (E. coli K-12-GR) carried a chromosomal insertion of the minitransposon MaGlowRedKm. Donor and recipient precultures were grown anaerobically and then cross-streaked onto Luria-Bertani plates and incubated under aerobic or anaerobic conditions at 37°C. After aerobic incubation, transconjugants were observed as bright green fluorescent cells when recipients carried pGLOW-Txn-Bs2 and as both green and red fluorescent cells when recipients contained pEvoGlowRed (Fig. 2 a and b). The same results were obtained with aerobically grown precultures (data not shown). Under anaerobic conditions, only green fluorescence was observed for both types of recipients (Fig. 2c and d). Similarly, green and red fluorescence in the presence, and only green fluorescence in the absence, of oxygen was detected when E. coli K-12-GR was used as the recipient (data not shown). The lack of red fluorescence under anaerobic conditions was expected and served as an internal control to confirm conditions of limited oxygen. Green color intensities and surface areas of fluorescent transconjugants were qualitatively similar under aerobic and anaerobic conditions. The two marked F′ plasmids showed identical results. Additional filter matings followed by plate counting confirmed that there was no obvious effect of the absence of oxygen on F′ plasmid transfer under these experimental conditions. The average frequencies of transconjugants per recipient under anaerobic and aerobic conditions were not significantly different (0.5 and 0.8; P < 0.05 [standard t test]).
FIG. 2.
Microscopy images of mating mixtures containing either E. coli MG1655Rif(F′::T7cat) (the donor strain) and MG1655Nal(pGLOW-Txn-Bs2) (recipient 1) (a and c) or the same donor and MG1655Nal(pEvoGlowRed) (recipient 2) (b and d). Mixtures were incubated for 48 h under aerobic conditions (a and b) and 72 h under anaerobic conditions (c and d). Panels: left, EcFbFP (green fluorescence); right, DsRed (red fluorescence); middle, overlay image of both channels.
We also validated the use of EcFbFP in thick cell layers by growing a colony of E. coli constitutively expressing EcFbFP and RFP on agar plates under alternating aerobic and anaerobic conditions (see Materials and Methods in the supplemental material). The cross-sectional micrograph shows that EcFbFP was expressed all the way down to the agar and that the RFP signal was detected only at the edge of the colony (see Fig. S3 in the supplemental material).
One caveat of our system is the requirement for recipient strains that contain either the reporter gene(s) or the T7 RNA polymerase gene, thus limiting its use in experiments aimed at monitoring plasmid transfer into indigenous bacterial populations. However, one can detect plasmid transfer from an exogenous donor if indigenous bacteria are first isolated, marked with a reporter or MaT7pol system, and then reintroduced into their ecosystem.
In conclusion, we present here the first fluorescent reporter system that allows in situ monitoring of plasmid population dynamics under both aerobic and anaerobic conditions with simultaneous verification of low-oxygen conditions through lack of red fluorescence. So far, all other reporter systems used to monitor plasmid transfer have been based on GFP and RFP and thus depend on the presence of oxygen.
Supplementary Material
Acknowledgments
The research was supported by NIH grant 1R01 GM73821; the Olympus FV1000 multiphoton confocal microscope was funded by the M. J. Murdock Charitable Trust (grant no. 2006-127JVZ022207) and an NIH small instrumentation grant (1S10RR02260601).
We thank L. Forney for helpful suggestions, A. Paszczynski and K. H. Kucharzyk for use of their anaerobic chamber, and L. Sherick and K. Turner for technical assistance. We are also grateful to J.-M. Ghigo for providing plasmid F′ and to T. T. Hoang for pBT20-Δbla-T7pol and pBT20-Δbla.
J.E.K. designed the dual reporter system, constructed all vectors, performed the plasmid transfer experiments, and wrote the manuscript draft. E.M.T., L.M.R., and S.M.K. oversaw the project, provided help with the experimental design and data interpretation, and assisted in writing the manuscript.
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baird, G. S., D. A. Zacharias, and R. Y. Tsien. 2000. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. U. S. A. 97:11984-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biebricher, C. K., and E. M. Duker. 1984. F and type 1 piliation of Escherichia coli. J. Gen. Microbiol. 130:951-957. [DOI] [PubMed] [Google Scholar]
- 3.Curtiss, R. I., L. G. Caro, D. P. Allison, and D. R. Stallions. 1969. Early stages of conjugation in Escherichia coli. J. Bacteriol. 100:1091-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drepper, T., T. Eggert, F. Circolone, A. Heck, U. Krausz, J.-K. Guterl, M. Wendorff, A. Losi, W. Gartner, and K.-E. Jaeger. 2007. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25:443-445. [DOI] [PubMed] [Google Scholar]
- 5.Elsas, J. D., and M. J. Bailey. 2002. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 42:187-197. [DOI] [PubMed] [Google Scholar]
- 6.Kang, Y., M. S. Son, and T. T. Hoang. 2007. One step engineering of T7-expression strains for protein production: increasing the host-range of the T7-expression system. Protein Expr. Purif. 55:325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 8.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122-S129. [DOI] [PubMed] [Google Scholar]
- 9.Reid, B. G., and G. C. Flynn. 1997. Chromophore formation in green fluorescent protein. Biochemistry 36:6786-6791. [DOI] [PubMed] [Google Scholar]
- 10.Shoemaker, N. B., C. Getty, E. P. Guthrie, and A. A. Salyers. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 12.Stallions, D. R., and R. I. Curtiss. 1972. Bacterial conjugation under anaerobic conditions. J. Bacteriol. 111:294-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strohmaier, H., R. Noiges, S. Kotschan, G. Sawers, G. Högenauer, E. L. Zechner, and G. Koraimann. 1998. Signal transduction and bacterial conjugation: characterization of the role of ArcA in regulating conjugative transfer of the resistance plasmid R1. J. Mol. Biol. 277:309-316. [DOI] [PubMed] [Google Scholar]
- 14.Top, E. M., and D. Springael. 2003. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14:262-269. [DOI] [PubMed] [Google Scholar]
- 15.Vos, J. C., I. De Baere, and R. H. Plasterk. 1996. Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev. 10:755-761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.