Abstract
In total, 782 Escherichia coli strains originating from various host sources have been analyzed in this study by using a highly discriminatory single-nucleotide polymorphism (SNP) approach. A set of eight SNPs, with a discrimination value (Simpson's index of diversity [D]) of 0.96, was determined using the Minimum SNPs software, based on sequences of housekeeping genes from the E. coli multilocus sequence typing (MLST) database. Allele-specific real-time PCR was used to screen 114 E. coli isolates from various fecal sources in Southeast Queensland (SEQ). The combined analysis of both the MLST database and SEQ E. coli isolates using eight high-D SNPs resolved the isolates into 74 SNP profiles. The data obtained suggest that SNP typing is a promising approach for the discrimination of host-specific groups and allows for the identification of human-specific E. coli in environmental samples. However, a more diverse E. coli collection is required to determine animal- and environment-specific E. coli SNP profiles due to the abundance of human E. coli strains (56%) in the MLST database.
Identification of the sources of microbial contamination in the environment is a prime objective of public health. Escherichia coli is an internationally recognized standard indicator microorganism for the initial assessment of human risk in water (8, 13, 33). Current methods used in routine laboratories are based on enumeration of E. coli colonies and do not provide any information regarding their sources. Because of this gap in information, researchers have developed microbial source tracking (MST) methods that are capable of differentiating between human and animal fecal pollution in environmental waters. Certain MST methods require the development of a library from known host groups followed by a comparison with unknown water samples in the environment. These methods are known as library-dependent methods. The most commonly used library-dependent methods are repetitive extragenic palindromic (REP) PCR (16), ribotyping (7), and pulsed-field gel electrophoresis (PFGE) (19); however, they are rarely used since they are difficult to develop and interpret (26, 32).
The most successful source tracking methods are library-independent methods, which are mainly PCR based and do not require the development of a library. Library-independent methods detect the presence of specific genes associated with certain groups of bacteria from human and animal sources (1). Library-independent methods have targeted Bacteroides species 16S rRNA clone groups (5), F+ RNA coliphage differentiation (12), and enteric viruses, such as polyomaviruses and adenoviruses (11). The advantage of these markers is that they appear to be host specific (2, 20, 21). Although certain studies reported the presence of human-specific markers in other animals, such as dogs (15) and fish (17), none of the MST methods could yet be regarded as a “gold standard.” Each of the methods has its advantages and disadvantages (10, 28). As outlined by Blanc (6), molecular typing methods can be regarded as library based or comparative. Library-based methods are applicable across the target species and provide an unambiguous type, while comparative methods are simply used to determine whether isolates are the same or different.
Multilocus sequence typing (MLST) is becoming more widely used as a genotyping method and has been applied successfully to Salmonella sp. MST (4); however, this sequencing procedure remains expensive and time consuming for routine water monitoring. It is reasonable to state that the ideal standardized molecular typing method would be a library-based method that indicates the position of the isolate within the species population structure and could also economically and conveniently serve as a high-resolution comparative method. Our approach of interrogating single-nucleotide polymorphisms (SNPs) located within MLST sequence data utilizes the discriminatory power of MLST and, at the same time, avoids the sequencing of a large number of E. coli strains. MLST has been applied primarily to pathogenic E. coli strains, while other fingerprinting methods, such as pulsed-field gel electrophoresis (PFGE) and denaturing gradient gel electrophoresis (DGGE) have been used to characterize highly diverse E. coli populations (7, 19, 29). The SNP typing method described in this paper is considered by the authors to be neither library dependent nor independent. In a previous study, the SNP genotyping method was used to fingerprint Campylobacter jejuni isolates based on microarray data (library independent) from comparative genome hybridization studies (22).
We hypothesize that there is a host-specific structure within the E. coli population. Some genetic variability among E. coli isolates can be explained by the host niche (18) compared to the E. coli community in surface water. By applying our SNP genotyping method, we attempted to answer the following questions. (i) What subset of polymorphisms needs to be interrogated to obtain a particular measurement of resolution? (ii) Do these polymorphisms provide a genetic fingerprint consistent with the population structure of E. coli?
Previous studies have shown that SNP typing has a discriminatory power similar to that of MLST, in particular when combined with virulence or flagellum genes (23). The aim of this study was to apply an SNP genotyping approach to differentiate E. coli isolates from six different animal species in Southeast Queensland (SEQ), Australia. We determined that the types defined by SNP genotyping were consistent with the natural groups that exist within the E. coli species. We compared the discriminatory power of SNP genotyping to that of MLST in distinguishing between closely related cattle, kangaroo, horse, dog, duck, and human E. coli strains and used this information to characterize them according to their sources.
MATERIALS AND METHODS
Identification of informative SNP sets.
The E. coli MLST database available at the NIH (http://www.shigatox.net) currently contains 668 E. coli strains that are grouped into 231 sequence types (STs).
Informative SNP sets were identified by using the software program called Minimum SNPs (25). The software processes the allelic sequences and MLST profile data together, executing a Simpson's index of diversity (D) function on the toolbox. The score of SNP sets with increasing D values is displayed as the name of the gene locus with the number of the informative nucleotide at that locus. Eight SNPs, with D values of 0.96, were determined by the program for the differentiation of E. coli isolates. The MLST database at http://www.shigatox.net is ideal for SNP analysis, since thousands of gene sequences have accumulated in the database and hundreds of STs are annotated in this database. The aligned sequences of aspC, clpX, fadD, icdA, lysP, mdh, and uidA were downloaded in the FASTA format and used as input data for the Minimum SNPs software program.
eBURST analysis.
The eBURST analysis program, accessed at http://www.eburst.mlst.net/, was used to visualize the relationship between STs (determined by MLST) (9) and the SNP profiles of SEQ isolates.
Assignment of SNP profiles to SEQ E. coli isolates. (i) Bacterial strains.
In total, 114 E. coli isolates were used in the current study for in vitro analysis (Table 1). Of these, 66 isolates were previously used for an MST study in Southeast Queensland, Australia (3), including the following: 16 cattle, 16 dog, 16 duck, 16 horse, and 2 kangaroo isolates. In addition, 34 clinical E. coli isolates, originating from human feces and urine specimens, were provided by Pathology Queensland. A further 14 E. coli isolates were isolated from kangaroos using the method described by Tutenel et al. (31) with modifications.
TABLE 1.
Primer sequences used for kinetic real-time PCRa
| SNP | Primer sequence (5′-3′) | ΔCT |
|---|---|---|
| fadD(234) | (F) GATTTCTCCWGTYTGCAYCTTTCY | 3 |
| (R) CCTTCCAGCAGATACTGTCCG | ||
| (R) CCTTCCAGCAGATACTGTCCT | ||
| (R) CCTTCCAGCAGATACTGTCCA | ||
| clpX(267) | (F) AGCGYGGKATTGTCTACATC | 3 |
| (R) ACCGGAAACGTCHCGDGTAACA | ||
| (R) ACCGGAAACGTCHCGDGTAACG | ||
| uidA(138) | (F) CCRGGAATGGTGATYACMGGC | 5 |
| (F) CCRGGAATGGTGATYACMGGT | ||
| (R) AGASRATYACGCTGCGATGG | ||
| clpX(177) | (F) AAACATYATTCAGAARCTGTTGCCA | 3 |
| (F) AAACATYATTCAGAARCTGTTGCCG | ||
| (R) GAGAAATYTTGTCRATYTMATCGAT | ||
| clpX(234) | (F) CTGTTGCARAARTGCGAYTAYG | 10 |
| (R) TTGTCYGACTTACGAGAAATYTTGTCG | ||
| (R) TTGTCYGACTTACGAGAAATYTTGTCA | ||
| lysP(198) | (F) GGTACAACTGGGCGGTGACYAGC | 15 |
| (F) GGTACAACTGGGCGGTGACYAGT | ||
| (R) CGGTGTATCCGGGAACCA | ||
| icdA(177) | (F) ATTCYCTTCCCRRAACATTGC | 3 |
| (F) ATTCYCTTCCCRRAACATTGT | ||
| (R) TTGCGTATTCRATCGCKGC | ||
| mdh(450) | (F) AACGYATCCAGAACGCG | 10 |
| (F) AACGYATCCAGAACGCA | ||
| (R) GGTTGCMGACCCRCCA |
SNPs are shown in bold. The difference in cycle threshold times between the matched and mismatched primer reactions (ΔCT) is depicted as numbers.
(ii) Isolation methods for fecal strains.
A 3-g amount of feces was added to 100 ml sterile PBS buffer and shaken vigorously. After homogenization, the sample was incubated for 4 h at 37°C in a water bath. A total of 0.1 ml of the suspension was plated onto mTEC agar (BD, NJ), and plates were incubated for 22 h at 44.5°C. Colonies with a bright red-magenta color were isolated and subcultured onto MacConkey no. 2 (Oxoid, United Kingdom) agar at 37°C for 24 h.
(iii) Primer design.
This was undertaken using the Primer Express 2.0 program (Applied Biosystems) on sequences that were aligned by ClustalX2 (30). As determined by Minimum SNPs, the specific SNP at each locus was incorporated into the primer design by positioning the polymorphism at the 3′ end of the primer. The aligned MLST allele sequences were screened visually for the presence of point mutations within the primer binding region. No more than three mismatches were acceptable in the primer sequences. Using the exclude function of the Minimum SNPs program, alternative SNPs were found to facilitate primer design. As a result, sets of bimorphic and trimorphic SNP primers were designed for allele-specific real-time PCR analysis. Primer sequences were analyzed for nonspecific binding by running the BLASTN program (http://ncbi.nih.gov). This confirmed gene-specific binding of primers only to E. coli MLST genes. Primers were obtained from Proligo (Brisbane, Australia).
(iv) DNA extraction.
To prepare genomic DNA, single lactose-fermenting colonies from MacConkey agar no. 2 (Oxoid, United Kingdom) were selected and incubated overnight in 5 ml nutrient broth (Oxoid, United Kingdom). Subsequently, 500 μl of culture was centrifuged at 10,000 × g for 1 min. Cell pellets were resuspended in 180 μl DNase/RNase-free water and used for DNA extraction on the Corbett X-tractorGene automated DNA extraction system (Corbett Robotics, Brisbane, Australia) using the DX kit, CorProtocol no. 14104 version 02. Purity of DNA extracts was determined spectrophotometrically on a DU 730 spectrophotometer (Beckman Coulter) by measuring the optical density (OD) ratio at 260 and 280 nm.
(v) Interrogation of SNPs using real-time PCR.
All reactions were performed using the Rotorgene 6000 real-time PCR instrument (previously Corbett Life Science, now Qiagen). Each 11-μl reaction, in duplicate, contained 5 μl of either 2× Platinum SYBR green qPCR supermix-UDG (Invitrogen, Carlsbad) or 2× SensiMix (Quantum Scientific, Australia), 3 pmol of each forward and reverse primer, and 2 μl DNA. Real-time PCR conditions were as follows: 50°C for 2 min and 95°C for 10 min and 35 cycles of 95°C for 15 s, 57°C for 20 s, and 72°C for 20 s, with a final melting step of 60°C to 95°C, increasing 1°C at each cycle. The difference in cycle threshold times (ΔCT) between the matched and mismatched reactions was calculated by subtracting the cycle time (CT) of the mismatched primer from the CT of the matched primer. A sufficient ΔCT for allele-specific real-time PCR is a criterion in which the default ΔCT value is more than three cycles. If the ΔCT value was <3 cycles between the matched and mismatched reactions, then these primers were modified by incorporating a subterminal mismatch at the 3′ end of the primer sequence as described previously (25). If a suitable ΔCT value was still not obtained after incorporation of a subterminal mismatch, new primers were designed (see “Primer design” above). All the SNP profiles obtained from SEQ isolates are referred to as “SEQ profiles.”
Assignment of SNP profiles to STs in the MLST database.
According to the information provided in the MLST database, 56% of the isolates were of human origin, 12% sourced from cows, 3% from food (beef), 2% from pigs, and 9% from various animals, such as rats, monkeys, deer, a Tasmanian devil, kangaroos, bats, dogs, possums, crocodiles, and sheep. Each of these animals represented less than 1% of the total MLST library. In terms of inanimate material, sand represented 2% of the total library and 16% of isolates were of unknown origin. Assignment of SNP profiles to STs was done by the “working backwards” application of the Minimum SNPs software program. This is done by determining the presence of a particular SNP in the MLST housekeeping gene and subsequently assigning these SNP profiles to STs. All the SNP profiles obtained from isolates stored in the MLST database are referred to as “MLST profiles.”
Validation of allele-specific real-time PCR SNP calling by sequencing.
Amplification and sequencing primers, together with amplification conditions, were obtained from the website at http://www.shigatox.net. PCR products from randomly chosen isolates were purified using the High Pure PCR product purification kit (Roche, Indianapolis, IN). Sequencing reactions were carried out using ABI Prism BigDye Terminator mix according to the manufacturer's instructions. Samples were submitted to the DNA Sequencing Facility at Griffith University (Brisbane, Australia). Obtained chromatograms were analyzed using the Vector NTI software program. These sequencing data were used to validate the correct SNP allocation by allele-specific real-time PCR.
Blind tests using SNP analysis.
Using the method described above (see “Isolation methods for fecal strains”), a set of six blind isolations were performed by a third party, with the host sources of the samples being kept from the researcher. The isolated colonies were subjected to SNP analysis, as described previously (see “Assignment of SNP profiles to SEQ E. coli isolates”). Wherever possible, multiple isolates (up to eight) were taken from each host source.
Discriminatory power analysis.
The discriminatory power (D) was estimated using Simpson's index of diversity as described previously (14) and provides the level of differentiation of each genotyping method. Calculations were performed using Excel from Microsoft Office 2003 (Seattle, WA).
RESULTS
High-D-value SNP profiles of E. coli isolates from SEQ and those defined by eBURST analysis.
The set of primers for allele-specific real-time PCR was developed to be used in highly discriminatory SNP interrogations of E. coli populations (Table 1). Eight SNPs, namely, fadD(234) (D = 0.63), clpX(267) (D = 0.8), uidA(138) (D = 0.88), clpX(177) (D = 0.92), clpX(234) (D = 0.93), lysP(198) (D = 0.94), icdA(177) (D = 0.95), and mdh(450) (D = 0.96), were selected based on the detection of a sufficient ΔCT. All SNPs were validated based on MLST sequencing of five selected SEQ isolates. The sequencing results showed that allele-specific real-time PCR successfully indicated the correct polymorphism at the SNP positions of all six housekeeping genes (data not shown).
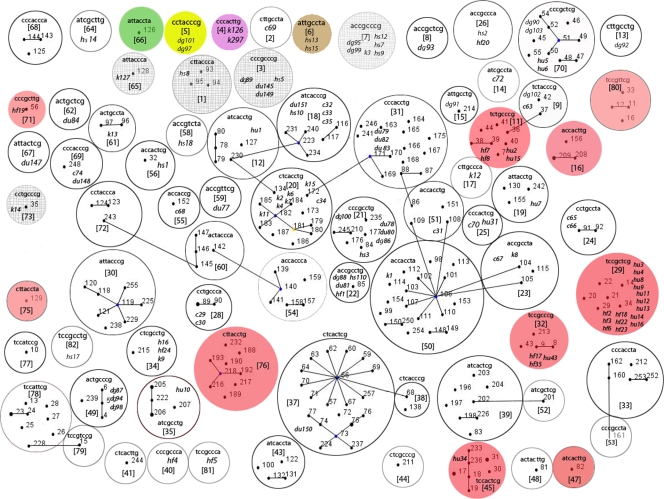
The current E. coli MLST database contains 668 E. coli strains, grouped into 231 STs with a D value of 0.95. Using the eBURST algorithm (Fig. 1), the 231 STs were combined into 76 clonal complexes (CCs), and 111 STs remained as singletons (not related to any other ST in the MLST database). On the other hand, the SNP genotyping method grouped these 231 STs into 55 SNP profiles with a D value of 0.90 (see Table S1 in the supplemental material).
FIG. 1.
Combined output of an eBURST analysis of E. coli STs from an MLST database and SNP profiles from SEQ isolates. Single-locus variants (STs that differ at 1/7 MLST alleles) are depicted by straight lines. Each circle represents a separate SNP profile. Each SNP profile is an 8-character “barcode” which indicates a particular SNP genotype (listed in an order of increasing discriminatory power): fadD(234) (D = 0.63), clpX(267) (D = 0.8), uidA(138) (D = 0.88), clpX(177) (D = 0.92), clpX(234) (D = 0.93), lysP(198) (D = 0.94), icdA(177) (D = 0.95), and mdh(450) (D = 0.96). SNP profile numbers were assigned to each individual barcode. All of the 114 SEQ isolates genotyped in this study are shown in italics and originated from cows (c), dogs (d), ducks (du), kangaroos (k), horses (h), human feces (hf), and human urine (hu). Host-specific SNP profiles are shaded in green for cattle, yellow for dogs, brown for horses, magenta for kangaroos, and pink for human E. coli. Animal-specific SNP profiles are depicted as hashed circles. SNP profiles with isolates originating from both animal and human sources were considered mixed and remain uncolored.
Using allele-specific real-time PCR, 50 different SNP profiles with a total D value of 0.97 were obtained for the 114 SEQ E. coli strains that originated from SEQ humans and various animals (Table 2). Finally, when the SNP profiles were merged, all 782 isolates from the MLST database and SEQ isolates were resolved into a total of 74 SNP profiles (Fig. 1).
TABLE 2.
SNP profiles from SEQ E. colia
| SEQ isolate(s) | No. of isolates in collection | fadD (234) | clpX (267) | uidA (138) | clpX (177) | clpX (234) | lysP (198) | icdA (177) | mdh (450) | SNP profile | SNP profile ID no. (assigned)b |
|---|---|---|---|---|---|---|---|---|---|---|---|
| hs8 | 1 | C | T | T | A | C | C | C | A | CTTACCCA | 1 |
| c69 | 1 | C | T | T | G | C | C | T | A | CTTGCCTA | 2 |
| c5, du145, du149, dg89 | 4 | C | C | C | G | C | C | C | G | CCCGCCCG | 3 |
| k126, k297 | 2 | C | C | C | A | C | T | T | G | CCCACTTG | 4 |
| dg97, dg101 | 2 | C | C | T | A | C | C | C | G | CCTACCCG | 5 |
| hs13, hs15 | 2 | A | T | T | G | C | C | T | A | ATTGCCTA | 6 |
| hs7, hs9, hs12, dg95, dg99, k3 | 6 | A | C | C | G | C | C | C | G | ACCGCCCG | 7 |
| dg93 | 1 | A | C | C | G | C | T | C | G | ACCGCTCG | 8 |
| c63, dg102 | 2 | T | C | T | A | C | C | C | G | TCTACCCG | 9 |
| hf7, hf8, hu2, hu15 | 4 | T | C | T | G | C | C | C | G | TCTGCCCG | 11 |
| hu1 | 1 | A | T | C | A | C | C | T | G | ATCACCTG | 12 |
| dg92 | 1 | C | T | T | G | C | C | T | G | CTTGCCTG | 13 |
| c72 | 1 | A | T | C | G | C | C | T | A | ATCGCCTA | 14 |
| dg91 | 1 | A | T | T | G | C | C | T | G | ATTGCCTG | 15 |
| k12 | 1 | C | T | T | G | C | C | C | A | CTTGCCCA | 17 |
| hs10, c32, du151 | 5 | A | T | C | A | C | C | C | G | ATCACCCG | 18 |
| hu7 | 1 | A | T | T | A | C | C | T | G | ATTACCTG | 19 |
| c34, k2, k4, k6, k7, k11, k15 | 8 | C | T | C | G | C | C | T | G | CTCACCTG | 20 |
| hs3, du78, du80, dg86, dg100 | 5 | C | C | C | G | C | C | T | G | CCCGCCTG | 21 |
| hf1, hs110, du81, dg88 | 4 | A | C | C | G | C | C | T | G | ACCGCCTG | 22 |
| c67, k8 | 2 | A | C | C | G | C | C | T | A | ACCGCCTA | 23 |
| c65, c66 | 2 | C | C | T | G | C | C | T | A | CCTGCCTA | 24 |
| hu31, c70 | 1 | C | C | C | A | C | T | C | G | CCCACTCG | 25 |
| hf20, hs2 | 2 | A | C | C | G | C | C | C | A | ACCGCCCA | 26 |
| c29, c30 | 2 | C | C | T | G | C | C | C | A | CCTGCCCA | 28 |
| hf2, hf3, hf6, hf18, hf22, hf23, hu3, hu4, hu8, hu9, hu11, hu12, hu13, hu14, hu16 | 15 | T | C | C | G | C | T | C | G | TCCGCTCG | 29 |
| du79 | 3 | C | C | C | A | C | C | T | G | CCCACCTG | 31 |
| hf17, hf35, hu43 | 3 | T | C | C | G | C | C | C | G | TCCGCCCG | 32 |
| hf24, hs16, k9 | 3 | C | T | C | G | C | C | T | G | CTCGCCTG | 34 |
| hu10 | 1 | A | T | C | G | C | C | T | G | ATCGCCTG | 35 |
| du150 | 1 | C | T | C | A | C | T | C | G | CTCACTCG | 37 |
| hf4 | 1 | C | C | C | G | C | C | C | A | CCCGCCCA | 40 |
| hu34 | 1 | T | C | C | A | C | T | C | G | TCCACTCG | 45 |
| dg94 | 3 | A | C | T | G | C | C | C | G | ACTGCCCG | 49 |
| c31 | 1 | A | C | C | A | C | C | T | G | ACCACCTG | 51 |
| c68, k1 | 2 | A | C | C | A | C | C | C | G | ACCACCCG | 55 |
| hs1 | 1 | A | C | C | A | C | T | C | G | ACCACTCG | 56 |
| hs18 | 1 | A | C | C | G | T | C | T | A | ACCGTCTA | 58 |
| du77 | 1 | A | C | C | G | T | T | C | G | ACCGTTCG | 59 |
| k13 | 1 | A | C | T | G | C | C | T | A | ACTGCCTA | 61 |
| du84 | 1 | A | C | T | G | C | T | C | G | ACTGCTCG | 62 |
| hs14 | 1 | A | T | C | G | C | T | T | G | ATCGCTTG | 64 |
| k127 | 1 | A | T | T | A | C | C | C | A | ATTACCCA | 65 |
| du147 | 1 | A | T | T | A | C | T | C | G | ATTACTCG | 67 |
| c74, du148 | 2 | C | C | C | A | C | C | C | G | CCCACCCG | 69 |
| hu5, hu6, dg90, dg103 | 4 | C | C | C | G | C | T | C | G | CCCGCTCG | 70 |
| hf19 | 1 | C | C | C | G | C | T | T | G | CCCGCTTG | 71 |
| k14 | 1 | C | C | T | G | C | C | C | G | CCTGCCCG | 73 |
| hf5 | 1 | T | C | C | G | C | C | C | A | TCCGCCCA | 81 |
| hs17 | 1 | T | C | C | G | C | C | T | G | TCCGCCTG | 82 |
Isolates originated from feces of horses (hs), cattle (c), ducks (du), dogs (dg), kangaroos (k), and human feces (hf) and human urine (hu).
ID, identification.
Application of SNP analysis to E. coli populations.
The SNP profiles were further analyzed against the known sources of the isolates in the MLST database and SEQ isolates to determine the ability of this method to interrogate a population of E. coli against a selected variable, in this case the variable being the host source of the ST. The MLST database defined 31 STs as host specific (13.4% of 231 STs); 26 STs were of human origin, three STs were from cattle, one ST was from a pig, and one ST was from a rabbit. By comparison, SNP analysis of the MLST E. coli isolates resulted in 20 host-specific SNP profiles that can be defined as follows: 14 SNP profiles were unique for humans and the remaining six SNP profiles were unique for animals (three for cows, two for pigs, and one for a rabbit). The remaining SNP profiles had mixed host sources. It was decided that any profile containing five or fewer isolates would not be included as “host specific” in further analysis. Obviously the future inclusion of further isolates matching these underrepresented profiles will strengthen the case for their inclusion as host-specific SNP profiles.
Of the SEQ isolates' 53 SNP profiles, 28 were found to be host specific. Of these 28, four were unique to humans, and the remaining 24 were of animal origin (Table 2). As for the MLST SNP profiles, many of these profiles had fewer than five isolates and so were considered to be underrepresented for the sake of assigning host specificity.
When the SEQ E. coli profiles were combined with the MLST strain profiles, 74 SNP profiles with a D value of 0.92 were obtained (Fig. 1). A total of 19 of these SNP profiles were found to remain host specific (25%), of which 10 were unique to humans (D = 0.79) and 9 to animals. However, when profiles with less than five isolates were removed, there remained eight host-specific SNP profiles. Of these eight, seven (profiles 45, 29, 76, 32, 11, 47, and 16) were unique to humans (D = 0.79) and one SNP profile (profile 7) was unique to animals.
Blind test results.
Of all of the blind tests performed on hidden host source isolates, the most successful were the multiple isolates taken from sample 6, a human fecal sample. The tests were all grouped as SNP profile 29, which has been noted as a uniquely human profile. Other isolates were grouped with mixed host source profiles, but most were aligned with the dominant host in the mixed profile. This was true of isolates from sample 1 (host source cat and SNP profile 21, which is 60% animal), sample 2 (host source human and SNP profile 34, which is 67% human), and sample 7 (host source human and SNP profiles 70, which is 94% human, and 21, which is 60% animal). Less successful were sample 3 (animal source and SNP profile 23, which is 67% human) and sample 4 (animal source and SNP profile 80, which is uniquely human, and an unnumbered profile, which is also found in unknown environmental samples). As profile 80 is presently represented by only two human-sourced isolates, the addition of an animal-sourced isolate to this profile supports the decision not to assign host sources to profiles represented by a low number of isolates.
DISCUSSION
Methods for molecular typing are an area of rapid innovation, due primarily to expanding understanding of comparative genomics and the advances in available methodologies for nucleic acid analysis. Zhang et al. (33) studied the evolution of E. coli O157:H7 and identified 906 different SNPs in 523 chromosomal genes. The interrogation of such a large number of SNPs is an immense task and not amenable to routine genotyping studies. We are engaged in the development of rapid and robust genotyping methods that ideally would involve the interrogation of only a small number of polymorphic loci and therefore be suitable for high-throughput routine use. The purpose of the study reported here was to determine the following. (i) What subset of polymorphisms needs to be interrogated to obtain a particular measurement of resolution? Previously, we have reported a bioinformatics approach for the identification of groups of SNPs with a high discriminatory power (25). The Minimum SNPs software program is designed to take an entire MLST database as input and provide as output groups of SNPs with a high discriminatory power (high-D-value SNPs). Seven housekeeping genes from the E. coli MLST database were selected for SNP analysis using Minimum SNPs software to characterize the E. coli population. The numbers of housekeeping genes were found to be similar in different organisms for clonal complex studies, as these genes exhibit low levels of horizontal gene transfer. We found eight high-D-value SNPs to be sufficient at discriminating E. coli STs as depicted by the eBURST algorithm. The identification of these eight high-D-value SNPs defines groups of isolates that are consistent with the E. coli population biology.
We used allele-specific real-time PCR to make the genotyping easier and quicker and to efficiently track E. coli in environmental samples. During our analysis we observed a high level of sequence diversity surrounding the target SNPs, which has also been the case for Campylobacter jejuni (23). Therefore, the exclude/include function of the Minimum SNPs software was used to find optimal primer binding sites. Also, it should be mentioned that three of eight high-D-value SNPs were identified in the clpX gene, possibly due to the sequence diversity of this gene being lower that those of the other MLST genes.
Previously, Hommais et al. (13) used 13 SNPs in 11 genes to characterize E. coli isolates and were able to define five phylogroups (A, B1, E, D, and B2) (13). In contrast, our method uses eight high-D-value SNPs to define 74 E. coli SNP profile groups that are concordant with the highly diverse E. coli population biology.
The second question we attempted to answer was the following. (ii) Do these polymorphisms provide a genetic fingerprint consistent with the population structure of E. coli, with respect to host source? The construction of phylogenetic trees from MLST databases may not provide meaningful results because of the effects of recombination between lineages. An alternative approach to revealing some aspects of the evolutionary history of a species is to use the eBURST program, which reveals groups of closely related STs and identifies the putative founder clones of these groups. An eBURST analysis was used in this study to determine the correlation between the SNP profiles and the population structure of E. coli as defined by MLST. It has been shown previously that a high-D-value SNP set provided good correlation with a highly diverse population structure defined by MLST for Staphylococcus aureus and Campylobacter jejuni (23, 27). Calculated D values for the E. coli population for MLST and SNP genotyping were 0.949 and 0.946, respectively, therefore demonstrating the equivalent discriminatory powers of interrogating only eight SNPs versus sequencing seven genes.
A total of 74 SNP profiles were found in the eBURST analysis of the combined MLST database and SEQ isolate populations (Fig. 1). A number of SNP profiles were found to be highly consistent with the host-derived population of E. coli, both those found in the MLST data and also those of the SEQ isolates. It was decided that only profiles with more than five members would be considered “significant” in terms of identifying host sources. This analysis demonstrated three broad types of results: those demonstrating 100% host specificity, those with various host sources, and those with fewer than 5 members and so considered to have too few to be assigned a unique host source at this time. Eight SNP profiles were specific to the host source and had more than 5 members (Table 3 ). SNP profile numbers 11, 29, 32, and 45 were found both in SEQ human-derived populations of E. coli and in MLST human-derived E. coli populations. These SNP profiles were furthermore considered to be consistent with a widespread human host source, since STs retrieved from the MLST database originated from different countries. For example, SNP profile 45 includes ST17, which was found in the United States and Brazil, ST18 from the United States, Portugal, Mexico, Indonesia, France, Germany, and the Congo, ST30 from Bangladesh, ST19 and ST31 from the Congo and the United States, and ST233 and ST236 from India. SNP profile 29 also included widely distributed STs, such as ST14, which is from Brazil and the United States, ST20 from Peru, ST2 and ST34 from Nigeria, ST22 from Bangladesh, and ST29 from the United States. This is a significant finding, as the results for host sources in the past, particularly those based on library-dependent methods, had a major flaw in that they were only locally applicable (24). The SNP genotyping method we have used has the capability to determine certain host-specific genotypes within the E. coli population. Such genotypes may be internationally used to detect human-sourced E. coli in environmental samples.
TABLE 3.
Combined MLST and SEQ SNP profiles in relation to human versus nonhuman clusters
| SNP profile | SNP profile ID no. (assigned)a | No. human-sourced isolates |
% Human-sourced isolates in cluster | No. animal/environment-sourced isolates |
% Animal/environment-sourced isolates in cluster | Total no. of isolates | ||
|---|---|---|---|---|---|---|---|---|
| MLST | SEQ | MLST | SEQ | |||||
| Profiles with 100% human or animal source and more than five members | ||||||||
| TCCACTCG | 45 | 39 | 1 | 100 | 0 | 40 | ||
| TCCGCTCG | 29 | 9 | 15 | 100 | 0 | 24 | ||
| CTTACCTG | 76 | 22 | 100 | 0 | 22 | |||
| TCCGCCCG | 32 | 10 | 3 | 100 | 0 | 13 | ||
| TCTGCCCG | 11 | 11 | 4 | 100 | 0 | 15 | ||
| ATCACTTG | 47 | 9 | 100 | 0 | 9 | |||
| ACCACTTG | 16 | 8 | 100 | 0 | 8 | |||
| ACCGCCCG | 7 | 0 | 6 | 100 | 6 | |||
| Profiles with mixed human and animal sources and more than five members | ||||||||
| ACCACCTG | 51 | 54 | 95 | 2 | 1 | 5 | 57 | |
| TCCATTCG | 78 | 31 | 94 | 2 | 6 | 33 | ||
| CCCGCTCG | 70 | 29 | 2 | 94 | 2 | 6 | 33 | |
| CTCACTCG | 37 | 146 | 94 | 9 | 1 | 6 | 156 | |
| ATTACCCG | 30 | 47 | 92 | 4 | 8 | 51 | ||
| ATCGCCTG | 35 | 9 | 1 | 91 | 1 | 9 | 11 | |
| ACTGCCTA | 61 | 6 | 86 | 1 | 14 | 7 | ||
| ACCACCTA | 50 | 42 | 84 | 7 | 1 | 16 | 50 | |
| ACTACCCA | 60 | 5 | 83 | 1 | 17 | 6 | ||
| ATCACCTG | 12 | 11 | 1 | 80 | 3 | 20 | 15 | |
| ATCACTCG | 39 | 10 | 77 | 3 | 23 | 13 | ||
| ACCGCCTA | 23 | 4 | 67 | 2 | 33 | 6 | ||
| CTCGCCTG | 34 | 3 | 1 | 67 | 2 | 33 | 6 | |
| CCCACCTG | 31 | 24 | 62 | 12 | 3 | 37 | 39 | |
| CTCACCTG | 20 | 16 | 59 | 4 | 7 | 41 | 27 | |
| CCTGCCTA | 28 | 3 | 50 | 1 | 2 | 50 | 6 | |
| ATTACCTG | 19 | 2 | 1 | 50 | 3 | 50 | 6 | |
| ACCACCCA | 54 | 5 | 50 | 5 | 50 | 10 | ||
| ATCACCCG | 18 | 11 | 48 | 7 | 5 | 52 | 23 | |
| CCCGCCTG | 21 | 4 | 40 | 1 | 5 | 60 | 10 | |
| ACTGCCCG | 49 | 3 | 38 | 2 | 3 | 63 | 8 | |
| ATCACCCG | 43 | 1 | 14 | 6 | 86 | 7 | ||
| Profiles with 5 or less members | ||||||||
| CCCGCCCA | 40 | 1 | 100 | 0 | 1 | |||
| CTCGCCCG | 44 | 1 | 100 | 0 | 1 | |||
| ACTACTTG | 48 | 1 | 100 | 0 | 1 | |||
| ATCGCTCG | 52 | 1 | 100 | 0 | 1 | |||
| CCCGCCTA | 53 | 1 | 100 | 0 | 1 | |||
| TCCATCCG | 77 | 1 | 100 | 0 | 1 | |||
| TCCGCCCA | 81 | 1 | 100 | 0 | 1 | |||
| CTTACCTA | 75 | 2 | 100 | 0 | 2 | |||
| CCCGCTTG | 71 | 1 | 1 | 100 | 0 | 2 | ||
| TCCGTTCG | 80 | 2 | 100 | 0 | 2 | |||
| CTTGCCTA | 2 | 0 | 1 | 100 | 1 | |||
| ACCGCTCG | 8 | 0 | 1 | 100 | 1 | |||
| CTTGCCTG | 13 | 0 | 1 | 100 | 1 | |||
| ATCGCCTA | 14 | 0 | 1 | 100 | 1 | |||
| CTTGCCCA | 17 | 0 | 1 | 100 | 1 | |||
| CCCACTCG | 25 | 0 | 1 | 100 | 1 | |||
| CTCACCCG | 38 | 0 | 1 | 100 | 1 | |||
| CTCACTTG | 41 | 0 | 1 | 100 | 1 | |||
| ACCGTCTA | 58 | 0 | 1 | 100 | 1 | |||
| ACCGTTCG | 59 | 0 | 1 | 100 | 1 | |||
| ACTGCTCG | 62 | 0 | 1 | 100 | 1 | |||
| ATCGCTTG | 64 | 0 | 1 | 100 | 1 | |||
| ATTACTCG | 67 | 0 | 1 | 100 | 1 | |||
| TCCGCCTG | 82 | 0 | 1 | 100 | 1 | |||
| CCCACTTG | 4 | 0 | 2 | 100 | 2 | |||
| CTTACCCA | 1 | 0 | 2 | 1 | 100 | 3 | ||
| CCTACCCG | 5 | 0 | 2 | 100 | 2 | |||
| ATTGCCTA | 6 | 0 | 2 | 100 | 2 | |||
| ATTACCCA | 65 | 0 | 1 | 1 | 100 | 2 | ||
| ATTACCTA | 66 | 0 | 2 | 100 | 2 | |||
| CCTGCCCG | 73 | 0 | 2 | 1 | 100 | 3 | ||
| CCCGCCCG | 3 | 0 | 4 | 100 | 4 | |||
| TCCGTCCG | 79 | 1 | 50 | 1 | 50 | 2 | ||
| ATTGCCTG | 15 | 1 | 50 | 1 | 50 | 2 | ||
| ACCGCCCA | 26 | 1 | 50 | 1 | 50 | 2 | ||
| ACCACCG | 55 | 1 | 50 | 1 | 50 | 2 | ||
| ACCGCCCA | 56 | 1 | 50 | 1 | 50 | 2 | ||
| TCTACCCG | 9 | 1 | 33 | 2 | 67 | 3 | ||
| CCCACCCG | 69 | 1 | 33 | 2 | 67 | 3 | ||
| CCTACCCA | 72 | 1 | 33 | 2 | 67 | 3 | ||
| CCCACCCA | 68 | 2 | 67 | 1 | 33 | 3 | ||
| CCTGCCTA | 24 | 1 | 25 | 1 | 2 | 75 | 4 | |
| ACCGCCTG | 22 | 1 | 25 | 3 | 75 | 4 | ||
| CCCACCTA | 33 | 1 | 20 | 4 | 80 | 5 | ||
ID, identification.
Additional uniquely human MLST E. coli SNP profiles with more than five isolates were SNP profiles 76, 47, and 16; however, none of these SNP profiles were found in SEQ, only in the MLST database. Also in Table 3, it is shown that SNP profile 7 was wholly represented by SEQ animal E. coli populations. Twenty-two SNP profiles with more than five members had two or more sources of E. coli represented. However, some profiles were observed to be predominantly human or animal sourced. For example, if an E. coli isolate had SNP profile 70, we observed that 94% of the isolates exhibiting this profile were from humans, and so there is a strong likelihood that this isolate originated from humans. Similarly, SNP profile 43 can be considered predominantly animal specific, with 86% of E. coli isolates with this profile being sourced from animals.
Some of the mixed profiles (e.g., profiles 28, 19, and 54) derive equally, or nearly so, from human and animal sources, and so SNP profiles did not successfully distinguish source identity for isolates with these profiles. There are no reports to indicate that some E. coli strains appear to be shared between animals and humans. It is possible that microbiota exchange may occur from time to time or that both hosts are consuming contaminated water from the same source, for example. Thus, dogs (or other household pets) may acquire human E. coli strains in situations of physical contact. These strains may then be transferred to other animals, such as cattle or horses, and then possibly transferred to wild animals, such as kangaroos, by means of contaminated grass as a food source or contaminated water sources. Such strains may show a lack of host specificity, regardless of the analytical tool used.
Twenty-one SNP genotypes were represented by only one isolate, and a further 23 SNP genotypes had between two and five isolates (Table 3). Due to the small number of isolates demonstrating each SNP profile, we may not conclusively designate source identity to these profiles. STs and SNP profiles containing only a single isolate were found from known host sources, and there does not appear to be any clear trend toward host or locality exhibited by the current SNP data for these isolates. The animal hosts for this group of isolates were diverse and included 25 cattle, 8 dogs, 2 kangaroos, and others. As noted, human-sourced single isolate STs and SNP profiles were from SEQ and the MLST database, and so these SNP profiles were not specifically local or international. Eight of the SNP profiles with between two and five isolates (profiles 4, 1, 5, 6, 65, 66, 73, and 3) were sourced 100% from animals, and three SNP profiles (profiles 75, 71 and 80) were sourced from humans. Further inclusion of isolates from known host sources with these SNP profiles would improve the significance of assigning these SNP profiles as host specific.
Blind testing demonstrated that while some E. coli sources could be correctly identified using the SNP typing method, only one source resulted in a unique host profile. Other isolates were found to be aligned with the mixed host source profiles, mostly with the majority host in the profile, but sometimes with the minority host. These results are sufficiently encouraging to warrant further investigation of the SNP analysis of E. coli populations, particularly with respect to identification of host sources.
In summary, there was good consistency between the SNP profiles found in this study and the eBURST-defined clonal complexes, with the major clonal complexes having distinct SNP profiles. Thus, highly discriminatory SNP interrogation using real-time PCR can be used as a preliminary method for the identification of new STs without sequencing. Here we report several human-specific SNP genotypes from human E. coli isolates. The majority of our isolated strains had SNP profiles 29 and 11. These SNP profiles are priority candidates to be detected in water in terms of potential human health risk. The rest of the human-specific SNP profiles determined from the MLST database were not frequently identified in any of the Australian environmental samples (unpublished data). However, five identified human-specific SNP profiles, profiles 11, 29, 32, 45, and 71, can be used for microbial source tracking worldwide. The numbers of isolates that have these SNP profiles are significant, and even if the human SNP groups gain additional isolates, they will remain human specific.
Conclusion.
The conventional method of testing the performance of a new molecular typing method is to devise a procedure that is likely to be discriminatory and then test the discrimination on actual isolates. This process can be streamlined if the typing methodology is based upon polymorphisms derived from a comparative sequence database. In this case, not only can the performance of the typing method be tested in silico using the database as a surrogate for the population structure, but also the method can be tested in silico against actual groups of isolates that have been subjected to sequence-based genotyping. We concluded that identifying the nucleotides at just eight positions provides a substantial fraction of the resolving power that is obtainable from identifying the nucleotides at >3,000 positions (full MLST determination). Identification of SNP sets on the basis of maximization of D yields sets that define groups of isolates consistent with the E. coli population.
The E. coli population is represented by unique, and also mixed, host-sourced SNP genotype clusters. Some types are widely distributed and can be found in different hosts. On the other hand, there are types that are in host-specific clusters. In particular, we found that 20% of all the E. coli isolates studied here (MLST and SEQ) belonged to SNP profile 37 and that about 5% of all E. coli isolates belonged to the human-specific profile 45. Since E. coli from animals is underrepresented in the MLST database, we observed host specificity mainly for human-originating E. coli. Human-specific SNP profiles were identified as unique profiles and may be used for microbial source tracking. The human-specific SNP profiles 11, 29, 32, and 45 are internationally distributed and may be useful as a global indicator of human fecal contamination in water. Our SNP typing method can be used for E. coli population investigations to which the MLST method is not applicable in terms of time and cost. Animal-specific SNP profiles were described in this study; however, due to low numbers of these isolates in the MLST database, further confirmation of the host specificity of these SNP profiles is required.
Supplementary Material
Acknowledgments
We thank W. Ahmed and N. George for providing the isolates. M. Pemberton provided assistance with statistical analyses. We are also grateful to P. Pinniger for valuable assistance with linguistic corrections during the writing of the manuscript.
M.S.S. is in receipt of a postgraduate studentship from the Institute of Sustainable Resources, Queensland University of Technology.
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahmed, W., A. Goonetilleke, D. Powell, K. Chauhan, and T. Gardner. 2009. Comparison of molecular markers to detect fresh sewage in environmental waters. Water Res. 43:4908-4917. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, W., A. Goonetilleke, D. Powell, and T. Gardner. 2009. Evaluation of multiple sewage-associated Bacteroides PCR markers for sewage pollution tracking. Water Res. 43:4872-4877. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed, W., R. Neller, and M. Katouli. 2005. Evidence of septic system failure determined by a bacterial biochemical fingerprinting method. J. Appl. Microbiol. 98:910-920. [DOI] [PubMed] [Google Scholar]
- 4.Alcaine, S. D., Y. Soyer, L. D. Warnick, W. L. Su, S. Sukhnanand, J. Richards, E. D. Fortes, P. McDonough, T. P. Root, N. B. Dumas, Y. Grohn, and M. Wiedmann. 2006. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl. Environ. Microbiol. 72:7575-7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc, D. S. 2004. The use of molecular typing for epidemiological surveillance and investigation of endemic nosocomial infections. Infect. Genet. Evol. 4:193-197. [DOI] [PubMed] [Google Scholar]
- 7.D'Elia, T. V., C. R. Cooper, and C. G. Johnston. 2007. Source tracking of Escherichia coli by 16S-23S intergenic spacer region denaturing gradient gel electrophoresis (DGGE) of the rrnB ribosomal operon. Can. J. Microbiol. 53:1174-1184. [DOI] [PubMed] [Google Scholar]
- 8.Eaton, A., L. S. Clesceri, E. W. Rice, and A. E. Greenberg (ed.). 2005. Standard methods for the examination of water and wastewater, centennial ed. American Public Health Association, Washington, DC.
- 9.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field, K. G., and M. Samadpour. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res. 41:3517-3538. [DOI] [PubMed] [Google Scholar]
- 11.Fong, T. T., and E. K. Lipp. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 69:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havelaar, A. H., K. Furuse, and W. M. Hogeboom. 1986. Bacteriophages and indicator bacteria in human and animal faeces. J. Applied Bacteriol. 60:255-262. [DOI] [PubMed] [Google Scholar]
- 13.Hommais, F., S. Pereira, C. Acquaviva, P. Escobar-Paramo, and E. Denamur. 2005. Single-nucleotide polymorphism phylotyping of Escherichia coli. Appl. Environ. Microbiol. 71:4784-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kildare, B. J., C. M. Leutenegger, B. S. McSwain, D. G. Bambic, V. B. Rajal, and S. Wuertz. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701-3715. [DOI] [PubMed] [Google Scholar]
- 16.Kon, T., S. C. Weir, E. T. Howell, H. Lee, and J. T. Trevors. 2009. Repetitive element (REP)-polymerase chain reaction (PCR) analysis of Escherichia coli isolates from recreational waters of southeastern Lake Huron. Can. J. Microbiol. 55:269-276. [DOI] [PubMed] [Google Scholar]
- 17.McLain, J. E. T., H. Ryu, L. Kabiri-Badr, C. M. Rock, and M. Abbaszadegan. 2009. Lack of specificity for PCR assays targeting human Bacteroides 16S rRNA gene: cross-amplification with fish feces. FEMS Microbiol. Lett. 299:38-43. [DOI] [PubMed] [Google Scholar]
- 18.McLellan, S. L. 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McQuaig, S. M., T. M. Scott, V. J. Harwood, S. R. Farrah, and J. O. Lukasik. 2006. Detection of human-derived fecal pollution in environmental waters by use of a PCR-based human polyomavirus assay. Appl. Environ. Microbiol. 72:7567-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogorzaly, L., A. Tissier, I. Bertrand, A. Maul, and C. Gantzer. 2009. Relationship between F-specific RNA phage genogroups, faecal pollution indicators and human adenoviruses in river water. Water Res. 43:1257-1264. [DOI] [PubMed] [Google Scholar]
- 22.Price, E. P., F. Huygens, and P. M. Giffard. 2006. Fingerprinting of Campylobacter jejuni by using resolution-optimized binary gene targets derived from comparative genome hybridization studies. Appl. Environ. Microbiol. 72:7793-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price, E. P., V. Thiruvenkataswamy, L. Mickan, L. Unicomb, R. E. Rios, F. Huygens, and P. M. Giffard. 2006. Genotyping of Campylobacter jejuni using seven single-nucleotide polymorphisms in combination with flaA short variable region sequencing. J. Med. Microbiol. 55:1061-1070. [DOI] [PubMed] [Google Scholar]
- 24.Ram, J. L., R. P. Ritchie, J. Fang, F. S. Gonzales, and J. P. Selegean. 2004. Sequence-based source tracking of Escherichia coli based on genetic diversity of beta-glucuronidase. J. Environ. Qual. 33:1024-1032. [DOI] [PubMed] [Google Scholar]
- 25.Robertson, G., F. Huygens, and G. Giffard. 2004. Identification and interrogation of highly informative single nucleotide polymorphism sets defined by bacterial multilocus sequence typing databases. J. Med. Microbiol. 53:35-45. [DOI] [PubMed] [Google Scholar]
- 26.Seurinck, S., W. Verstraete, and S. Siciliano. 2005. Microbial source tracking for identification of fecal pollution. Rev. Environ. Sci. Biotechnol. 4:19-37. [Google Scholar]
- 27.Stephens, A. J., F. Huygens, J. Inman-Bamber, E. P. Price, G. R. Nimmo, J. Schooneveldt, W. Munckhof, and P. M. Giffard. 2006. Methicillin-resistant Staphylococcus aureus genotyping using a small set of polymorphisms. J. Med. Microbiol. 55:43-51. [DOI] [PubMed] [Google Scholar]
- 28.Stoeckel, D., and V. Harwood. 2007. Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 73:2405-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoeckel, D. M., M. V. Mathes, K. E. Hyer, C. Hagedorn, H. Kator, J. Lukasik, T. L. O'Brien, T. W. Fenger, M. Samadpour, K. M. Strickler, and B. A. Wiggins. 2004. Comparison of seven protocols to identify fecal contamination sources using Escherichia coli. Environ. Sci. Technol. 38:6109-6117. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tutenel, A. V., D. Pierard, J. Uradzinski, E. Jozwik, M. Pastuszczak, J. V. Hende, M. Uyttendaele, J. Debevere, T. Cheasty, J. V. Hoof, and L. D. Zutter. 2002. Isolation and characterization of enterohaemorrhagic Escherichia coli O157:1H7 from cattle in Belgium and Poland. Epidemiol. Infect. 129:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan, T., and M. J. Sadowsky. 2007. Determining sources of fecal bacteria in waterways. Environ. Monit. Assess. 129:97-106. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, W., W. Qi, T. J. Albert, A. S. Motiwala, D. Alland, E. K. Hyytia-Trees, E. M. Ribot, P. I. Fields, T. S. Whittam, and B. Swaminathan. 2006. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 16:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.