Abstract
Gluconobacter oxydans N44-1, an obligatory aerobic acetic acid bacterium, oxidizes glucose primarily in the periplasm to the end products 2-ketogluconate and 2,5-diketogluconate, with intermediate formation of gluconate. Only a minor part of the glucose (less than 10%) is metabolized in the cytoplasm after conversion to gluconate or after phosphorylation to glucose-6-phosphate via the only functional catabolic routes, the pentose phosphate pathway and the Entner-Doudoroff pathway. This unusual method of glucose metabolism results in a low growth yield. In order to improve it, we constructed mutants of strain N44-1 in which the gene encoding the membrane-bound glucose dehydrogenase was inactivated either alone or together with the gene encoding the cytoplasmic glucose dehydrogenase. The growth and product formation from glucose of the resulting strains, N44-1 mgdH::kan and N44-1 ΔmgdH sgdH::kan, were analyzed. Both mutant strains completely consumed the glucose but produced neither gluconate nor the secondary products 2-ketogluconate and 2,5-diketogluconate. Instead, carbon dioxide formation of the mutants increased by a factor of 4 (N44-1 mgdH::kan) or 5.5 (N44-1 ΔmgdH sgdH::kan), and significant amounts of acetate were produced, presumably by the activities of pyruvate decarboxylase and acetaldehyde dehydrogenase. Most importantly, the growth yields of the two mutants increased by 110% (N44-1 mgdH::kan) and 271% (N44-1 ΔmgdH sgdH::kan). In addition, the growth rates improved by 39% (N44-1 mgdH::kan) and 78% (N44-1 ΔmgdH sgdH::kan), respectively, compared to the parental strain. These results show that the conversion of glucose to gluconate and ketogluconates has a strong negative impact on the growth of G. oxydans.
As the Gram-negative acetic acid bacterium Gluconobacter oxydans is able to oxidize sugars and sugar alcohols regioselectively, it is a valuable and versatile biocatalyst and has been used in industry for a long time, e.g., for the production of vitamin C via Reichstein synthesis (32) and of 1-deoxynojirimycin, a precursor of the antidiabetic drug miglitol (35). Both processes are combined biotechnological-chemical syntheses carried out on a large scale (10, 35). The key biotechnological reactions in these two examples are the regioselective oxidation of d-sorbitol to l-sorbose and N-formyl-1-amino-1-deoxy-d-sorbitol to N-formyl-6-amino-6-deoxy-l-sorbose, respectively. The latter conversion is performed in whole-cell biotransformations with resting cells of G. oxydans as a catalyst.
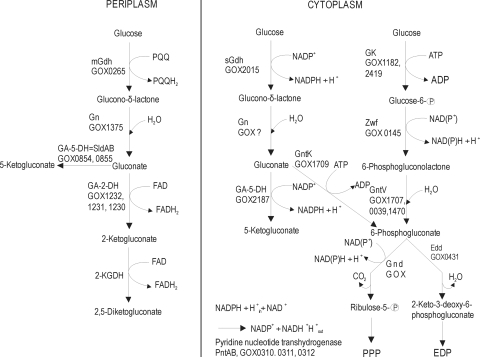
A characteristic trait of G. oxydans is the presence of parallel but spatially separated pathways for the oxidation of nonphosphorylated substrates and intermediates in both the periplasmic and cytoplasmic compartments (Fig. 1). G. oxydans is known for its incomplete oxidation of a wide range of carbohydrates and alcohols. The corresponding products (aldehydes, ketones, and organic acids) are secreted almost completely into the medium. The natural habitats of G. oxydans are sugary environments, such as flowers and fruits (13). The organism is able to grow in highly concentrated sugar solutions and at low pH values (31). The only two functional central metabolic pathways are the pentose phosphate pathway (PPP) and the Entner-Doudoroff pathway (EDP). The Embden-Meyerhof-Parnas pathway is inactive due to the lack of phosphofructokinase. A functional citric acid cycle is also absent due to the absence of succinate dehydrogenase (31).
FIG. 1.
Scheme of glucose oxidation of G. oxydans. Gn, glucono-δ-lactonases; Ga-2-DH, membrane-bound gluconate 2-dehydrogenase; Ga-5-DH, membrane-bound gluconate 5-dehydrogenase (= SldAB); 2-KGDH, membrane-bound 2-ketogluconate dehydrogenase; GK, glucose kinase; Gnt K, gluconokinase; Zwf, glucose-6-phosphate dehydrogenase; GntV, 6-phophoglucono-δ-lactonase; Gnd, 6-phosphogluconate dehydrogenase; Edd, 6-phosphogluconate dehydratase. The GOX numbers refer to the analogous genes/enzymes in G. oxydans 621H. For the 2-KGDH-encoding gene in G. oxydans N44-1, no analog exists in G. oxydans 621H.
When the organism is growing on glucose, the major part of this sugar is oxidized in the periplasm. A membrane-bound pyrroloquinoline quinone (PQQ)-dependent glucose dehydrogenase (mGDH) catalyzes the initial reaction linked to the generation of a proton motive force (25). The direct oxidation product, glucono-δ-lactone, is converted to gluconate, either spontaneously or enzymatically catalyzed by glucono-δ-lactonase (39). Further oxidation of gluconate by membrane-bound and respiratory-chain-coupled quinoprotein or flavoprotein dehydrogenases leads to the formation of 5-ketogluconate (5-KGA), 2-ketogluconate (2-KGA), and 2,5-diketogluconate (2,5-DKGA) (25). Another part of the glucose is oxidized in a similar way in the cytoplasm by the NADP+-dependent dehydrogenases soluble glucose dehydrogenase (sGDH) and gluconate-5-dehydrogenase (18, 26, 30).
A minor part of the glucose is assimilated into cell material. This involves an initial phosphorylation of glucose or gluconate to glucose-6-phosphate or 6-phosphogluconate, respectively, and subsequent metabolism of these intermediates via the PPP or EDP (29, 31, 40). The fact that the majority of the carbon source is incompletely oxidized to ketoacids as products rather than being metabolized via the PPP and EDP results in very low biomass yields, in the range of 0.10 g cell dry weight (CDW) per g of glucose consumed (29).
The low biomass yields limit the industrial exploitation of the many interesting metabolic capabilities of G. oxydans, for instance, in processes employing biotransformation with resting cells. In this work, we applied metabolic engineering in order to generate strains with an improved growth yield on glucose. For this purpose, the genes encoding the membrane-bound glucose dehydrogenase and the soluble glucose dehydrogenase were inactivated. In fact, the growth rates and biomass yields of the resulting mutant strains were significantly improved.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The Escherichia coli strains were cultivated in Luria-Bertani (LB) medium or on LB agar plates at 37°C (33). When required, kanamycin (50 μg ml−1), ampicillin (100 μg ml−1), or gentamicin (10 μg ml−1) was added to the final concentration indicated. G. oxydans N44-1 (39) and G. oxydans N44-1 mgdH::kan were obtained from DSM Nutritional Products (Basel, Switzerland) and routinely cultivated on medium no. 5 (yeast extract [YE]) having the following composition per liter: 50 g glucose, 15 g yeast extract, 2.5 g MgSO4·7 H2O, 0.5 g glycerol, 15 g CaCO3. G. oxydans possesses a natural resistance to cefoxitin; as a precaution to prevent bacterial contamination, cefoxitin was added to the media at a concentration of 50 μg ml−1. For preparation of electroporation-competent cells of G. oxydans, cells were cultivated in EP medium containing the following components per liter: 80 g mannitol, 15 g yeast extract, 2.5 g MgSO4·7 H2O, 0.5 g glycerol, 1.5 g CaCl2. The pH was adjusted to pH 6.0 with HCl. G. oxydans was grown at 30°C either in 50 ml medium no. 5 (YE) in 500-ml shaking flasks at 200 rpm agitation (the shaking-flask cultures were inoculated at an optical density at 600 nm [OD600] of 0.25) or in a bioreactor (Dasgip, Jülich, Germany) with pH and aeration control (CaCO3 was omitted from the medium; the bioreactor cultures were inoculated to an OD600 of 0.30). The working volume of the each of the four parallel Dasgip bioreactors was 200 ml medium; the stirrer velocity was set to 700 rpm and the temperature to 30°C. Fifty microliters of antifoam 204 (Sigma, Taufkirchen, Germany) was added to each culture. By supplying a controlled ratio of O2, CO2, and N2 at a flow rate of 12 standard liters/h, partial pressures of O2 and CO2 were maintained at constant values of 15% or 30% O2 and 0.05% CO2. The pH was kept constant at a value of 6 by titration with 4 M NaOH.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Properties | Reference(s)/source |
|---|---|---|
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 14 |
| HB101 | F−hsdS20 (rB− mB−) supE44 ara-14 galK-2 lacY1 proA2 rpsL20 xyl-5 mtl-1 recA13 Kanr oriColE1 RK2-Mob+ RK2-Tra+ mH-1 with plasmid pRK2013 | 4, 11, 22 |
| G. oxydans | ||
| N44-1 | Cefr; derived from G. oxydans IFO 3293 | 42 |
| N44-1 mgdH::kan | Cefr Kanr; chromosomal inactivation of mgdH | DSM Nutritional Products, Kaiseraugst, Switzerland |
| N44-1 ΔmgdH | Cefr; chromosomal deletion of mgdH | This study |
| N44-1 ΔmgdH sgdH::kan | Cefr Kanr; chromosomal inactivation of mgdH and sgdH | This study |
| Plasmids | ||
| pBBR1MCS2 | KmrlacI lacZ Plac; pBBR1 replicon | 19 |
| pBBR1MCS4 | AprlacI lacZ Plac, pBBR1 replicon | 20 |
| pSUP202 | Apr Cmr Tcr; oriT from RP4; cannot replicate in G. oxydans | 41 |
| pSUP202 mgdH::kan | Apr Cmr Tcr Kmr; pSUP202 derivative containing an mgdH gene in which an internal part is replaced by a kanamycin resistance gene | DSM Nutritional Products, Kaiseraugst, Switzerland |
| pSUP202 sgdh::kan | Apr Cmr Tcr Kmr; pSUP202 derivative containing an sgdH gene in which an internal part is replaced by a kanamycin resistance gene | This study |
| pK19mobsacB | Kmr; vector for allelic exchange; cannot replicate in G. oxydans | 34 |
| pK19mobsacBΔmgdH | Kmr; pK19mobsacB derivative used for marker-free deletion of mgdH gene; contains an overlap extension PCR product covering the regions up- and downstream of mgdH | |
| Oligonucleotides | ||
| sgdh_GoxKpnIF | 5′-GGA GAG GTG AGG TAC CAT GCC TGC CCC-3′ | Amplification of sgdH; KpnI site; proof of sgdH disruption |
| sgdh_GoxSacIR | 5′-GGA TCA TCA GGA GCT CGT CTG TCC AGA CTG G-3′ | Amplification of sgdH; SacI site; proof of sgdH disruption |
| coPCR-gdh-5F | 5′-GCA CGG CCG GGC ATG CCA CTT GTG GC-3′ | Disruption of mgdH; SphI site |
| coPCR-gdh-5R | 5′-CCC ATC CAC TAA ACT TAA ACA CGT GCC GCC GAT CTG GCG GGA GAC G-3′ | Disruption of mgdH; inner primer |
| coPCR-gdh-3F | 5′-TGT TTA AGT TTA GTG GAT GGG CTC TCC ACG GCT GGC AAC CTC GGC TTC C-3′ | Disruption of mgdH; inner primer |
| coPCR-gdh-3R | 5′-CCG GTT GTG AGG CGGTCG ACT GGC AGT CGG TGG-3′ | Disruption of mgdH; SalI site |
| gdh-control-5 (for) | 5′-CGG GCA GGA ATG TTG AGC TGT AGG-3′ | Proof of mgdH disruption |
| gdh-control-3 (rev) | 5′-CCC TCG CCG TAA ACG ATT GGT CC-3′ | Proof of mgdH disruption |
Cloning and DNA techniques.
For DNA manipulation, standard methods were used, as described by Sambrook and Russell (33). Gene amplification was done by standard PCR. Restriction enzymes and DNA-modifying enzymes were purchased from Roche Diagnostics (Mannheim, Germany), and oligonucleotides were obtained from Operon (Cologne, Germany). Competent cells of E. coli were prepared with RbCl according to the method of Cohen et al. (7) and transformed as described by Hanahan (14). Transformation of G. oxydans was carried out using a modified electroporation protocol according to the method of Choi et al. (6). For this purpose, cells were pregrown in EP medium at 30°C for 15 h. The preculture was then used for inoculation of 50 ml EP medium in a 500-ml shake flask. Cultivation was carried out at 30°C and 200 rpm on a rotary shaker until an OD600 of 0.4 was reached. The cells were harvested by centrifugation at room temperature for 1 to 2 min at 16,000 × g. The cell pellets were washed twice with 1 ml of 300 mM sucrose at room temperature and then resuspended in 1 ml of the sucrose solution. For electroporation, 500 ng of nonreplicative plasmid DNA was mixed with 100 μl of electrocompetent cells and transferred into an electroporation cuvette with a 2-mm gap width. After a pulse (settings, 2.5 kV, 25 μF, 200 Ω on a Bio-Rad GenePulserXcell; Bio-Rad) was applied, 1 ml of EP medium (room temperature) was added and the cells were regenerated for 3 h at 30°C. They were then plated on selective agar. Colonies formed after 2 to 3 days of incubation at 30°C.
Construction of G. oxydans mutant strains.
A mutant of strain N44-1 with an in-frame deletion of the mgdH gene (GOX0265 in G. oxydans strain 621H) was constructed by the method described by Link et al. (23) using the vector pK19mobsacB (34). Briefly, the upstream and downstream regions of mgdH (26 bp and 33 bp) were amplified and fused by overlap extension PCR using the primers coPCR-gdh-5F, coPCR-gdh-5R, coPCR-gdh-3F, and coPCR-gdh-3R (Table 1). The resulting PCR product, in which the central part of mgdH (codons 114 to 729) was absent, was cloned into pK19mobsacB via SphI and SalI restriction sites. The resulting plasmid, pK19mobsacB-ΔmgdH, was transferred by electroporation into G. oxydans N44-1, and selection for the first and second recombination event was performed as described by Niebisch and Bott (28). Kanamycin-sensitive and sucrose-resistant clones with the desired deletion of mgdH were identified by PCR with chromosomal DNA and the primers gdh-control-3 and gdh-control-5. In order to additionally inactivate the sgdH gene (GOX2015 in G. oxydans strain 621), plasmid pSUP202 sgdH::kan was constructed. This plasmid contained an sgdH gene in which the central part (codons 45 to 139) was replaced by a kanamycin resistance gene from plasmid pDRIVE (Qiagen, Hilden, Germany). The plasmid was transferred into G. oxydans N44-1 ΔmgdH by electroporation, and kanamycin-resistant cells containing a disrupted sgdH gene were isolated and controlled by PCR with the primers sgdh-GoxKpnIF and sgdh-GoxSacIR.
Determination of cell dry weight.
Quantitative determination of cell dry weight was performed by membrane filtration. Cellulose nitrate filters with a pore diameter of 0.45 μm (Millipore, Schwalbach, Germany) were dried at 110°C for 24 h, subsequently cooled to room temperature in an desiccator, and weighed. For dry-weight determination, 10-ml aliquots were taken from a growing culture of G. oxydans and filtered. The filters were then washed with 200 ml cold distilled water. After drying (110°C; 24 h) and cooling to room temperature in an exsiccator, the filters were weighed again, and the net weight of the dried biomass was calculated. A correlation of cell dry weight and optical density was calculated from a calibration curve: cell dry weight (g liter−1) = 0.2345 × OD600.
Preparation of cell extract and enzyme assays.
Aliquots (25 ml) of an exponentially growing culture of G. oxydans were harvested by centrifugation (7 min at 2,000 × g and 25°C), followed by a washing step with 25 ml 50 mM potassium phosphate buffer, pH 7.0. Then, the cells were resuspended in 1.5 ml of the same buffer, and “complete” protease inhibitor (Roche, Mannheim, Germany) was added. The cells were disrupted by sonication with a UP200S sonifier (Hielscher, Teltow, Germany) (4°C; 5 min; 70% power; 50% pulse). Intact cells and cell debris were removed by centrifugation (15 min at 16,000 × g and 4°C). The supernatant was used as a cell extract.
For the activity measurement of pyruvate decarboxylase, an assay system modified by the method of Bringer-Meyer et al. (5) was used. Acetaldehyde dehydrogenase (ALDH) activity was assayed according to a modified method described by Bernt and Gutmann (3), following the formation of NADPH at 340 nm in 1 ml of 50 mM Tris-HCl buffer (pH 8.3) containing 10 mM dithiothreitol (DTT), 2 mM NADP+, and 22 mM acetaldehyde as a substrate. One unit of ALDH activity corresponds to 1 μmol NADH formed per minute at 30°C.
Quantification of substrate and products.
Glucose, gluconate, 2-KGA, and 2,5-DKGA were determined by high-performance liquid chromatography (HPLC) as reported by Herrmann et al. (15). The substances were separated with a Shodex DE 613 reversed-phase HPLC column (Phenomenex, Aschenburg, Germany) and 2 mM HClO4 as the eluant at a flow rate of 0.5 ml min−1. The acetate concentration was determined enzymatically using a kit from R-Biopharm (Darmstadt, Germany) and by gas chromatography (HP-6890A gas chromatograph; Agilent, Waldbronn, Germany) using a Carbowax Permabond 20 M column (50 m by 0.32 mm [inside diameter]; Macherey-Nagel, Düren, Germany).
RESULTS
Growth phenotype of G. oxydans N44-1 mgdH::kan in shake flasks.
When G. oxydans grows on glucose, a major part of the sugar is converted in the periplasm to different ketogluconates as products. Consequently, this carbon cannot be used for biomass formation, but only for the generation of proton motive force. In order to make a larger fraction of glucose available for biomass, we analyzed a mutant of strain N44-1 in which the mgdH gene encoding the membrane-bound glucose dehydrogenase was inactivated. This enzyme catalyzes the PQQ-dependent oxidation of glucose to gluconate, with the reaction taking place in the periplasm (25, 30) (Fig. 1). The parental and mutant strains were cultivated in shaking flasks with medium no. 5, which contained 50 g glucose liter−1. Like G. oxydans N44-1, the mgdH::kan strain showed reproducible growth without an initial lag phase but reached a final OD600 of 10.1 ± 0.3, significantly higher than that of its parent (OD600, 3.70 ± 0.2), and a higher maximal growth rate (0.16 h−1 versus 0.11 h−1). This result supported the assumption that the periplasmic oxidation of the substrate limits biomass formation from glucose.
Construction of G. oxydans N44-1 ΔmgdH sgdH::kan.
As described in the introduction, G. oxydans can oxidize unphosphorylated glucose, not only in the periplasm, but also in the cytoplasm. Thus, it was of interest to construct a strain in which both the membrane-bound and the cytoplasmic glucose dehydrogenases were absent. Since only a limited number of antibiotics are applicable as selection markers for G. oxydans, we first constructed a strain containing a marker-free deletion of the mgdH gene, as described in Materials and Methods. One of the clones with the desired deletion, designated N44-1 ΔmgdH, was subsequently used to inactivate the sgdH gene for the soluble glucose dehydrogenase by replacing an internal part of the gene with a kanamycin resistance gene using plasmid pSUP202 sgdH::kan. The resulting kanamycin-resistant double mutant was named G. oxydans N44-1 ΔmgdH sgdH::kan.
Growth phenotypes of strains N44-1 ΔmgdH and N44-1 ΔmgdH sgdH::kan in shake flasks.
In a first series of experiments, the growth of the newly constructed strains was analyzed in shake flasks with medium no. 5 containing 50 g glucose liter−1. The single mutant N44-1 ΔmgdH showed growth behavior similar to that of N44-1 mgdH::kan and reached a final OD600 of 10.7. The double mutant N44-1 ΔmgdH sgdH::kan reached an even higher OD600 of 13.0 (data not shown). Thus, the additional deletion of sgdH further improved biomass formation from glucose.
Growth and product formation of strain N44-1 and its glucose dehydrogenase mutants under controlled conditions in a bioreactor.
To gain more information about the metabolic consequences of the gdh gene inactivations, the parent strain, N44-1, and the mutant strains, N44-1 mgdH::kan and N44-1 ΔmgdH sgdH::kan, were cultivated in Dasgip bioreactors containing 200 ml medium without CaCO3 under pH and aeration control. Strain N44-1 mgdH::kan was used instead of N44-1 ΔmgdH to allow better comparison with strain N44-1 ΔmgdH sgdH::kan. All three strains contained the vector pBBR1MCS4, required in subsequent studies.
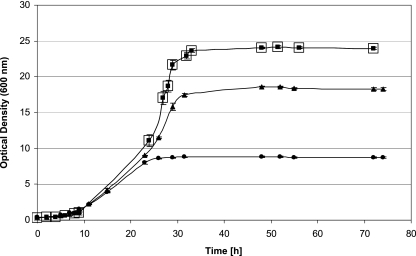
Presumably, due to the improved aeration in the bioreactor, the biomass yields of all three tested strains increased significantly compared to the shake flask cultivations. Final OD600 values of 9.0 (N44-1), 18.7 (N44-1 mgdH::kan), and 24.4 (ΔmgdH sgdH::kan) were reached (Fig. 2). As summarized in Table 2, the growth rates of the mutant strains mgdH::kan and ΔmgdH sgdH::kan were 39% and 78% higher than that of the parental strain, N44-1. Thus, elimination of glucose oxidation to gluconate improved both the growth rate and the biomass yield. There was no difference in growth or substrate consumption when cultures were grown with 15% (dissolved-oxygen [DO]) O2 or 30% (DO) O2 (Fig. 2). Hence, 15% (DO) O2 was sufficient for growth of G. oxydans under the conditions used, which is in accordance with the results of an investigation on the influence of increased oxygen partial pressures on gluconic acid formation and growth of G. oxydans (43).
FIG. 2.
Growth of G. oxydans strains with glucose (275 mM; 50 g liter−1). Cultivation was carried out in duplicate (points in the graph represent averages) in bioreactors (Dasgip, Juelich, Germany) under pH and aeration control (15% dissolved oxygen). •, G. oxydans N44-1 pBBR1MCS4; ▴, G. oxydans N44-1 mgdH::kan pBBR1MCS4; squares, G. oxydans N44-1 ΔmgdH sgdH::kan pBBR1MCS4, with 15% dissolved oxygen (▪) and with 30% dissolved oxygen (□).
TABLE 2.
Growth parameters of the G. oxydans N44-1 strains in fermentors with 15% dissolved oxygen and 50 g liter−1 glucosea
| G. oxydans strain | Final OD600 | CDW (g liter−1) | μb (h−1) | tdc (h) | Yx/s (g CDW g glucose−1)d |
|---|---|---|---|---|---|
| N44-1 pBBR1MCS4 | 9.0 ± 0.11 | 2.1 ± 0.05 | 0.18 ± 0.02 | 3.9 | 0.09 ± 0.005 |
| N44-1 mgdH::kan pBBRMCS4 | 18.7 ± 0.19 | 4.4 ± 0.09 | 0.25 ± 0.05 | 2.8 | 0.13 ± 0.005 |
| N44-1 ΔmgdH sgdH::kan pBBR1MCS4 | 24.4 ± 0.19 | 5.7 ± 0.07 | 0.32 ± 0.03 | 2.2 | 0.14 ± 0.002 |
Cultivation was carried out in bioreactors (Dasgip, Jülich, Germany) under pH and aeration control (Fig. 2).
μ, growth rate.
td, doubling time.
Final dry weight per liter divided by the substrate consumed: initial glucose (g liter−1) − residual glucose (g liter−1). Yx/s, growth yield coefficient.
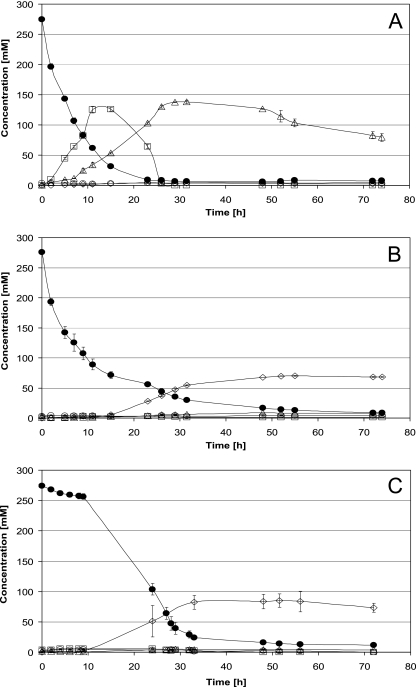
As shown in Fig. 3A, strain N44-1 converted a large part of the glucose first to gluconate (up to 140 mM in the medium), which was subsequently oxidized to 2-ketogluconate and 2,5-diketogluconate (about 140 mM). In contrast, neither gluconate nor ketogluconates was detectable in significant concentrations in the mutant strains N44-1 mgdH::kan (Fig. 3B) and N44-1 ΔmgdH sgdH::kan (Fig. 3C). Further differences were observed with respect to glucose consumption. Whereas the parent strain, N44-1, consumed the glucose almost completely within 15 h, the gdh mutants required about 30 h (Fig. 3). Whereas the mgdH::kan mutant showed rapid initial glucose consumption, the double mutant ΔmgdH sgdH::kan displayed an initial lag phase of about 10 h (Fig. 3). This difference indicates that in the absence of mGDH, glucose is initially oxidized by sGDH.
FIG. 3.
Substrate consumption and product formation of G. oxydans strains growing with glucose (275 mM; 50 g liter−1). Cultivation was carried out in bioreactors (Dasgip, Jülich, Germany) under pH and aeration control. (A) G. oxydans N44-1 pBBR1MCS4, mean values and standard deviations from two biological experiments. (B) G. oxydans N44-1 mgdH::kan pBBR1MCS4, mean values and standard deviations from two biological experiments. (C) G. oxydans N44-1 ΔmgdH sgdH::kan pBBR1MCS4, mean values and standard deviations from four biological experiments. The concentrations of glucose (•), gluconate (□), 2-KGA/2,5-DKGA (▵), and acetate (⋄) are shown.
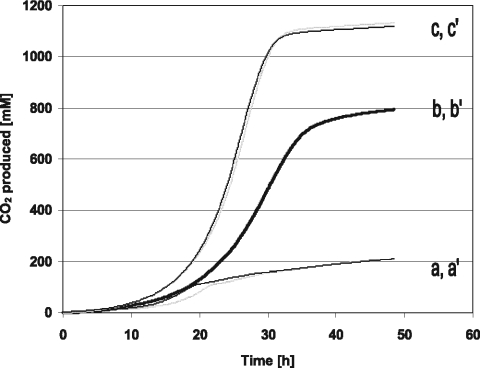
As the gdh mutant strains did not accumulate gluconate or ketogluconates, other products (besides biomass) had to be formed. As shown in Fig. 4, the mgdH::kan and ΔmgdH sgdH::kan strains produced 4-fold and 5.5-fold more CO2 than the parent strain, N44-1. This indicates that a significant part of the glucose is catabolized via the pentose phosphate pathway. In addition and in contrast to the parent strain, both gdh mutants produced up to 80 mM acetate (Fig. 3).
FIG. 4.
Carbon dioxide production (mmol liter medium−1) by G. oxydans strains during growth with glucose (275 mM; 50 g liter−1). Cultivation was carried out in duplicate in bioreactors (Dasgip, Jülich, Germany) under pH and aeration control. a and a′, G. oxydans N44-1 pBBR1MCS4; b and b′, G. oxydans N44-1 mgdH::kan pBBR1MCS4; c and c′, G. oxydans N44-1 ΔmgdH sgdH::kan pBBR1MCS4 cultivated with either 15% dissolved oxygen (c) or 30% dissolved oxygen (c′).
Pathway of acetate formation from glucose by the gdh mutants.
Acetic acid production by Acetobacteriaceae is usually based on the oxidation of ethanol (24). The medium used in our work did not contain ethanol, so acetate had to be formed differently. In bacteria, several pathways are known in which acetate is formed from pyruvate. After oxidative decarboxylation to acetyl-coenzyme A (CoA) by the pyruvate dehydrogenase complex (EC 1.2.4.1, 2.3.1.12, and 1.8.1.4), acetyl-CoA can be converted to acetate by the concerted actions of phosphotransacetylase (EC 2.3.1.8) and acetate kinase (EC 2.7.2.1). G. oxydans (strain 621H) contains genes for the pyruvate dehydrogenase complex (GOX2289 to GOX2292), but not for phosphotransacetylase or acetate kinase (31). Another pathway is offered by pyruvate: quinone oxidoreductase (also named pyruvate oxidase, EC 1.2.2.2), which converts pyruvate to acetate, transferring the reducing equivalents to ubiquinone or menaquinone. However, no corresponding gene occurs in the G. oxydans genome. Still another possibility for acetate formation from pyruvate is via pyruvate decarboxylase (EC 4.1.1.1) and acetaldehyde dehydrogenase (EC 1.2.1.4). It has been known for a long time that acetic acid bacteria possess pyruvate decarboxylase (5, 9). While pyruvate decarboxylase is widely distributed in plants (44), in bacteria it has been detected in only a few species (37), such as Zymomonas mobilis (8), Sarcina ventriculi (2, 12), and the acetic acid bacteria (9). In the genome of G. oxydans, genes encoding pyruvate decarboxylase (GOX1081) and acetaldehyde dehydrogenase (GOX2018) are present, and the two enzymes have been characterized (1, 17, 36). Based on the situation described above, we assumed that acetate formation in the gdh mutant strains occurred via this pathway and therefore assayed the corresponding enzymatic activities in cell extracts of G. oxydans N44-1 and G. oxydans N44-1 ΔmgdH sgdH::kan. Whereas the activities of pyruvate decarboxylase were similar in the two strains (0.77 ± 0.11 U mg protein−1 in N44-1 and 0.79 ± 0.09 U mg protein−1 in the mutant), the mutant contained an almost 4-fold-higher activity of acetaldehyde dehydrogenase (23.16 ± 0.01 U mg protein−1) than the parent strain, N44-1 (6.80 ± 0.01 U mg protein−1). This difference was most likely due to induction of the enzyme by acetaldehyde, as was also observed in Z. mobilis (1), and supports our assumption that strain N44-1 ΔmgdH sgdH::kan catalyzes acetate formation with this enzyme. The acetaldehyde dehydrogenase of G. oxydans N44-1 and the gdh mutant was strongly NADP dependent and showed almost no activity with NAD as a cofactor (data not shown).
DISCUSSION
In the present study, we analyzed the effects on growth and metabolism of G. oxydans N44-1 caused by inactivation of the genes mgdH and sgdH encoding membrane-bound glucose dehydrogenase (mGDH) and soluble glucose dehydrogenase (sGDH), respectively. Deletion of mgdH alone was sufficient to completely abolish gluconate formation from glucose (Fig. 3A). This indicates that the gluconate that accumulates in the medium in the first phase of glucose catabolism is synthesized almost exclusively by mGDH, not by sGDH. Since no ketogluconates were formed in the mgdH mutant, gluconate formed by sGDH is apparently neither exported and oxidized by membrane-bound gluconate dehydrogenases nor oxidized by the soluble gluconate-5-dehydrogenase. Comparison of strain mgdH::kan with strain ΔmgdH sgdH::kan revealed a number of differences, such as in the kinetics of glucose consumption, CO2 formation, growth rate, and growth yield (Table 2). These differences clearly indicated that sGDH was present and active. Presumably, the gluconate formed by sGDH is almost completely phosphorylated by gluconate kinase (EC 2.7.1.12; GOX1709) and then metabolized in the pentose phosphate or Entner-Doudoroff pathway.
In the strain lacking mGDH, glucose has to be taken up by the cells in order to be metabolized. In the genome of G. oxydans, some components of the phosphoenolpyruvate-dependent phosphotransferase system are encoded, but no sugar-specific proteins (EIIB and EIIC) (31). Therefore, a secondary transporter, whose identity is not yet known, probably takes up glucose. Once inside the cell, either of two putative glucokinases (GOX1182 or GOX2419) can phosphorylate glucose. Cell extracts of strain N44-1 showed a high specific glucokinase activity of ∼0.5 U mg protein−1 (21), which presumably is not limiting for glucose metabolism.
Both the mgdH single mutant and the mgdH sgdH double mutant formed much more CO2 than the parent strain, N44-1, consistent with the fact that a major part of the glucose was metabolized in the pentose phosphate pathway rather than in the Entner-Doudoroff pathway. Moreover, both mutants accumulated acetate in the medium. As outlined above, acetate formation probably occurs via the combined actions of pyruvate decarboxylase and acetaldehyde dehydrogenase, as no alternative pathway appears to exist and because the latter enzyme was found to be inducible by acetaldehyde in the gdh mutant. The fact that this pathway was active in the gdh mutants but not in the parent strain can be explained by the fact that G. oxydans does not possess either a functional tricarboxylic acid (TCA) cycle or phosphotransacetylase and acetate kinase. Consequently, the increased amount of acetyl-CoA in the central metabolism of the gdh mutants could not be metabolized and accumulated. Acetyl-CoA, then, might allosterically inhibit the pyruvate dehydrogenase complex, as shown, e.g., for the Escherichia coli enzyme (38), and thus switches the carbon flux to pyruvate decarboxylase and acetaldehyde dehydrogenase. Along with acetate formation, the organism gains additional reduction power in the form of NADPH for biosynthetic purposes.
The primary aim of this study was to generate strains with an improved growth yield on glucose by inactivating the genes for glucose dehydrogenase. This aim was reached, as, e.g., the cell yield of strain N44-1 ΔmgdH sgdH::kan was increased by 271% compared to the parent strain and the growth rate was increased by 78% (Fig. 3 and Table 2). Therefore, this strategy should improve the industrial usage of G. oxydans, especially in production processes employing whole-cell biotransformation. In the case of miglitol production, the modified polyol N-formyl-1-amino-1-deoxy-d-sorbitol did not promote growth of G. oxydans. Therefore, the biotransformation reaction had to be carried out as a two-step process. In the first step, G. oxydans biomass was produced by cultivation with d-sorbitol, yeast extract, and salts, giving rise to a large-volume wastewater stream with a high biological oxygen demand. In the second step, N-formyl-1-amino-1-deoxy-d-sorbitol was oxidized with G. oxydans biomass as a catalyst under “resting-cell” conditions (35). Thus, the use of an engineered strain would avoid wasteful incomplete oxidation of the substrate during the production of biomass. Of course, mutations must not affect membrane dehydrogenases needed for the biotransformation, i.e., the substrate for growth optimization should be different from that to be biotransformed. However, the growth yield of strain N44-1 ΔmgdH sgdH::kan (0.14 g CDW g glucose−1) is still low compared, e.g., to that reached by E. coli (0.5 g CDW g glucose−1) (27), which possesses functional glycolytic and citric acid cycle pathways.
As the conversion of glucose to gluconate and ketogluconates obviously has a negative impact on the growth rate and the growth yield of G. oxydans, what is the benefit of these pathways? Gluconacetobacter diazotrophicus, a plant growth-promoting acetic acid bacterium, is known to exert a number of beneficial effects on plants, among others, by solubilizing phosphorus and zinc, thus providing the plant with these minerals. Evidence was recently provided that gluconic acid production is the most important pathway for the solubilization of both nutrients (16). Incomplete oxidation of sugars and sugar alcohols to sugar acids by Gluconobacter species is likely to have similar effects in the natural habitat of these organisms. The advantage of promoting plant growth is that the plant provides a wealth of nutrients, in particular, carbon sources, for the bacteria.
Acknowledgments
We thank DSM Nutritional Products, Kaiseraugst, Switzerland, for financial support.
We acknowledge Cornelia Gaetgens for her excellent technical assistance and Hans-Peter Hohmann, Tanja Hanke, and Stefanie Schweikert for critical reading of the manuscript.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Barthel, T., R. Jonas, and H. Sahm. 1989. NADP+-dependent acetaldehyde dehydrogenase from Zymomonas mobilis. Isolation and partial characterization. Arch. Microbiol. 153:95-100. [Google Scholar]
- 2.Bauchop, T., and E. A. Dawes. 1959. Metabolism of pyruvic and formic acids of Zymosarcina ventriculi. Biochim. Biophys. Acta 36:294-296. [DOI] [PubMed] [Google Scholar]
- 3.Bernt, E., and I. Gutmann. 1974. Aethanol, p. 1545-1548. In H. U. Bergmeyer (ed.), Methoden der enzymatischen Analyse, vol. 2. Verlag Chemie, Weinheim, Germany. [Google Scholar]
- 4.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 5.Bringer-Meyer, S., K.-L. Schimz, and H. Sahm. 1986. Pyruvate decarboxylase from Zymomonas mobilis. Isolation and partial characterization. Arch. Microbiol. 146:105-110. [Google Scholar]
- 6.Choi, K.-H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. N., A. C. Y. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. U. S. A. 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawes, E. A., D. W. Ribbons, and P. J. Large. 1966. The route of ethanol formation in Zymomonas mobilis. Biochem. J. 98:795-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLey, J., and J. Schell. 1962. Lactate and pyruvate catabolism in acetic acid bacteria. J. Gen. Microbiol. 29:589-601. [DOI] [PubMed] [Google Scholar]
- 10.Deppenmeier, U., M. Hoffmeister, and C. Prust. 2002. Biochemistry and biotechnological applications of Gluconobacter strains. Appl. Microbiol. Biotechnol. 60:233-242. doi: 10.1007/s00253-002-1114-5. [DOI] [PubMed] [Google Scholar]
- 11.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finn, R. K., S. Bringer, and H. Sahm. 1984. Fermentation of arabinose to ethanol by Sarcina ventriculi. Appl. Microbiol. Biotechnol. 19:161-166. [Google Scholar]
- 13.Gupta, A., V. K. Singh, G. N. Qazi, and A. Kumar. 2001. Gluconobacter oxydans: its biotechnological applications. J. Mol. Microbiol. Biotechnol. 3:445-456. [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann, U., M. Merfort, M. Jeude, S. Bringer-Meyer, and H. Sahm. 2004. Biotransformation of glucose to 5-keto-D-gluconic acid by recombinant Gluconobacter oxydans DSM 2343. Appl. Microbiol. Biotechnol. 64:86-90. doi: 10.1007/s00253-003-1455-8. [DOI] [PubMed] [Google Scholar]
- 16.Intorne, A. C., M. Vinicius, V. de Oliveira, M. L. Lima, J. F. da Silva, F. L. Olivares, and G. A. de Souza Filho. 2009. Identifcation and characterization of Gluconacetobacter diazotrophicus mutants defective in the solubilisation of phosphorus and zinc. Arch. Microbiol. 191:477-483. doi: 10.1007/s00203-009-0472-0. [DOI] [PubMed] [Google Scholar]
- 17.King, T. E., and V. H. Cheldelin. 1954. Pyruvic carboxylase of Acetobacter suboxydans. J. Biol. Chem. 208:821-831. [PubMed] [Google Scholar]
- 18.Klasen, R., S. Bringer-Meyer, and H. Sahm. 1995. Biochemical characterization and sequence analysis of the gluconate:NADP 5-oxidoreductase gene from Gluconobacter oxydans. J. Bacteriol. 177:2637-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 20.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 21.Krajewski, V. 2008. Modifikation des Glucosestoffwechsels in Gluconobacter oxydans. Ph.D. thesis. University of Düsseldorf, Düsseldorf, Germany.
- 22.Lam, S. T., B. S. Lam, and G. Strobel. 1985. A vehicle for the introduction of transposons into plant-associated pseudomonads. Plasmid 13:200-204. [DOI] [PubMed] [Google Scholar]
- 23.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madigan, M. T., J. M. Martinko, and J. Parker. 1997. Brock biology of microorganisms, 8th ed. Prentice Hall, Upper Saddle River, NJ.
- 25.Matsushita, K., H. Toyama, and O. Adachi. 1994. Respiratory chains and bioenergetics of acetic acid bacteria. Adv. Microbiol. Physiol. 36:247-301. [DOI] [PubMed] [Google Scholar]
- 26.Merfort, M., U. Herrmann, S. Bringer-Meyer, and H. Sahm. 2006. High-yield 5-keto-D-gluconic acid formation is mediated by soluble and membrane-bound gluconate-5-dehydrogenases of Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 73:443-451. doi: 10.1007/s00253-006-0467-6. [DOI] [PubMed] [Google Scholar]
- 27.Ng, H. 1969. Effect of decreasing growth temperature on cell yield of Escherichia coli. J. Bacteriol. 98:232-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niebisch, A., and M. Bott. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175:282-294. [DOI] [PubMed] [Google Scholar]
- 29.Olijve, W., and J. J. Kok. 1979. Analysis of growth of Gluconobacter oxydans in glucose containing media. Arch. Microbiol. 121:283-290. [Google Scholar]
- 30.Pronk, J. T., P. R. Levering, W. Olijve, and J. P. van Dijken. 1989. Role of NADP-dependent and quinoprotein glucose dehydrogenases in gluconic acid production by Gluconobacter oxydans. Enzyme Microb. Technol. 11:160-164. [Google Scholar]
- 31.Prust, C., M. Hoffmeister, H. Liesegang, A. Wiezer, W. F. Fricke, A. Ehrenreich, G. Gottschalk, and U. Deppenmeier. 2005. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 23:195-200. doi: 10.1038/nbt1062. [DOI] [PubMed] [Google Scholar]
- 32.Reichstein, T., and A. Grüssner. 1934. Eine ergiebige Synthese der l-Ascorbinsäure (C-Vitamin). Helvetica Chimica Acta 17:311-328. [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 35.Schedel, M. 2000. Regioselective oxidation of aminosorbitol with Gluconobacter oxydans, key reaction in the industrial 1-deoxynojirimycin synthesis, p. 295-311. In H.-J. Rehm and G. Reed (ed.), Biotechnology II. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- 36.Schweiger, P., S. Volland, and U. Deppenmeier. 2007. Overproduction and characterization of two distinct aldehyde-oxidizing enzymes from Gluconobacter oxydans 621H. J. Mol. Microbiol. Biotechnol. 13:147-155. [DOI] [PubMed] [Google Scholar]
- 37.Scrutton, M. C. 1971. Assay of enzymes of CO2 metabolism, p. 479-541. In J. R. Morris and D. W. Ribbons (ed.), Methods in enzymology, vol. 6A. Academic Press, New York, NY. [Google Scholar]
- 38.Shen, L. C., and D. E. Atkinson. 1970. Regulation of pyruvate dehydrogenase from Escherichia coli—interactions of adenylate energy charge and other regulatory parameters. J. Biol. Chem. 245:5974-5978. [PubMed] [Google Scholar]
- 39.Shinagawa, E., Y. Ano, T. Yakushi, O. Adachi, and K. Matsushita. 2009. Solubilization, purification and properties of membrane-bound D-glucono-δ-lactone hydrolase from Gluconobacter oxydans. Biosci. Biotechnol. Biochem. 73:241-244. [DOI] [PubMed] [Google Scholar]
- 40.Shinjoh, M., Y. Setoguchi, T. Hoshino, and A. Fujiwara. 1990. l-Sorbose dissimilation in 2-keto-l-gulonic acid-producing mutant UV10 derived from Gluconobacter melanogenus IFO 3293. Agric. Biol. Chem. 54:2257-2263. [Google Scholar]
- 41.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784-791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 42.Sugisawa, T., T. Hoshino, S. Masuda, S. Nomura, Y. Setoguchi, M. Tazoe, M. Shinjoh, S. Someha, and A. Fujiwara. 1990. Microbial production of 2-keto-L-gulonic acid from L-sorbose and L-sorbitol by Gluconobacter melanogenus. Agric. Biol. Chem. 45:1201-1209. [Google Scholar]
- 43.Träger, M., U. Müller, and U. Onken. 1987. Einfluss erhöhter Sauerstoff-Partialdrucke auf die Bildung von Gluconsäure durch Gluconobacter oxydans. Chem. Ing. Tech. 59:940-944. [Google Scholar]
- 44.Vennesland, B. 1951. Keto acid decarboxylases, p. 183-215. In P. D. Boyer, H. Lardy, and K. Myrbick (ed.), The enzymes, vol. 2. Academic Press, New York, NY. [Google Scholar]