Abstract
Coxiella burnetii is an obligate intracellular bacterium that causes the zoonotic disease Q fever. Because C. burnetii is highly infectious, can survive under a variety of environmental conditions, and has been weaponized in the past, it is classified as a select agent and is considered a potential bioweapon. The agent is known to be present in domestic livestock and in wild animal populations, but the background levels of C. burnetii in the environment have not been reported. To better understand the amount of C. burnetii present in the environment of the United States, more than 1,600 environmental samples were collected from six geographically diverse parts of the United States in the years 2006 to 2008. DNA was purified from these samples, and the presence of C. burnetii DNA was evaluated by quantitative PCR of the IS1111 repetitive element. Overall, 23.8% of the samples were positive for C. burnetii DNA. The prevalence in the different states ranged from 6 to 44%. C. burnetii DNA was detected in locations with livestock and also in locations with primarily human activity (post offices, stores, schools, etc.). This study demonstrates that C. burnetii is fairly common in the environment in the United States, and any analysis of C. burnetii after a suspected intentional release should be interpreted in light of these background levels. It also suggests that human exposure to C. burnetii may be more common than what is suggested by the number of reported cases of Q fever.
The Gram-negative obligate intracellular bacterium Coxiella burnetii can infect humans and cause Q fever, an acute febrile illness (15, 17). Most cases of Q fever have fairly nonspecific symptoms, such as high fever, headache, myalgia, cough, and fatigue (29). Over one-third of patients may show signs of pneumonia or hepatitis (17). Acute cases typically resolve in 1 to 2 weeks, but a small percentage of Q fever cases result in a chronic infection that can present as endocarditis and be life-threatening (12).
Q fever occurs worldwide, and numerous natural outbreaks have been reported in the United States (2, 23, 25) and other countries (5, 11, 18, 20, 22, 24). An ongoing natural outbreak in the Netherlands resulted in more than 2,000 cases of Q fever from 2007 to 2009 (27). In the United States Q fever became a nationally notifiable disease in 1999, and increasing numbers of cases have been reported to the CDC in recent years. However, the highest number of annual cases in the United States so far has been 171, reported in 2007 (8). Although this is a fairly small number of reported cases, it is possible that the number of actual cases in the United States is much higher. The relatively nonspecific nature of Q fever symptoms makes the disease difficult to diagnose, and people infected with C. burnetii are likely to show a diversity of symptoms with variable severity. The idea that Q fever is underreported is supported by our recent data using serum samples from the National Health and Nutrition Examination Survey (NHANES) to determine that the seroprevalence in the United States among people who are ≥20 years old is 3.1% (1).
A common mechanism for people to become infected with C. burnetii is the inhalation of aerosolized bacteria. Potential sources for aerosolized C. burnetii are livestock and other animals. It is known that many herds of livestock are infected with C. burnetii and that seroprevalence rates in a variety of wild animal species can be quite high (17). Infected livestock herds do not typically show clinical signs of infection, but surges in abortion rates have been reported, particularly with goats (9, 10, 17). It is known that C. burnetii can replicate to high levels in the placenta of infected animals and that infectious C. burnetii can be spread to humans during parturition (9). The prevalence of C. burnetii in animals makes contact with animals a likely risk factor for Q fever. For example, the ongoing Q fever outbreak in the Netherlands has been linked to Q fever infections in goat farms (27), and we have recently found that 22.2% of a group of 508 veterinarians had antibodies against C. burnetii, a much higher seroprevalence than in the general U.S. population (31).
C. burnetii exists as a replicating large-cell variant (LCV), but nonreplicating bacteria can form a more stable small-cell variant (SCV) (4). Although it is not an endospore, the SCV form of Coxiella is known to be very stable under a variety of conditions (16). C. burnetii is also highly infectious, with a dose of 1 to 10 organisms capable of causing Q fever in humans (30). These unique features of C. burnetii, along with its aerosol route of transmission, have led to the designation of C. burnetii as a category B bioterrorism weapon and inclusion on the list of select agents. The potential for the use of C. burnetii as a bioweapon was explored in detail by the U.S. bioweapons program of the 1950s and 1960s (26). Although not typically lethal, C. burnetii is considered a threat due to its ability to cause widespread debilitating illness. Indeed, many U.S. soldiers returning from Iraq between 2005 and 2008 suffered from Q fever while deployed (6, 7). These cases are suspected to be naturally acquired infections.
The potential for both intentional releases and natural outbreaks makes it important to understand the presence of C. burnetii in the environment. Investigations of the source of Q fever cases will include a determination of the presence of C. burnetii in the environment from which the bacteria may have been acquired. The purpose of this study was to analyze a large number of samples across a wide geographic distribution in the United States and to establish a baseline for the presence of C. burnetii in different regions of the country.
MATERIALS AND METHODS
Samples were collected in cooperation with state and local health departments. CDC staff visited the different sites (18 total sites) and, with the guidance of the local public health officials, went to the specific locations (nine locations per site) chosen for sampling. At each site, an effort was made to represent areas of primarily human activity (post offices, grocery stores, etc.) in about 50% of the locations and to represent areas with expected animal activity (dairy farms, ranches, etc.) in the remainder. Samples were collected by bulk sampling, HEPA vacuuming, or by sponge or swab wipes of solid surfaces. Bulk sampling consisted of collecting 10 to 50 g of material from the ground, and this was typically soil. HEPA vacuuming was used primarily in indoor areas where there was a solid surface that contained dust and other particulates. Sponge or swab wipes also sampled particulates found on solid surfaces. Samples were transported back to the CDC for DNA extraction and PCR analysis.
Full details of the methods used to extract DNA from the environmental samples will be published in a separate manuscript (6a), but a brief description will be provided here. Five grams of each environmental sample was mixed with 10 to 30 ml of phosphate-buffered saline (PBS), depending on the absorbency of the sample. Samples were incubated at room temperature with rocking for 1 h and then centrifuged for 5 min at 200 × g to remove the larger particles. The supernatants were saved and then centrifuged for 15 min at 20,000 × g to concentrate the microorganisms. After this spin, supernatants were discarded, and pellets were resuspended in less than 1 ml of PBS. A total of 700 μl of this microbial suspension was then used to purify DNA using either a QIAamp DNA mini-kit (tissue protocol) or a MoBio Ultraclean soil kit. The samples were run through the kits according to the manufacturers' instructions, and the DNA was eluted in water or in the elution buffer provided with the kits. The DNA eluate was then evaluated for its ability to inhibit a PCR by adding 1 μl to a quantitative PCR that amplified the IS1111 gene from 200 genome equivalents (GE) of C. burnetii (strain Nine Mile Phase 1) DNA (14). Any environmental DNA sample that caused an increase of at least one cycle required to reach threshold was considered inhibitory.
DNA eluates that were found to be inhibitory were further purified using a second kit. Samples that were originally purified using the MoBio kit were run through the QIAamp kit for the second round, and samples originally run through the QIAamp kit were run through the MoBio kit for the second round. A few samples were run through a Qiagen DNA stool kit for the second round. After a second round of purification, the samples were analyzed for PCR inhibition as described above. The combination of two purification kits removed inhibition in more than 99% of the samples, and the few samples that still inhibited PCR after the second purification kit were excluded from the study.
Samples that were free of PCR inhibition were analyzed for the presence of C. burnetii using a quantitative TaqMan PCR assay that amplified the IS1111 sequence from C. burnetii (14). In the Nine Mile Phase 1 strain of C. burnetii, which has 20 copies of the IS1111 sequence, this assay can detect 0.2 genome equivalents per reaction and is highly specific for C. burnetii based on a negative result on DNA from 11 near-neighbor bacterial species (unpublished data). PCR mixtures of 25 μl were set up with 1 μl of the DNA eluate in each reaction mixture. Duplicates were run for each environmental sample. Positive controls consisted of a dilution series of known amounts of genomic C. burnetii (strain Nine Mile Phase 1) DNA, and negative controls were reaction mixtures without template. The PCRs were run for 40 cycles, and samples with a threshold cycle (CT) of less than 40 were considered positive. Samples where one duplicate had a threshold cycle below 40 and the other duplicate did not were analyzed in a second PCR using 5 μl of the DNA eluate. It was first demonstrated that the 5 μl did not inhibit the PCR, and then the IS1111 PCR was run in duplicate using 5 μl of template. If both reactions did not have a threshold cycle below 40, the sample was considered negative. The PCR was run on an Applied Biosystems 7900 instrument, and the data were analyzed using Applied Biosystems SDS, version 2.3, software. For statistical analysis, correlation coefficients were calculated in Microsoft Excel, and Pearson's chi-square values were determined using an interactive online calculator (Quantitative Skills, Hilversum, Netherlands).
For real-time PCR of the com1 gene, a TaqMan PCR assay was performed using the primers COM1 TaqMan fwd (5′-AATAAAAACCTCCGCGTTGTCTT-3′) and COM1 TaqMan rev (5′-TTGGCAGCGTATTGCGATT-3′) and COM1 probe (5′-AAAGAACTGCCCATTTTTGGCGGC-3′). Cycling conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. For sequencing, PCR of the com1 gene was done according to the protocol of Zhang et al. using the nested primer sets OMP1/OMP2 and OMP3/OMP4 (32).
For injection into mice, 5 g of environmental sample was mixed with 10 ml of PBS for 1 h, centrifuged at 200 × g to remove large particles, and then pelleted at 20,000 × g for 15 min. The pellets were resuspended in 1 ml of PBS, and then 100 μl of this mixture was injected intraperitoneally (i.p.) into each of two male BALB/c mice. The mice were sacrificed 3 weeks later, and spleens and serum were collected. Serum was analyzed for the presence of anti-Coxiella antibodies using an in-house immunofluorescence assay (IFA). The number of C. burnetii organisms in the spleen was estimated by performing a com1 real-time PCR on spleen genomic DNA and comparing the threshold cycles to a standard curve of Nine Mile Phase 1 genomic DNA. All animal studies were performed using procedures approved by the CDC IACUC.
RESULTS
Samples were collected in six different states, which are identified by their geographic regions in Table 1. Approximately 270 samples were taken in each state, with a total of 1,622 samples included in the analysis. Out of these 1,622 samples, 386 were positive by a PCR assay that specifically detects the IS1111 insertion sequence of C. burnetii (14). Thus, 23.8% of the samples were positive for C. burnetii DNA. The percentage of positive samples in each state varied widely. The highest percentage was found at the sites in the Rocky Mountains region, where 44.6% of the samples analyzed indicated the presence of C. burnetii. On the low end, the East Coast state had only 6% of samples positive. Overall, these data indicate that background levels of C. burnetii in the environment are quite high and that C. burnetii DNA is found over a broad geographic distribution.
TABLE 1.
Determination of the presence of C. burnetii DNA in environmental samples from six states
| State geographic region | No. of samples | No. of positive results | % Positive | Date of collection |
|---|---|---|---|---|
| Rocky Mountains | 271 | 121 | 44.6 | November 2006 |
| South-central | 285 | 103 | 36.1 | December 2006 |
| Upper Midwest | 271 | 67 | 24.7 | November 2007 |
| West Coast | 271 | 37 | 13.7 | April 2008 |
| East Coast | 266 | 16 | 6.0 | April 2008 |
| Deep South | 258 | 42 | 16.3 | June 2008 |
| Total | 1,622 | 386 | 23.8 |
To confirm that these samples were positive for C. burnetii, 23 of the IS1111-positive samples were analyzed using a real-time PCR assay that targets the C. burnetii com1 gene (32). Because com1 is a single-copy gene, this assay is less sensitive than the IS1111 PCR assay, so it was expected that many of the IS1111-positive samples would be negative by the com1 assay due to limits of detection. In this analysis, 15 out of 23 samples were positive for C. burnetii com1. This suggests that the IS1111 assay is reliably detecting C. burnetii genomic DNA in these environmental samples. The samples that were positive for IS1111 but negative for com1 all had IS1111 CT values that were greater than 30.7 and most likely did not have a sufficient number of genomes to be detected by the less sensitive com1 assay. A 438-bp section of the com1 gene was also amplified from three of the environmental DNA samples by PCR, and these com1 gene fragments were sequenced. In all three cases, the environmental com1 gene fragment had 100% identity to published C. burnetii com1 sequences (3, 28).
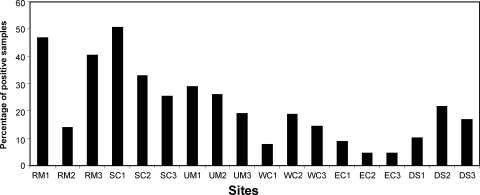
Within each state, samples were taken at three different sites. The sites were roughly equivalent to a county and were the source of 90 samples. Each site consisted of nine locations (small farm, office building, school, etc.) from which 10 samples each were collected. We considered the possibility that the percentage of positive samples in some states could be strongly influenced by only one of the sites having a very high or very low percentage of positive samples. However, the percentage of positive samples taken in the different sites was fairly consistent within a state (Fig. 1), the largest exception being a relatively low percentage of positive samples in one of the Rocky Mountain sites. These results suggest that factors affecting the presence of C. burnetii at a given site are not site specific and influence a fairly large geographic area.
FIG. 1.
Percentage of Coxiella-positive samples at each of the 18 sampling sites. Environmental samples were evaluated for the presence of C. burnetii DNA using an IS1111 PCR assay. Approximately 90 samples were acquired at each of the 18 sites, and the percentages with C. burnetii DNA are shown. RM, Rocky Mountains; SC, south-central; UM, Upper Midwest; WC, West Coast; EC, East Coast; DS, Deep South.
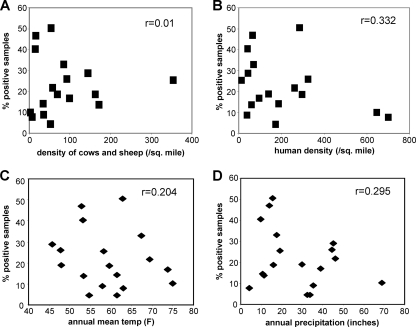
The presence of C. burnetii in herds of livestock and the known association of animal contact with human exposure (2, 5, 24, 27) raise the possibility that the numbers of livestock at a given site could influence the ability to find Coxiella-positive samples. To investigate this possibility, the percentage of positive samples at each site was compared to the density of cows and sheep at each site. As shown in Fig. 2, there was a very poor correlation between livestock density and the percentage of positive samples. This indicates that the overall numbers of livestock at a given site are not a major factor in the ability of Coxiella to infiltrate the local environment. This suggests that factors influencing much larger geographic areas are more important, as indicated by the differences between states. Inclusion of goat populations in this analysis did not influence the results, as goat populations are small compared to populations of sheep and cows. Analysis of goat densities alone also did not correlate with the percentage of Coxiella-positive samples (data not shown).
FIG. 2.
Comparison of the percentages of Coxiella-positive samples to environmental factors. The percentages of samples with C. burnetii DNA at each of the 18 sampling sites were plotted against livestock density (A), human population density (B), annual mean temperature (C), and annual mean precipitation (D). Livestock density was based on the number of cows and sheep per square mile in the county of the sampling site. Cow and sheep numbers were determined from the 2002 USDA Census of Agriculture. Human population density was determined from 2000 U.S. Census data. Temperature and precipitation data were taken from the National Climatic Data Center annual climatological summary for 2007. The correlation coefficients (r) for each data set are indicated in the top right of each plot.
Other site-specific factors were compared to the percentage of positive samples at the sites. The human population density (Fig. 2B), mean annual temperature (Fig. 2C), and annual precipitation (Fig. 2D) did not correlate with the ability to detect positive samples at the sites. Other factors such as median household income, percentage of Hispanic persons, and percentage of residents living in poverty also had a very weak correlation with the presence of C. burnetii DNA (correlation coefficients less than 0.52 [data not shown]). This further supports the idea that the characteristics of the regional environment are not a large factor in determining the prevalence of C. burnetii at these sites.
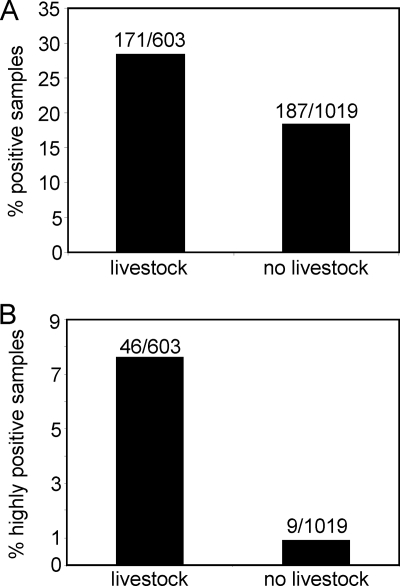
Although the general environment of the sites analyzed did not make a substantial difference, the known association of C. burnetii with wild and domesticated animals (2, 5, 24, 27) led us to consider the effect of livestock directly at the sampling locations on the ability to detect C. burnetii DNA at those locations. For this analysis, all of the samples were categorized as coming from a location where livestock were likely to be encountered (dairy farm, veterinary hospital, etc.) or from a location where livestock were unlikely to be found (post office, grocery store, etc.). When the percentages of positive samples coming from these two types of locations were calculated, the samples from locations with animals were positive 28.4% of the time, whereas samples from locations without animals were positive only 18.4% of the time (Fig. 3A). This is a statistically significant difference (Pearson's chi-square of 22.06; P of <0.00001) and shows that the presence of livestock at the place where the samples were taken can make the presence of C. burnetii DNA more likely. However, this analysis also shows that a large number of positive samples (187) came from locations where livestock are not normally found. There was no evidence that the presence of livestock in the surrounding area or the local climate influenced the presence of C. burnetii in the collected samples.
FIG. 3.
Percentage of Coxiella-positive samples that were taken from locations that had livestock or taken from locations without livestock. This analysis was done on all samples (A) and on highly positive samples (B). Samples were defined as highly positive if they had a CT value in the IS1111 PCR assay of less than 34. For statistical analysis, the data were entered into a two-by-two table, and the Pearson's chi-square values were 22.06 with a P of <0.00001 for panel A and 52.62 with a P of <0.00001 for panel B. The values above the bars indicate the number of positive samples/number of samples tested.
Because a quantitative assay was used to detect C. burnetii, a more stringent threshold could be placed on the samples to look only at samples considered to be “highly positive.” If a cutoff for highly positive samples is set at a threshold cycle of less than 34, then 55 samples out of 1,622 total samples (3.4%) are highly positive. If these 55 highly positive samples are segregated based on the presence of livestock at the sampling location, a fairly striking segregation is found: 46 of the 55 highly positive samples are found at locations where livestock are expected (Fig. 3B). Thus, the presence of livestock makes a very significant difference in whether highly positive samples will be found at a particular location.
If the locations where highly positive samples were found are listed, the locations that had livestock and also highly positive samples are not surprising. These locations are dairies (six), farms (three), veterinary hospitals (two), ranches (two), goat facilities, a cattle feedlot, fairgrounds, an experiment station, and a livestock auction, i.e., places where the presence of C. burnetii-positive animals would not be surprising and where contamination of the local environment could be easily explained. A bit more unexpected was the presence of highly positive samples at the locations where close contact with livestock is not expected: a bank, a co-op/general store, a state government building, a community center, a county administration building, a high school, a retail store, and a city hall. The source of C. burnetii DNA at these locations is unknown, but the organisms could be carried there by human foot traffic, the wind, birds, or other small animals.
To determine if any viable C. burnetii was present in these environmental samples, six samples (5 g each) positive by both IS1111 and com1 PCR were prepared in the same way as described for a DNA extraction. However, in this case the high-speed PBS pellet was resuspended in 1 ml of PBS, and 100 μl of this mixture was injected into each of two BALB/c mice by the intraperitoneal route. Four of the samples were from the West Coast state, one was from the East Coast state, and one was from the southeastern state. The com1 PCR from these samples enabled a rough estimate of the number of genome equivalents that were injected into the mice. These ranged from 16,000 to over 1 × 106 (Table 2). For two of the samples (both from the West Coast state), the mice became ill soon after the injection and had to be sacrificed. For the other four samples, the mice were sacrificed at 3 weeks after the injection of the environmental sample. Analysis of these mice indicated that they had developed antibody titers against C. burnetii Nine Mile Phase 1 and Phase 2 and had a mild splenomegaly. com1 PCR of spleen DNA indicated that the injected C. burnetii had expanded significantly in vivo (Table 2). These results suggest that viable C. burnetii can be found in this collection of environmental samples.
TABLE 2.
Testing for viability in BALB/c mice
| Samplea | Estimated no. of GEb injected | Estimated no. of GE in spleen at 3 weeks | Nine Mile Phase 1 titer | Nine Mile Phase 2 titer | Spleen mass/body mass |
|---|---|---|---|---|---|
| PBS | NAc | None | None | None | 0.006 |
| WC-21 | 50,000 | NA | NA | NA | NA |
| WC-36 | 180,000 | 220,000 | ≥1:256 | ≥1:256 | 0.008 |
| WC-25 | 900,000 | 3,300,000 | ≥1:256 | ≥1:256 | 0.018 |
| WC-35 | 1,100,000 | NA | NA | NA | NA |
| SE-101 | 50,000 | 5,100,000 | ≥1:256 | ≥1:256 | 0.012 |
| EC-217 | 16,000 | 1,040,000 | ≥1:256 | ≥1:256 | 0.010 |
WC, West Coast; EC, East Coast; SE, Southeast.
GE, genome equivalents.
NA, not available.
DISCUSSION
The purpose of this study was to determine the level of C. burnetii in the environment of the United States. Samples were taken from a variety of locations although an effort was made to have approximately half of the locations in places where animal activity was expected. Overall, 23.8% of the 1,622 samples analyzed were positive for C. burnetii DNA, with variation among states from 44.6% to 6%. The locations that contained C. burnetii DNA were quite diverse. The presence of C. burnetii in domesticated animal populations has been well established (2, 5, 10, 24, 27), so it is not surprising that C. burnetii DNA was found at dairy farms, cattle feed lots, veterinary hospitals, and goat-breeding facilities. However, many samples positive for C. burnetii DNA were collected from locations that had no livestock present. These locations included a wide variety of places with fairly large amounts of human activity: high schools, retail stores, grocery stores, football stadiums, banks, and post offices. Thus, C. burnetii has spread across a wide geographic area, and any outbreak investigation should take into account the fairly high prevalence of C. burnetii DNA in normal locations of daily life. Factors responsible for the distribution of C. burnetii DNA will require further investigation.
The IS1111 assay used to detect C. burnetii DNA was a quantitative TaqMan PCR assay (14). Although IS1111 can be present at variable copy numbers (7 to 110 copies) in different strains of C. burnetii (13), the assay does allow a rough estimation of the amount of C. burnetii present in these samples by comparison to a standard curve of genomic DNA from C. burnetii strain Nine Mile Phase 1 (20 copies of IS1111). Most of the samples analyzed had a fairly low number of C. burnetii bacteria detected. These would fall into the range of 100 to 1,000 genome equivalents (GE) in each environmental sample. About 10% of the positive samples were much stronger and could have had from 103 to 106 GE present. Testing of the methods used for isolation of DNA from the environmental samples has shown that yields of C. burnetii DNA were low, with only 1 to 3% of C. burnetii DNA recoverable from these samples (6a). Although it was not possible to determine the yield for each sample, if we assume that yields were typically around 2%, the actual number of GE in each sample prior to processing was likely to be 30- to 50-fold higher than the numbers listed above. This would be about 103 GE on the low end, and the majority of samples would fall between 103 GE and 104 GE. The most concentrated samples would be on the order of 107 GE per sample.
With complex environmental samples that may contain viable C. burnetii bacteria, it is difficult to directly assess the viability of the organisms. Although the recent development of host-cell-free growth of C. burnetii may provide new methods for viability testing in the near future (19), currently a host cell is required to test viability. However, placement of the environmental sample directly onto cultured host cells is likely to result in contamination of the culture by a variety of microbes present in the sample. Such contamination will preclude an evaluation of the growth of potential C. burnetii cells. Because of these difficulties, viability is best determined by injection of environmental materials into mice. Mice have the ability to quickly clear most of the contaminating microbes, whereas C. burnetii will grow slowly in mice. Thus, analysis of mice 3 weeks after the injection of C. burnetii can reveal whether viable C. burnetii bacteria were present and can greatly facilitate isolation. The data presented above using six PCR-positive environmental samples suggest that many of the strongly positive samples are viable although it will not be possible to test all of the samples for viability. For many of the samples, all of the collected material was consumed in the process of DNA isolation. As seen in our viability studies, mice are not always able to clear the microbes present in soil samples. Treatment of samples with moderate heat and/or fungicides may increase the probability of survival after injection of environmental samples. The environmental samples that have material remaining will be useful for further studies to optimize isolation methodologies.
The data demonstrate that C. burnetii is commonly found in the environment and that many of these samples may contain viable bacteria. Previous studies have demonstrated that C. burnetii can be highly infectious, with only a few organisms sufficient for infection (21). Taken together, these data could lead to the expectation that cases of Q fever are common in the United States. However, Q fever has been a nationally reportable disease for almost 10 years, and there have never been more than 200 cases reported in the United States in any given year. Our recent analysis of seroprevalence in the United States showed that 3.1% of the population has antibodies against C. burnetii (1). This suggests that many more people have been exposed to C. burnetii than is apparent from the number of reported cases. The seroprevalence rate is much more congruent with the prevalence of C. burnetii in the environment. So the question of why Q fever is not more often reported remains. At least part of the answer could be that the nonspecific symptoms of Q fever are not recognized as a bacterial infection, and the disease is allowed to progress without testing or treatment. It is also possible that many people may experience fairly mild symptoms and never seek medical attention.
The data in this study demonstrate that C. burnetii can be quite common in the environment, with approximately 50% of samples positive at some sites. The possibility of high, naturally occurring background levels must be taken into account when any analysis of C. burnetii in the environment is undertaken during investigations of natural outbreaks or intentional releases. The detection of C. burnetii was linked with the presence of livestock at the sampling location; however, animal presence was clearly not required for C. burnetii detection. Furthermore, livestock density did not correlate with the presence of C. burnetii, suggesting that other factors control the distribution of C. burnetii in the environment.
Acknowledgments
We thank the numerous state and local public health officials that helped with determining the locations for collection and with the collection itself: from the Texas Department of State Health Services, James Alexander, Kathy Parker, Erika Quinones, Dudley Moore, and Keith Stevenson; from the Lubbock City Health Department, Rick West; from the San Angelo/Tom Green County Health Department, Mike Loving; from the Texas AgriLife Extension Service, Rick Auckerman; from the Texas Tech University Health Sciences Center, Ron Warner; from the Florida Department of Health, Danielle Stanek, Jean Munden, Paul Myers, Frank Kruppa, Alina Alonso, John O'Malley, Mary Echols, Sericea Smith, Pedro Castellon, Robert Palussek, and Carina Blackmore; from the Virginia Department of Health, Julia Murphy, Tabatha Mamorno, Aubrey Meekins, Harry Bennett, Ed Dunn, Paul Freed, Marsha Duckstein, Matthew Cloud, Renee Hoover, and Kevin D. Yost; from the Colorado Department of Public Health and Environment, James L. Beebe, Kate Lujan, Al Scott, and Bill Ray; from the California Department of Public Health, James Glover; from the County of San Diego Animal Disease Diagnostic Laboratory, Gundula Dunne, Daniel Barbour, and Daniel Golson.
A.J.C., N.E.P., T.M.W., and K.A.F. were supported by an appointment to the Emerging Infectious Diseases Fellowship program administered by the Association of Public Health Laboratories and funded by the Centers for Disease Control and Prevention.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC or the Department of Health and Human Services.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Anderson, A. D., D. Kruszon-Moran, A. D. Loftis, G. McQuillan, W. L. Nicholson, R. A. Priestley, A. J. Candee, N. E. Patterson, and R. F. Massung. 2009. Seroprevalence of Q fever in the United States, 2003-2004. Am. J. Trop. Med. Hyg. 81:691-694. [DOI] [PubMed] [Google Scholar]
- 2.Bamberg, W. M., W. J. Pape, J. L. Beebe, C. Nevin-Woods, W. Ray, H. Maguire, J. Nucci, R. F. Massung, and K. Gershman. 2007. Outbreak of Q fever associated with a horse-boarding ranch, Colorado, 2005. Vector Borne Zoonotic Dis. 7:394-402. [DOI] [PubMed] [Google Scholar]
- 3.Beare, P. A., N. Unsworth, M. Andoh, D. E. Voth, A. Omsland, S. D. Gilk, K. P. Williams, B. W. Sobral, J. J. Kupko III, S. F. Porcella, J. E. Samuel, and R. A. Heinzen. 2009. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect. Immun. 77:642-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman, S. A., E. R. Fischer, D. Howe, D. J. Mead, and R. A. Heinzen. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol. 186:7344-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupuis, G., J. Petite, O. Peter, and M. Vouilloz. 1987. An important outbreak of human Q fever in a Swiss Alpine valley. Int. J. Epidemiol. 16:282-287. [DOI] [PubMed] [Google Scholar]
- 6.Faix, D. J., D. J. Harrison, M. S. Riddle, A. F. Vaughn, S. L. Yingst, K. Earhart, and G. Thibault. 2008. Outbreak of Q fever among US military in western Iraq, June-July 2005. Clin. Infect. Dis. 46:e65-e68. [DOI] [PubMed] [Google Scholar]
- 6a.Fitzpatrick, K. A., G. J. Kersh, and R. F. Massung. 2010. Practical method for extraction of PCR-quality DNA from environmental soil samples. Appl. Environ. Microbiol. 76:4571-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gleeson, T. D., C. F. Decker, M. D. Johnson, J. D. Hartzell, and J. R. Mascola. 2007. Q fever in US military returning from Iraq. Am. J. Med. 120:e11-e12. [DOI] [PubMed] [Google Scholar]
- 8.Hall-Baker, P. A., J. Enrique Nieves, R. A. Jajosky, D. A. Adams, P. Sharp, W. J. Anderson, J. J. Aponte, G. F. Jones, A. E. Aranas, A. Rey, B. Lane, and M. S. Wodajo. 2009. Summary of notifiable diseases, United States, 2007. MMWR Morb. Mortal. Wkly. Rep. 56(53):1-94. [Google Scholar]
- 9.Hatchette, T., N. Campbell, R. Hudson, D. Raoult, and T. J. Marrie. 2003. Natural history of Q fever in goats. Vector Borne Zoonotic Dis. 3:11-15. [DOI] [PubMed] [Google Scholar]
- 10.Hatchette, T. F., R. C. Hudson, W. F. Schlech, N. A. Campbell, J. E. Hatchette, S. Ratnam, D. Raoult, C. Donovan, and T. J. Marrie. 2001. Goat-associated Q fever: a new disease in Newfoundland. Emerg. Infect. Dis. 7:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawker, J. I., J. G. Ayres, I. Blair, M. R. Evans, D. L. Smith, E. G. Smith, P. S. Burge, M. J. Carpenter, E. O. Caul, B. Coupland, U. Desselberger, I. D. Farrell, P. J. Saunders, and M. J. Wood. 1998. A large outbreak of Q fever in the West Midlands: windborne spread into a metropolitan area? Commun. Dis. Public Health 1:180-187. [PubMed] [Google Scholar]
- 12.Karakousis, P. C., M. Trucksis, and J. S. Dumler. 2006. Chronic Q fever in the United States. J. Clin. Microbiol. 44:2283-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klee, S. R., J. Tyczka, H. Ellerbrok, T. Franz, S. Linke, G. Baljer, and B. Appel. 2006. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loftis, A. D., W. K. Reeves, D. E. Szumlas, M. M. Abbassy, I. M. Helmy, J. R. Moriarity, and G. A. Dasch. 2006. Rickettsial agents in Egyptian ticks collected from domestic animals. Exp. Appl. Acarol. 40:67-81. [DOI] [PubMed] [Google Scholar]
- 15.Maurin, M., and D. Raoult. 1999. Q fever. Clin. Microbiol. Rev. 12:518-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCaul, T. F., and J. C. Williams. 1981. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J. Bacteriol. 147:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McQuiston, J. H., and J. E. Childs. 2002. Q fever in humans and animals in the United States. Vector Borne Zoonotic Dis. 2:179-191. [DOI] [PubMed] [Google Scholar]
- 18.McQuiston, J. H., R. V. Gibbons, R. Velic, W. L. Nicholson, L. Castrodale, S. H. Wainright, T. J. Vanniewenhoven, E. W. Morgan, L. Arapovic, A. Delilic, M. O'Reilly, and T. Bajrovic. 2003. Investigation of a focus of Q fever in a nonfarming population in the Federation of Bosnia and Herzegovina. Ann. N. Y. Acad. Sci. 990:229-232. [DOI] [PubMed] [Google Scholar]
- 19.Omsland, A., D. C. Cockrell, D. Howe, E. R. Fischer, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, and R. A. Heinzen. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 106:4430-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oren, I., Z. Kraoz, Y. Hadani, I. Kassis, N. Zaltzman-Bershadsky, and R. Finkelstein. 2005. An outbreak of Q fever in an urban area in Israel. Eur. J. Clin. Microbiol. Infect. Dis. 24:338-341. [DOI] [PubMed] [Google Scholar]
- 21.Ormsbee, R., M. Peacock, R. Gerloff, G. Tallent, and D. Wike. 1978. Limits of rickettsial infectivity. Infect. Immun. 19:239-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panaiotov, S., M. Ciccozzi, N. Brankova, V. Levterova, M. Mitova-Tiholova, M. Amicosante, G. Rezza, and T. Kantardjiev. 2009. An outbreak of Q fever in Bulgaria. Ann. Ist. Super. Sanita 45:83-86. [PubMed] [Google Scholar]
- 23.Pinsky, R. L., D. B. Fishbein, C. R. Greene, and K. F. Gensheimer. 1991. An outbreak of cat-associated Q fever in the United States. J. Infect. Dis. 164:202-204. [DOI] [PubMed] [Google Scholar]
- 24.Porten, K., J. Rissland, A. Tigges, S. Broll, W. Hopp, M. Lunemann, U. van Treeck, P. Kimmig, S. O. Brockmann, C. Wagner-Wiening, W. Hellenbrand, and U. Buchholz. 2006. A super-spreading ewe infects hundreds with Q fever at a farmers' market in Germany. BMC Infect. Dis. 6:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauch, A. M., M. Tanner, R. E. Pacer, M. J. Barrett, C. D. Brokopp, and L. B. Schonberger. 1987. Sheep-associated outbreak of Q fever, Idaho. Arch. Intern. Med. 147:341-344. [PubMed] [Google Scholar]
- 26.Rosebury, T., E. A. Kabat, and M. H. Boldt. 1947. Bacterial warfare, a critical analysis of the available agents, their possible military applications, and the means for protection against them. J. Immunol. 56:7-96. [PubMed] [Google Scholar]
- 27.Schimmer, B., F. Dijkstra, P. Vellema, P. M. Schneeberger, V. Hackert, R. ter Schegget, C. Wijkmans, Y. van Duynhoven, and W. van der Hoek. 2009. Sustained intensive transmission of Q fever in the south of the Netherlands, 2009. Euro Surveill. Pii:19210. [DOI] [PubMed] [Google Scholar]
- 28.Seshadri, R., I. T. Paulsen, J. A. Eisen, T. D. Read, K. E. Nelson, W. C. Nelson, N. L. Ward, H. Tettelin, T. M. Davidsen, M. J. Beanan, R. T. Deboy, S. C. Daugherty, L. M. Brinkac, R. Madupu, R. J. Dodson, H. M. Khouri, K. H. Lee, H. A. Carty, D. Scanlan, R. A. Heinzen, H. A. Thompson, J. E. Samuel, C. M. Fraser, and J. F. Heidelberg. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 100:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terheggen, U., and P. A. Leggat. 2007. Clinical manifestations of Q fever in adults and children. Travel Med. Infect. Dis. 5:159-164. [DOI] [PubMed] [Google Scholar]
- 30.Tigertt, W. D., A. S. Benenson, and W. S. Gochenour. 1961. Airborne Q fever. Bacteriol. Rev. 25:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitney, E. A., R. F. Massung, A. J. Candee, E. C. Ailes, L. M. Myers, N. E. Patterson, and R. L. Berkelman. 2009. Seroepidemiologic and occupational risk survey for Coxiella burnetii antibodies among US veterinarians. Clin. Infect. Dis. 48:550-557. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, G. Q., S. V. Nguyen, H. To, M. Ogawa, A. Hotta, T. Yamaguchi, H. J. Kim, H. Fukushi, and K. Hirai. 1998. Clinical evaluation of a new PCR assay for detection of Coxiella burnetii in human serum samples. J. Clin. Microbiol. 36:77-80. [DOI] [PMC free article] [PubMed] [Google Scholar]