Abstract
Tick-borne encephalitis (TBE), a viral infection of the central nervous system, is endemic in many Eurasian countries. In Switzerland, TBE risk areas have been characterized by geographic mapping of clinical cases. Since mass vaccination should significantly decrease the number of TBE cases, alternative methods for exposure risk assessment are required. We established a new PCR-based test for the detection of TBE virus (TBEV) in ticks. The protocol involves an automated, high-throughput nucleic acid extraction method (QIAsymphony SP system) and a one-step duplex real-time reverse transcription-PCR (RT-PCR) assay for the detection of European subtype TBEV, including an internal process control. High usability, reproducibility, and equivalent performance for virus concentrations down to 5 × 103 viral genome equivalents/μl favor the automated protocol compared to the modified guanidinium thiocyanate-phenol-chloroform extraction procedure. The real-time RT-PCR allows fast, sensitive (limit of detection, 10 RNA copies/μl), and specific (no false-positive test results for other TBEV subtypes, other flaviviruses, or other tick-transmitted pathogens) detection of European subtype TBEV. The new detection method was applied in a national surveillance study, in which 62,343 Ixodes ricinus ticks were screened for the presence of TBE virus. A total of 38 foci of endemicity could be identified, with a mean virus prevalence of 0.46%. The foci do not fully agree with those defined by disease mapping. Therefore, the proposed molecular test procedure constitutes a prerequisite for an appropriate TBE surveillance. Our data are a unique complement of human TBE disease case mapping in Switzerland.
Tick-borne encephalitis (TBE) is a zoonotic arbovirus infection of the central nervous system affecting humans (10). With approximately 3,000 cases annually in Europe and about 11,000 cases annually in Russia, it is the most important tick-borne viral disease of humans in Eurasia (17, 19). TBE is caused by the tick-borne encephalitis virus (TBEV), a member of the genus Flavivirus within the Flaviviridae family (18).
TBEV was first isolated in 1937 in far-eastern Russia (44). Based on the sequence of the envelope gene, the virus is taxonomically classified into European, Siberian, and Far Eastern subtypes (11). Whereas encephalitis caused by European subtype viruses is usually mild with a fatal outcome of 1 to 5% (14), Far Eastern strains cause severe encephalitis with a case fatality rate of 20 to 60%. Siberian subtype isolates produce a less severe disease, but with a tendency for development of chronic infections (16, 17).
TBEV is typically transmitted by tick bites. Ixodes ricinus, a three-host tick with larval, nymphal, and adult male or female stages, is known to be the principal vector of European subtype TBEV (30). Rarely, alimentary routes of transmission have been described (15). Ticks are infected chronically throughout their life cycle (17). In addition to this transstadial transmission, the virus is spread transovarially (6) or between ticks feeding on the same host (21). As they are more numerous than adults, nymphs are thought to be the most important stage in the transmission of the virus (39).
Between 1990 and 2007, an increase of 317.8% in registered TBE cases in Europe was observed. This disease spread may have been favored by many factors, including climate change and social, political, ecological, economic, and demographic factors (40). As it was the case in many European countries, an increase in TBE morbidity was also observed in Switzerland and Germany, where the European subtype TBE virus was found to circulate within foci of endemicity limited to strict regions (2, 25, 42). Except for several studies focusing on restricted geographic areas (2, 43) the actual rate of TBEV infection of ticks in Switzerland is not known, and current Swiss distribution maps of TBEV are based on human disease cases (13). However, since mass vaccination programs have been introduced in Switzerland, alternative methods for predicting foci of endemicity are required.
Molecular biological methods are a convenient tool for studies of the prevalence of tick-borne pathogens. So far, however, such methods have not been standardized (8). Reverse transcription-PCR (RT-PCR) assays to detect TBEV RNA in ticks have been described previously (28, 33, 34, 36, 37, 43). Many of the described protocols exhibit unsatisfactory sensitivity and specificity (7) and do not include an internal process control (IPC) which would detect presumed inhibitors.
Likewise, numerous approaches for nucleic acid (NA) extraction from ticks using conventional precipitation or commercial purification kits based on silica gel or magnetic particle technology (2, 5, 38, 43) have been described. In addition, sample preparations using Chelex resin have been specified, allowing (RT-)PCR detection of pathogens without NA purification (33). However, most of these protocols describe a single-tube assay not suitable for large-scale surveillance studies.
As the endemicity of TBEV in Switzerland is largely unknown, we intended to perform a national screening of ticks for the presence of this virus using a high-throughput method. Thus, the aims of the present study were as follows: first, to develop and validate a one-step duplex real-time RT-PCR system for the specific detection of European subtype TBEV in ticks, including an internal process control; second, to establish and evaluate an automated high-throughput nucleic acid extraction method using the magnetic particle-based QIAsymphony SP system (Qiagen); and third, to assess the TBEV prevalence in ticks collected in areas of Switzerland of potential endemicity and in areas where the virus is not endemic.
MATERIALS AND METHODS
Viral strains and cell culture conditions.
Tick-borne encephalitis virus (TBEV) strains used for real-time RT-PCR and sample preparation method development were kindly provided by Daniel Růžek (University of South Bohemia, České Budějovice, Czech Republic) or were acquired from the National Collection of Pathogenic Viruses (NCPV) (Wiltshire, United Kingdom) (Table 1). Viral strains classified as biosafety level 4 pathogens (as defined by the Swiss Agency for the Environment, Forests and Landscape) were supplied in an inactivated form. We used the porcine kidney stable (PS) cell line for the propagation of all TBEV strains and other viruses of the TBEV complex. The cell line was maintained in L-15 medium (Leibowitz, Biochrom AG, Berlin, Germany) supplemented with 1% glutamine, 5% fetal calf serum, 1% penicillin-streptomycin, and 0.5% neomycin. Cells were cultured in 25-cm2 Corning culture flasks (Sigma-Aldrich, Basel, Switzerland) at 37°C. Thirty minutes before inoculation, the culture medium of PS cell monolayers was renewed with 2.5 ml of fresh medium. Subsequently, cells were inoculated with 100 μl of either virus culture supernatant or tick homogenate and incubated for 1 h at room temperature. At 1 h postinfection, we added 12.5 ml of culture medium to a total volume of 15 ml, and cultures were incubated for 6 days at 37°C.
TABLE 1.
Specificity of the one-step duplex real-time RT-PCR assay
| Taxon | Strain (subtype)a | Sourceb | BSLc | PCR resultd |
|---|---|---|---|---|
| Tick-borne encephalitis virus | Neudörfl (E), NCPV 364 | 2 | 3 | + |
| Absettarov (E), NCPV 344 | 2 | 3 | + | |
| Hypr (E) | 1 | 3 | + | |
| Hanzalova (E) | 1 | 3 | + | |
| Soukup (E) | 1 | 3 | + | |
| Isolate 43 (E) | 1 | 3 | + | |
| Isolate 94 (E) | 1 | 3 | + | |
| Isolate 117 (E) | 1 | 3 | + | |
| Isolate 465 (E) | 1 | 3 | + | |
| Isolate 8641 (E) | 1 | 3 | + | |
| Aina (S) | 1 | 4 | ns | |
| Vasilchenko (S) | 1 | 4 | ns | |
| Sofjin (FE) | 1 | 4 | − | |
| Louping ill virus | NCPV 212 | 1 | 3 | − |
| Powassan virus | 1 | 3 | − | |
| Eyach virus | 1 | 2 | − | |
| Tribec virus | 1 | 2 | − | |
| Tahyna virus | 1 | 2 | − | |
| Uukuniemi virus | 1 | 2 | − | |
| Dengue virus 1 | TC 974, NCPV 670 | 2 | 3 | − |
| Dengue virus 2 | New Guinea, NCPV 151 | 2 | 3 | − |
| Dengue virus 3 | H 87, NCPV 153 | 2 | 3 | − |
| Dengue virus 4 | H 241, NCPV 152 | 2 | 3 | − |
| Yellow fever virus | 17 D, NCPV 507 | 2 | 2 | − |
| Westnile virus | NY 99, NCPV 398 | 2 | 3 | − |
| Japanese encephalitis virus | Nakayama, NCPV 502 | 2 | 3 | − |
| St. Louis encephalitis virus 1 | NCPV 052 | 2 | 3 | − |
| Borrelia burgdorferi sensu stricto | 3 | 2 | − | |
| Borrelia afzellii | 3 | 2 | − | |
| Borrelia bissettii | 3 | 2 | − | |
| Borrelia garinii | 3 | 2 | − | |
| Borrelia spielmanii | 3 | 2 | − | |
| Borrelia valaisiana | 3 | 2 | − | |
| Rickettsia helvetica | 4 | 3 | − | |
| Rickettsia slovaca | 4 | − | − | |
| Francisella tularensis subsp. tularensis | 8 | 3 | − | |
| Babesia divergens | 5 | 2 | − | |
| Babesia microti | 5 | 2 | − | |
| Mengovirus | vMC0 | 7 | + | |
| Enterovirus 71 | 6 | 2 | − | |
| Polio virus type 1 | NCPV 140 | 2 | 2 | − |
E, European; S, Siberian; FE, Far Eastern.
1, Daniel Růžek, University of South Bohemia, České Budějovice, Czech Republic; 2, NCPV, Wiltshire, United Kingdom; 3, Lise Gern, University of Neuchâtel, Switzerland; 4, Olivier Péter, Institut Central des Hôpitaux Valaisans ICHV, Sion, Switzerland; 5, Bruno Gottstein, Vetsuisse Faculty, University of Bern, Switzerland; 6, Kathrin Mühlemann, University of Bern, Switzerland; 7, Maria Isabel Costafreda, University of Barcelona, Spain; 8, National Collection of Type Cultures (NCTC), Colindale, United Kingdom.
BSL, biosafety level (classification according to the Swiss Agency for the Environment, Forests and Landscape).
+, positive test result; −, negative test result; ns, nonsignificant (reaction efficiency of <0.18).
We used an avirulent-phenotype mutant strain of the infectious mengovirus (vMC0; Cardiovirus, Picornaviridae) (9, 23) as an internal process control (IPC); the plasmid pMC0 and the virus derived from it were kindly provided by Maria Isabel Costafreda (University of Barcelona, Spain). The virus was propagated in HeLa cell cultures (ATCC CCL-2) as described before (9, 23). Cell cultures showing a cytopathic effect of 75% were frozen and thawed once and diluted in Dulbecco's phosphate-buffered saline (PBS) without Ca2+ and Mg2+ (D-PBS) (Biochrom AG) to yield a concentration of 5 × 106 viral genome equivalents/ml. Aliquots were stored at −80°C for further use as IPC.
Primer and probe design.
The specific primers and hydrolysis probes for the detection of the European subtype TBEV (envelope gene; 5′-FAM and 3′-BHQ-1) and mengovirus mutant strain vMC0 (5′-noncoding region; 5′-JOE and 3′-BHQ-1) were designed using Primer Express software v3.0 (Applied Biosystems, Foster City, CA) and purchased from Microsynth (Balgach, Switzerland).
RNA standards.
Two RNA standards were generated for the validation of both the TBEV- and the mengovirus vMC0-specific real-time RT-PCRs. Restriction enzyme digestion, cloning, subcloning, and DNA electrophoresis were done using standard techniques (35). Specific cDNA sequences were synthesized and PCRs were performed using Herculase II Fusion DNA polymerase (Stratagene, Zurich, Switzerland). The synthetic fragments were digested with 5′ EcoRI and 3′ SalI and ligated into a pGEM3zfp cloning vector (Promega, Madison, WI) using T4 DNA ligase (New England Biolabs, Bioconcept, Alschwil, Germany). TOP10 Escherichia coli (One Shot Top10 Electrocomp E. coli; Invitrogen Life Technologies, Basel, Switzerland) was transformed by electroporation. Plasmid DNA was prepared with the Qiagen plasmid maxi kit (Qiagen, Düsseldorf, Germany) and linearized with the restriction enzymes SalI and XbaI. Two micrograms of the linearized DNA was gel purified with the QIAquick gel extraction kit (Qiagen) and used for in vitro transcription with a riboprobe in vitro transcription system with T7 RNA polymerase (Promega). The resulting RNA was purified using NucAway spin columns (Applied Biosystems/Ambion, Rotkreuz, Switzerland). All steps were performed by Solvias (Basel, Switzerland). The copy numbers of the standard RNAs were calculated using the Mongo Oligo mass calculator v2.06 (Jef Rozenski, University of Utah).
One-step real-time RT-PCR and cycling conditions.
Single and duplex one-step real-time RT-PCRs were performed using the QuantiFast probe RT-PCR kit (Qiagen). The real-time RT-PCR conditions were as follows: 12.5 μl of 2× QuantiFast probe RT-PCR master mix, 0.25 μl of QuantiFast RT mix, 2 μl of each primer stock (10 μM), 0.5 μl of the probe stock (10 μM), 2 μl of sample, and RNase-free water to adjust the volume to 25 μl. Pipetting was performed with the CAS-1200 liquid-handling system (CAS Robotics 4 version 4.7.98 software; Corbett Robotics Pty Ltd., Queensland, Australia) into 96-well Twin.tec real-time PCR plates (Eppendorf AG, Hamburg, Germany). Plates were sealed with a 230-V heat sealer (Eppendorf), and amplification was performed using a Mastercycler ep realplex S (Eppendorf). The cycling conditions for the amplification were as follows: reverse transcription at 50°C for 10 min, an initial PCR activation step at 95°C for 5 min, and 45 cycles of two-step cycling for 10 s at 95°C and 30 s at 60°C.
Real-time RT-PCR specificity.
To confirm the specific detection of European subtype TBEV as well as mengovirus vMC0 in the duplex assay, we tested a total of 40 microorganisms, including other flaviviruses, other tick-transmitted pathogens, and two members of the Picornaviridae family (Table 1).
Real-time RT-PCR sensitivity.
The limit of detection (LOD) is defined as the minimal amount of standard RNA that can be detected with a probability of 95%. We prepared 20× 10 and 20 copies/μl of TBEV and vMC0 standard RNAs diluted in Tris-EDTA buffer solution (Sigma-Aldrich) to identify the LOD of the RT-PCR.
Real-time RT-PCR efficiency, linearity, and effective range.
The linearity of the real-time RT-PCR was determined by assaying 10 replicates of Tris-EDTA buffer solution spiked with 10-fold dilutions of standard RNA ranging from 101 to 107 copies/μl. A standard curve (linear regression) was calculated based on the mean quantification cycle (Cq) values. Amplification efficiency was calculated from the log-linear portion of the standard curve using the equation efficiency = 10−1/slope − 1.
Real-time RT-PCR accuracy and precision.
The accuracy of the duplex real-time RT-PCR was defined as the percentage of false-positive and false-negative results. We analyzed Tris-EDTA buffer solution (Sigma Aldrich) spiked with 0 (known true-negative sample), 10, 20, and 50 copies/μl of the TBEV and mengovirus vMC0 standard RNAs. At the same time, the recovery rates were determined using samples spiked with 100, 1,000, and 10,000 copies/μl. The assay was repeated four times, with each concentration in five replicates. The recovery rates, in percent, were calculated as the assessed copy number divided by the spiked copy number multiplied by 100. Interassay precision (reproducibility), which indicates the variability between different runs, was assessed by analyzing five replicates of 100, 1,000, and 10,000 standard RNA copies/μl in four independent experiments. Intra-assay precision (repeatability), defining variability within the same run, was determined by simultaneously assaying dilution series of 10 replicates with 100, 1,000, and 10,000 standard RNA copies/μl.
Homogenization of ticks.
Tick samples were prepared from frozen tick pools of 10 nymphs or 5 adult female or male ticks. Six hundred microliters of buffer solution kept at 4°C (see “Optimization of the automated nucleic acid extraction” below) was added to each frozen tick pool. Samples were immediately homogenized using the TissueLyser system (Qiagen). One 3-mm tungsten carbide bead (Qiagen) was added to each tube (collection microtubes; Qiagen), and tick pools were homogenized for 4 min at 30 Hz. After a short centrifugation step (5 s at 3,220 × g), the supernatants were collected in separate collection microtubes for further use.
Automated nucleic acid extraction using the QIAsymphony SP system.
We performed automated nucleic acid (NA) extraction using the QIAsymphony SP system (Qiagen). For this purpose, 200 μl of tick homogenate supernatant or cell culture supernatant was inactivated in 800 μl of AVL viral lysis buffer (Qiagen). The AVL buffer was supplemented with 3 μg of carrier RNA (Qiagen) and a defined amount of the IPC mengovirus vMC0 (104 viral genome equivalents/sample). NA extraction was performed using the QIAsymphony Virus/Bacteria Midi kit (Qiagen) and a specially adapted protocol (CP Complex 920 FIX v1; Qiagen). RNA was eluted in a final volume of 60 μl and either directly used for downstream applications or stored at −80°C for further use.
Manual total RNA extraction procedure.
The efficiency of the automated NA purification was compared to that of a modified guanidinium thiocyanate-phenol-chloroform extraction procedure (4) with Trizol reagent (Invitrogen Life Technologies), which was performed according to the manufacturer's instructions.
Comparison of the manual and the automated nucleic acid extraction procedures.
To compare the efficiencies of the automated and the manual NA extractions, various samples were prepared and processed using both methods. In all experiments, we added 5 × 103 viral genome equivalents of IPC (mengovirus vMC0) to each sample prior to the NA extraction procedure. Viral genome equivalents were defined by real time RT-PCR analysis using quantified standard RNA. Pools of 10 laboratory-bred Ixodes ricinus nymphal ticks (kindly provided by Daniel Růžek) were homogenized in 600 μl of D-PBS and spiked with serial dilutions of TBE virus strain Hypr (GenBank accession no. U39292) (27) ranging from 5 × 105 to 5 × 102 viral genome equivalents/μl. In parallel, we prepared serial dilutions ranging from 5 × 104 to 50 viral genome equivalents/μl in pure D-PBS solution in order to exclude any matrix effects. Each virus dilution was done in four, replicates and the experiment was repeated on three days. Samples were extracted using both the QIAsymphony SP system and the modified guanidinium thiocyanate-phenol-chloroform protocol, and the RNA recovery rates were compared.
Optimization of the automated nucleic acid extraction.
To optimize the NA recovery using the QIAsymphony SP system, four different D-PBS solutions were compared for homogenization of laboratory-bred ticks. We compared the standard procedure using pure D-PBS to protocols using D-PBS supplemented either with InhibitEX tablets (one tablet in 20 ml of buffer; Qiagen), with Complete protease inhibitor cocktail mini tablets (one tablet in 10 ml of buffer; Roche Diagnostics, Rotkreuz, Switzerland), or with bovine serum albumin (BSA) (100 μg/ml; Sigma-Aldrich). Homogenates were spiked with serial dilutions of TBEV (strain Hypr) ranging from 5 × 105 to 5 × 102 viral genome equivalents/μl, and RNA was extracted using the QIAsymphony SP system. In parallel, we spiked serial dilutions ranging from 5 × 104 to 50 viral genome equivalents/μl into pure solutions (not containing any tick cell debris) in order to exclude any matrix effects. Each sample extraction was performed in duplicate, and the experiment was repeated three times. We compared the RNA recovery rates and selected an optimal homogenization buffer.
Comparison of the manual and the optimized automated procedures for the analysis of naturally infected ticks.
Both the optimized automated NA purification protocol and the manual total RNA extraction procedure were finally applied to a total of 162 pooled tick samples (257 adults and 960 nymphs). We collected these ticks in a known area of TBE endemicity of Switzerland. Ticks were homogenized in D-PBS supplemented with InhibitEX at a concentration of one tablet/20 ml as described above. Two hundred microliters of each sample was used for NA purification with the QIAsymphony SP system, and 100 μl was used for the manual total RNA extraction procedure. We also added 5 × 103 viral genome equivalents of IPC (mengovirus vMC0) to each sample. After extraction, viral RNA was quantified using the one-step duplex real-time RT-PCR assay.
Exclusion of inhibitory or toxic effects of InhibitEX on TBEV propagation in PS cells.
Since TBEV-PCR-positive tick homogenates will be applied to cell culture in order to isolate and propagate viable viruses, any toxic or inhibitory effect of the D-PBS homogenization buffer solution supplemented with InhibitEX had to be excluded. Therefore, PS cell monolayers (cultured in 25-cm2 Corning culture flasks [Sigma-Aldrich]) were supplemented with 100, 200, or 1,000 μl of InhibitEX solution (one tablet/20 ml of D-PBS) and inoculated with TBE virus (strain Hypr) as described above, with each approach performed in triplicate. The cell cultures were incubated at 37°C for 4 days. To monitor virus propagation, we extracted RNA using the QIAsymphony SP system and quantified the viral load by one-step real-time RT-PCR every day.
Tick sampling.
The required sample size for estimating the TBEV prevalence in a cross-sectional study was calculated using the formula n = [(1.96 × SD)/L]2, with SD being the standard deviation of the expected prevalence and L being the desired precision. With an expected virus prevalence of 0.5% (10, 25, 31), 765 ticks have to be analyzed in order to obtain a prevalence estimate with a 95% level of confidence (CL) and a precision of ±0.50%. On average, about 400 ticks were collected in one area. This sample size allows for a prevalence estimation with a 95% CL and a precision of ±0.70% (n = 390) if the prevalence turns out to be the expected value of 0.5%. A total of 62,343 questing ticks were collected at 165 collection sites throughout Switzerland by flagging low vegetation. Ticks were randomly identified based on morphological characteristics and immediately stored at −80°C. Subsequently, ticks were washed once in 75% alcohol and twice in deionized water, dried on paper towels, and sorted into pools of 10 nymphs or 5 adult male or female ticks. Pooled samples were stored at −80°C until further processing. The biostatistical rationale of pooling has been described elsewhere (1).
Screening for the presence of European subtype TBEV in ticks.
Tick pools were screened for the presence of European subtype TBEV using the optimized automated NA extraction protocol and the established one-step duplex real-time RT-PCR assay (including IPC). The resulting data show an estimate of the TBEV prevalence in ticks for the 165 collection sites in Switzerland. For all samples giving a TBEV-positive PCR test result, we performed virus isolation experiments as described above.
Data analysis.
We calculated the one-step duplex real-time RT-PCR efficiencies using the LinRegPCR software version 11.5.0.0 (29). Efficiencies and Cq values of real-time RT-PCRs were compared by two-way analysis of variance (ANOVA) using the software GraphPad Prism version 4.0. (GraphPad Software, Inc., La Jolla, CA); a P value of <0.05 was regarded as significant. To compare the virus propagations in cell cultures treated with different concentrations of InhibitEX solution we performed a one-way ANOVA, and a P value of <0.05 was regarded as significant. Maximum-likelihood estimators of TBEV prevalence at each collection site were calculated using an online calculation tool of the Australian Biosecurity Cooperative Research Centre for Emerging Infectious Diseases (http://www.abcrc.org.au). The tool uses a generalized linear model to calculate the maximum-likelihood estimate and confidence limits of the estimated prevalence for variable pool sizes, assuming perfect test sensitivity and specificity (12). Stage-specific prevalences were calculated according to the formula r = 1 − (1 − Xp/np)1/c with r being the estimated prevalence, Xp being the number of test-positive pools, np being the number of analyzed pools, and c being the pool size.
RESULTS
Robustness of the real-time RT-PCR.
The one-step duplex real-time RT-PCR appeared to be highly robust. An elongation of the reverse transcription step to 20 or 30 min and use of various primer concentrations (0.4 to 1.0 μM) did not improve the results of the assay (data not shown). Performing the one-step duplex reaction using the QuantiTect multiplex RT-PCR kit (Qiagen) instead of the QuantiFast probe RT-PCR kit yielded slightly inferior reaction efficiencies and somewhat higher Cq values, mainly for low RNA concentrations (50 and 500 copies/μl) (data not shown).
Analytical specificity of the one-step duplex real-time RT-PCR assay.
The primers and hydrolysis probes used in the one-step duplex real-time RT-PCR assay are listed in Table 2. All European subtype TBEV isolates were successfully detected (mean reaction efficiency, 0.815). Minor, nonsignificant reactions with TBEV Vasilchenko and TBEV Aina (both Siberian subtype) were recorded, with reaction efficiencies of 0.053 and 0.178, respectively (data not shown). We did not observe any nonspecific amplification of all tested flaviviruses, other tick-transmitted bacteria and parasites, or members of the genus Picornaviridae (Table 1).
TABLE 2.
One-step real-time RT-PCR primers and hydrolysis probes specific to European subtype TBEV and IPC mengovirus vMC0
| Target and description | Primer name | Sequence (5′ → 3′) | Position on genea | Product size (bp) |
|---|---|---|---|---|
| TBEV E gene | ||||
| Forward | TBEE-F6 | GGCTTGTGAGGCAAAAAAGAA | 1329-1349 | 87 |
| Reverse | TBEE-R2 | TCCCGTGTGTGGTTCGACTT | 1397-1416 | |
| Probe | TBEE-P4 | FAM-AAGCCACAGGACATGTGTACGACGCC-BHQ-1 | 1349-1374 | |
| Mengovirus vMC0 5′ noncoding region | ||||
| Forward | Mengo-F1 | GACTACCCACTCCCCCTTTC | 64-84 | 103 |
| Reverse | Mengo-R1 | GCTTCGGCCAGTAATGTGAT | 147-167 | |
| Probe | Mengo-P1 | JOE-TGAAGGCTACGATAGTGCCAGGGC-BHQ-1 | 88-112 |
For the TBEV E gene, positions of genes are according to accession number U27495.1. For the mengovirus vMC0 5′ noncoding region, positions are according to http://virology.wisc.edu/acp/Plasmids/PlasmidFiles/pMC0.gb.txt.
Analytical sensitivity of the real-time RT-PCR assay.
The LOD of the one-step duplex real-time RT-PCR assay is 10 standard RNA copies/μl for the detection of both TBEV and the IPC.
Real-time RT-PCR efficiency, linearity, and effective range.
The assay was linear over the range of 101 to 107 copies/μl. The fitted linear model for TBEV quantification had a correlation coefficient (r2) of 0.9997, with a P value of 2.46 × 10−10. The regression analysis for quantification of mengovirus vMC0 was slightly inferior, with a P value of 1.444 × 10−7 and an r2 of 0.9966. The estimated efficiencies were 0.97 for the TBEV-specific reaction and 0.92 for the mengovirus-specific reaction.
Real-time RT-PCR accuracy and precision.
The false-positive and false-negative rates of the assay are 0%. The recovery rates for 100, 1,000, and 10,000 standard RNA copies/μl are given in Table 3. The data on reproducibility and repeatability of the one-step duplex real-time RT-PCR are summarized in Table 4.
TABLE 3.
Recovery rates of the one-step duplex real-time RT-PCR
| Target and copies/μla | Recovery rate (%) | CV (%)b |
|---|---|---|
| TBEV E gene | ||
| 100 | 169.80 | 19.59 |
| 1000 | 160.45 | 18.59 |
| 10,000 | 139.04 | 17.54 |
| Mengovirus vMC0 5′ noncoding region | ||
| 100 | 221.06 | 23.79 |
| 1000 | 177.72 | 27.39 |
| 10,000 | 180.78 | 28.45 |
Number of copies of TBEV or IPC mengovirus vMC0 standard RNA.
Calculated on the basis of assessed copy numbers.
TABLE 4.
Precision of the one-step duplex real-time RT-PCR
| Target and copies/μla | CV (%)b |
|
|---|---|---|
| Intra-assay precision | Interassay precision | |
| TBEV E gene | ||
| 100 | 25.40 | 35.05 |
| 1000 | 9.16 | 36.65 |
| 10,000 | 17.11 | 29.89 |
| Mengovirus vMC0 5′ noncoding region | ||
| 100 | 37.99 | 41.18 |
| 1000 | 24.83 | 41.47 |
| 10,000 | 26.35 | 37.61 |
Number of copies of TBEV or IPC mengovirus vMC0 standard RNA.
Calculated on the basis of assessed copy numbers.
Comparison of the manual and automated nucleic acid extraction procedures.
Analyzing buffer samples spiked with 5 × 104 to 50 viral genome equivalents/μl, we did not observe any significant difference between the two NA extraction protocols (P > 0.05, Bonferroni posttest following two-way ANOVA). On the other hand, the precipitation method showed a higher RNA recovery for tick samples spiked with 5 × 103 and 5 × 102 TBE viral genome equivalents/μl (P < 0.001, comparison of Cq values using Bonferroni posttest following two-way ANOVA) (Tables 5 and 6). PCR efficiency was not significantly influenced by the RNA purification protocol or by the amount of spiked TBEV (P > 0.05 for all pairwise comparisons, Bonferroni posttest following two-way ANOVA) (data not shown).
TABLE 5.
Cq values and SDs of TBEV RNA quantification following NA extraction using different protocols
| Sample type and amt (viral genome equivalents/μl) of spiked TBEV | Precipitation |
QIAsymphony SP system |
QIAsymphony SP system with InhibitEX |
|||
|---|---|---|---|---|---|---|
| Mean Cqa | SD | Mean Cqb | SD | Mean Cqb | SD | |
| Buffer samples | ||||||
| 5 × 104 | 23.47 | 2.25 | 22.49 | 2.25 | 20.22 | 0.63 |
| 5 × 103 | 26.77 | 1.52 | 26.65 | 1.31 | 23.80 | 0.85 |
| 5 × 102 | 30.48 | 1.67 | 30.11 | 1.45 | 27.94 | 0.82 |
| 5 × 101 | 34.64 | 2.59 | 34.47 | —c | 31.41 | 1.31 |
| Tick homogenates | ||||||
| 5 × 105 | 20.52 | 0.96 | 20.90 | 1.10 | 20.35 | 0.64 |
| 5 × 104 | 24.28 | 1.28 | 25.36 | 1.33 | 23.75 | 0.57 |
| 5 × 103 | 26.51 | 1.88 | 29.00 | 1.33 | 27.13 | 0.41 |
| 5 × 102 | 28.63 | 3.49 | 31.25 | 1.15 | 30.92 | 1.10 |
n = 24 measurements.
n = 12 measurements.
There was only one valid measurement.
TABLE 6.
P values of Bonferroni posttests comparing Cq values of TBEV RNA quantification following RNA extraction using different protocols
| Sample type and amt (viral genome equivalents/μl) of spiked TBEV |
P value |
||
|---|---|---|---|
| Precipitation vs QIAsymphony SP system | Precipitation vs QIAsymphony SP system with InhibitEX | QIAsymphony SP system vs QIAsymphony SP system with InhibitEX | |
| Buffer samples | |||
| 5 × 104 | NSa | <0.01 | NS |
| 5 × 103 | NS | <0.05 | NS |
| 5 × 102 | NS | NS | NS |
| 5 × 101 | NS | <0.01 | NS |
| Tick homogenates | |||
| 5 × 105 | NS | NS | NS |
| 5 × 104 | NS | NS | <0.05 |
| 5 × 103 | <0.001 | NS | <0.05 |
| 5 × 102 | <0.001 | <0.01 | NS |
NS, nonsignificant.
Optimization of the automated nucleic acid extraction.
To optimize the automated NA extraction protocol, we compared the RNA recovery rates with the precipitation method and the automated NA extraction protocol (QIAsymphony SP system) for tick homogenates and pure matrix samples spiked with serial dilutions of TBE virus (strain Hypr). While Complete protease inhibitor cocktail mini tablets (Invitrogen Life Technologies) and BSA did not enhance the automated NA extraction (data not shown), RNA recovery from tick homogenates was improved by supplementing D-PBS-buffer with InhibitEX. Thereby, a level of sensitivity equivalent to that of the precipitation method (down to 5 × 103 viral genome equivalents/μl) could be attained (Tables 5 and 6). For pure InhibitEX-D-PBS solutions, the performance of the optimized extraction protocol was significantly improved compared to that of the precipitation method for buffer samples spiked with 5 × 104, 5 × 103, and 50 TBE viral genome equivalents/μl. While improving RNA extraction efficiency, however, InhibitEX did not enhance PCR efficiency compared to the precipitation method and the standard automated protocol (P > 0.05 for all pairwise comparisons, Bonferroni posttest following two-way ANOVA) (data not shown). IPC mengovirus vMC0 RNA was successfully extracted using all protocols, excluding inhibition during the process of NA extraction (data not shown).
Comparison of the manual and optimized automated procedures for the analysis of naturally infected ticks.
One of the 162 investigated pooled samples tested positive for TBEV with a viral load of 5 × 105 viral genome equivalents/μl and could clearly be identified using both methods. However, the precipitation method yielded a total of 11 questionable results (confirmed to be TBEV-negative by gel electrophoresis [data not shown]). Furthermore, the automated system provided much more reproducible results than the precipitation procedure (coefficient of variation [CV] for IPC quantification of 3.7% for the automated system versus 19% for the precipitation method [data not shown]).
Exclusion of inhibitory effects of InhibitEX on TBEV propagation in PS cells.
One-way ANOVA revealed no significant difference (P = 0.4175) in virus propagation in cultures treated with different concentrations of InhibitEX (i.e., 0, 100, 200, and 1,000 μl of InhibitEX solution) (data not shown).
Screening for the presence of European subtype TBEV in ticks.
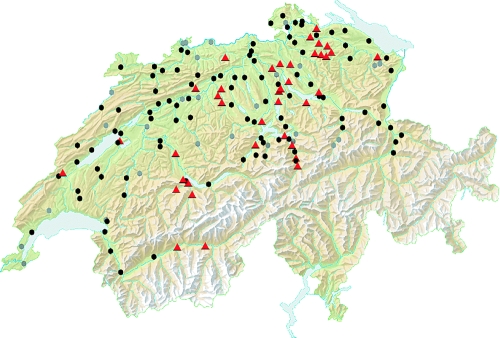
A total of 62,343 questing ticks were screened for TBE virus presence. The IPC was successfully detected in all samples, excluding false-negative test results due to inhibition. Among the 165 collection sites, TBEV was found to be endemic in 38, with a mean estimated prevalence of 0.46% (Table 7; Fig. 1). The overall mean virus prevalence was higher in female (1.21%) and male (0.74%) adults than in nymphal ticks (0.20%). Virus isolates from 63 out of the 71 PCR-positive samples could successfully be amplified on cell culture. Eight isolates did not proliferate but were detectable by real-time RT-PCR in the virus culture supernatant at unvarying low concentrations throughout the incubation period.
TABLE 7.
Maximum-likelihood estimators of TBEV prevalence in areas of endemicity in Switzerlanda
| Commune (canton)b | Altitude (m above sea level) | Sample sizec | Prevalence (%) | 95% CL (%) |
|
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Zofingen (AG) | 575 | 457 | 0.44 | 0.07 | 1.35 |
| Brittnau (AG) | 540 | 455 | 1.09 | 0.39 | 2.34 |
| Gipf-Oberfrick (AG) | 410 | 446 | 0.22 | 0.01 | 0.95 |
| Belp (BE) | 520 | 467 | 0.21 | 0.01 | 0.93 |
| Reichenbach i.K. (BE) | 720 | 384 | 0.52 | 0.09 | 1.6 |
| Erlenbach i.S. (BE) | 715 | 449 | 0.90 | 0.28 | 2.08 |
| Thun (BE) | 590 | 332 | 0.30 | 0.02 | 1.31 |
| Spiez, Rustwald (BE) | 660 | 462 | 0.43 | 0.07 | 1.33 |
| Dagmarsellen (LU) | 540 | 545 | 0.54 | 0.13 | 1.4 |
| Ebikon (LU) | 440 | 441 | 0.23 | 0.01 | 0.99 |
| Alpnach (OW) | 455 | 384 | 0.26 | 0.01 | 1.13 |
| Mörschwil (SG) | 525 | 523 | 0.38 | 0.06 | 1.17 |
| Stein am Rhein (SH) | 525 | 526 | 0.19 | 0.01 | 0.82 |
| Oensingen (SO) | 525 | 561 | 0.53 | 0.13 | 1.38 |
| Gersau (SZ) | 465 | 546 | 0.18 | 0.01 | 0.8 |
| Freienbach (SZ) | 415 | 434 | 0.44 | 0.07 | 1.36 |
| Wängi (TG) | 530 | 721 | 0.85 | 0.34 | 1.71 |
| Frauenfeld (TG) | 450 | 451 | 0.22 | 0.01 | 0.96 |
| Lommis (TG) | 680 | 678 | 0.45 | 0.11 | 1.15 |
| Aadorf (TG) | 530 | 533 | 0.19 | 0.01 | 0.82 |
| Thundorf (TG) | 620 | 504 | 0.19 | 0.01 | 0.85 |
| Silenen (UR) | 620 | 358 | 0.28 | 0.02 | 1.21 |
| Sisikon (UR) | 530 | 430 | 0.47 | 0.08 | 1.45 |
| Schattdorf (UR) | 410 | 411 | 0.49 | 0.08 | 1.49 |
| Rances (VD) | 610 | 555 | 0.36 | 0.06 | 1.10 |
| Cudrefin (VD) | 430 | 50 | 1.89 | 0.11 | 8.05 |
| Salgesch (VS) | 570 | 135 | 0.8 | 0.05 | 3.46 |
| Raron (VS) | 640 | 466 | 0.21 | 0.01 | 0.93 |
| Steinhausen (ZG) | 485 | 474 | 0.85 | 0.26 | 1.96 |
| Unterengstringen (ZH) | 470 | 599 | 0.17 | 0.01 | 0.73 |
| Aeugst am Albis (ZH) | 675 | 430 | 0.45 | 0.08 | 1.39 |
| Langnau am Albis (ZH) | 890 | 411 | 0.47 | 0.08 | 1.44 |
| Elgg (ZH) | 710 | 712 | 0.14 | 0.01 | 0.62 |
| Bassersdorf (ZH) | 470 | 723 | 0.28 | 0.05 | 0.85 |
| Rümlang (ZH) | 490 | 417 | 0.23 | 0.01 | 1.02 |
| Oberstammheim (ZH) | 440 | 90 | 1.02 | 0.06 | 4.41 |
| Rüti ZH (ZH) | 600 | 535 | 0.19 | 0.01 | 0.82 |
| Zollikon (ZH) | 540 | 461 | 0.44 | 0.07 | 1.35 |
Confidence interval = 95%.
AG, Aargau; BE, Bern; LU, Lucerne; OW, Obwalden; SG, St. Gallen; SH, Schaffhausen; SO, Solothurn; SZ, Schwyz; TG, Thurgau; UR, Uri; VD, Vaud; VS, Valais; ZG, Zug; ZH, Zurich.
Total number of analyzed ticks, i.e., nymphs, adult male, and adult female ticks. Ticks were analyzed in pools of 10 (nymphs) or 5 (adult male or adult female ticks).
FIG. 1.
TBE risk map based on tick surveillance data. Foci of endemicity are marked with red triangles, and collection sites where TBEV is not endemic are marked with circles (black, n ≥ 200; gray, n < 200).
DISCUSSION
Despite the necessity for tick surveys in national TBE surveillance systems, no standardized tool for this purpose has been available so far (8). Here we present a validated PCR-based, high-throughput analysis system for the detection of TBE viruses in ticks. Its application in a cross-sectional national surveillance study proved the validity and efficiency of the novel procedure.
High specificity of the one-step duplex real-time RT-PCR assay was achieved by designing primers and hydrolysis probes hybridizing with subtype-specific positions of the envelope gene. Interestingly, a primer and probe combination with a one-base overlap between the forward primer and probe allowed the most sensitive and specific detection of European subtype TBEV (Table 2). A U.S. patent published in 2008 (22) also describes an overlapping primer and probe yielding an efficient PCR. Although minor and particularly inefficient reactions with two Siberian subtype strains of TBEV were recorded, the specificity of the assay was confirmed by negative test results when assaying other flaviviruses as well as other tick-transmitted bacteria and parasites.
LODs of TBEV-specific RT-PCRs have previously been evaluated by assaying serial dilutions of mouse brain suspensions (34) or by preparing serial dilutions of in vitro transcripts of a cloned TBEV fragment (37, 43). We validated the analytical sensitivity but also other key issues of our one-step duplex real-time RT-PCR assay, i.e., the efficiency, linearity, effective range, accuracy, and precision, using self-constructed RNA standards. These quality criteria are a prerequisite to evaluate the performance of the real-time RT-PCR assay and thus to improve the interpretation of test results. Given that the TBEV concentration per infected tick lies between 102 and 108 PFU, with a mean virus concentration below 103 PFU (20), an LOD of 10 RNA copies/μl guarantees reliable detection of TBEV-positive ticks. However, the overestimated recovery rates obtained for the quantification of TBEV (between 139.04% and 169.80%) should be addressed by estimating the effective TBEV concentration in a tick sample.
Inhibitors are known to be present in many environmental samples and to hamper the interpretation of test results. Therefore, the addition of an internal process control at the sample preparation step is mandatory in order to monitor the presence of inhibitory substances and thus to prevent false-negative test results. Whereas most of the described TBEV-specific RT-PCR protocols do not include an IPC (28, 34, 36), the method described here allows the simultaneous quantification of European subtype TBEV and IPC mengovirus vMC0 RNA.
A special emphasis should be placed on the benefit of simultaneously extracting both RNA and DNA from tick samples using the Virus/Bacteria Midi kit (Qiagen), which enables the monitoring of any tick-borne pathogen of interest (3, 41). This is a clear advantage over protocols which, though automated, do not simultaneously extract both DNA and RNA (24) or procedures which, while purifying both RNA and DNA, constitute single-tube approaches (5).
We compared the RNA extraction efficiency of a standard protocol using the QIAsymphony SP system based on magnetic particle technology to that of the modified guanidinium thiocyanate-phenol-chloroform extraction procedure (4) using Trizol reagent. Whereas both protocols showed similar extraction efficiencies for spiked buffer samples, larger amounts of RNA (as concluded from lower Cq values) were recovered from tick samples using the precipitation method (Tables 5 and 6). The lower RNA recovery from tick samples could be explained by the presence of tick residues that could impair binding of NAs to the magnetic particle in the QIAsymphony protocol. These inhibitory effects can be reduced by addition of counteracting substances. Guanidinium thiocyanate, for instance, as a component of lysis buffers, inhibits RNA-degrading enzymes by its chaotropic effect. The performance of experiments involving virus amplification from tick homogenates as a sideline of the proposed analysis system, however, prohibits the application of such cell-toxic substances in the homogenization step. As an alternative, protocols with nontoxic substances such as InhibitEX (Qiagen) were considered. InhibitEX is an adsorption resin provided in tablet form that has previously been shown to remove inhibitors and thus improve pathogen detection in animal samples (26).
As estimated, homogenizing ticks in a buffer solution supplemented with InhibitEX improved NA extraction using the QIAsymphony SP system (Tables 5 and 6). This effect, unexpectedly, was even more considerable when processing virus dilution series prepared in pure solutions. Since pure solutions are expected not to contain any inhibitory substances, we suppose that InhibitEX directly affects the subsequent real-time RT-PCR. Further experiments quantifying serial dilutions of viral RNA and standard RNA in pure D-PBS solution or D-PBS supplemented with InhibitEX (both processed in the QIAsymphony SP system and used as diluents) (data not shown) confirmed the positive effect on the amplification step. Although there was no significant improvement of PCR efficiency, the lowered Cq values let us hypothesize that InhibitEX improves RNA accessibility, possibly by reducing the formation of secondary structures. Some components of the absorptive resin thus seem to remain in the eluate of magnetic particle-purified samples and affect the real-time RT-PCR.
Despite the improvement attributable to the use of InhibitEX tablets, the optimized protocol still performed worse than the precipitation procedure for low virus concentrations (5 × 102 virus genome equivalents/μl). However, this turned out to be of minor importance when applying the method in our large-scale survey; all TBEV-positive samples reached Cq values of between 16.84 and 24.64 (data not shown), corresponding to concentrations obviously higher than 5 × 102 viral genome equivalents/μl. In addition, the low CVs for the quantification of the IPC confirmed the high reproducibility of the automated method.
The established molecular test procedure proved to be appropriate for tick surveys. We applied the protocol in a national study on the prevalence of TBEV in ticks. The assessed mean virus prevalence of 0.46% in areas of endemicity (Table 7; Fig. 1) is in agreement with the average virus prevalence of 0.5 to 5% found in foci of endemicity in Europe (10, 25, 31). While we could confirm endemicity in several regions, no TBEV-positive ticks could be detected in some areas with confirmed human cases of TBE (national surveillance data on human disease cases from 1984 to 2008 were kindly provided by H. P. Zimmermann, Federal Office for Public Health, Switzerland). However, since areas of TBE endemicity are limited to strict regions (foci) (2, 10), the prevalence data derived from the samples in our cross-sectional study may not be generalized. They merely provide an indication of the tick infection rate in the very discrete area under investigation and are valid for only the time of the study. Nevertheless, the data are an important completion of risk assessment and monitoring of TBE in Switzerland. Interestingly, we could identify two new TBE foci in a southern region of Switzerland where TBE was so far not known to be endemic (Canton Valais, communes Salgesch and Raron). We could also detect TBEV-infected ticks in communes (Freienbach [Schwyz], Gersau [Schwyz], and Oensingen [Solothurn]) where only isolated disease cases have been reported. Furthermore, the virus could be detected in a commune (Steinhausen, Zug), where the last disease case was reported 10 years ago. All regions of endemicity were situated between 410 (Gipf-Oberfrick, Aargau) and 890 (Langnau am Albis, Zurich) meters above sea level. For a total of 33 collection sites, the reduced sample size (n < 200) could lead to a false interpretation of an area with an effective virus prevalence of 0.5% or smaller.
Sixty-three of 71 TBE virus isolates detected by PCR could successfully be propagated on cell culture. Eight isolates, however, though present at low concentrations throughout the incubation period, did not proliferate. It has been suggested that TBE viruses exist as a heterogeneous population that contains virus variants most adapted to reproduction in either ticks or mammals (32). Probably the ratio of these variants was very high in favor of tick-adapted quasispecies in these eight isolates, whereas virus reproduction in porcine kidney stable cells was inefficient. Further cultivation of the isolates would possibly select mammal-adapted quasispecies, shifting the ratio of the variants and thus enhancing virus propagation in cell culture. This hypothesis has to be confirmed by indirect immunofluorescence, which allows for the quantification of TBEV-infected cells. However, the fact that all virus isolates could be recovered in cell culture excludes false-positive PCR test results. This conclusion is also supported by the successful detection of the internal process control in all tested samples.
In summary, we have developed and validated an analytical system based on PCR which can be applied in tick-borne encephalitis surveillance. The efficiency of the method was confirmed in a tick survey, where more than 60,000 ticks were screened for the presence of TBE virus. For the present, we focused on sensitive detection of TBEV by PCR and subsequent propagation of the virus. Further work will concentrate on the molecular characterization of all TBEV isolates by specific gene sequencing reactions and whole-genome sequencing for detailed taxonomic identification. In addition, all tick samples will be analyzed for the presence of other tick-borne pathogens, including species of Rickettsia, Francisella, and Ehrlichia/Anaplasma.
Acknowledgments
We thank Lise Gern (Institute of Zoology, University of Neuchâtel, Switzerland) for her technical advice concerning tick collection and for providing strains of Borrelia. Special thanks go to Daniel Růžek (Biology Centre of the Academy of Sciences of the Czech Republic, University of South Bohemia, České Budějovice, Czech Republic) for providing TBEV strains, for helpful advice concerning viral cultures, and for valuable discussions. Many thanks go to Hans Peter Zimmermann (Federal Office for Public Health) for detailed information on human disease case reporting (1984 to 2008). Tick collection was supported by the Spiez Laboratory and the Swiss Army. We particularly thank Michael Hächler and Mario Burger for permitting this project, as well as technical officers Daniel Kohler and Patrick Müller for coordinating the collection activities. Many thanks go to Fritz Wüthrich, Björn Hagmann, Johanna Signer, and Jasmine Portmann for their excellent laboratory assistance. We also thank Marcus Doherr (Vetsuisse Faculty, University of Bern) for his advice on statistical evaluation of epidemiological data. Finally, we thank André Pignolet (Spiez Laboratory) for his support in generating the map of the tick collection sites.
This work was supported by a research grant (353000874) from the Federal Office for Civil Protection (FOPC), Federal Department of Defense, Civil Protection and Sports (DDPS).
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Abel, U., R. Schosser, and J. Suss. 1999. Estimating the prevalence of infectious agents using pooled samples: biometrical considerations. Zentralbl. Bakteriol. 289:550-563. [DOI] [PubMed] [Google Scholar]
- 2.Casati, S., L. Gern, and J. C. Piffaretti. 2006. Diversity of the population of tick-borne encephalitis virus infecting Ixodes ricinus ticks in an endemic area of central Switzerland (Canton Bern). J. Gen. Virol. 87:2235-2241. [DOI] [PubMed] [Google Scholar]
- 3.Charrel, R. N., H. Attoui, A. M. Butenko, J. C. Clegg, V. Deubel, T. V. Frolova, E. A. Gould, T. S. Gritsun, F. X. Heinz, M. Labuda, V. A. Lashkevich, V. Loktev, A. Lundkvist, D. V. Lvov, C. W. Mandl, M. Niedrig, A. Papa, V. S. Petrov, A. Plyusnin, S. Randolph, J. Suss, V. I. Zlobin, and X. de Lamballerie. 2004. Tick-borne virus diseases of human interest in Europe. Clin. Microbiol. Infect. 10:1040-1055. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Crowder, C., M. Rounds, C. Phillipson, J. Picuri, H. Matthews, J. Halverson, S. Schutzer, D. Ecker, and M. Eshoo. 2010. Extraction of total nucleic acids from ticks for the detection of bacterial and viral pathogens. J. Med. Entomol. 47:89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danielova, V., J. Holubova, M. Pejcoch, and M. Daniel. 2002. Potential significance of transovarial transmission in the circulation of tick-borne encephalitis virus. Folia Parasitol. (Praha) 49:323-325. [PubMed] [Google Scholar]
- 7.Donoso Mantke, O., S. W. Aberle, T. Avsic-Zupanc, M. Labuda, and M. Niedrig. 2007. Quality control assessment for the PCR diagnosis of tick-borne encephalitis virus infections. J. Clin. Virol. 38:73-77. [DOI] [PubMed] [Google Scholar]
- 8.Donoso Mantke, O., R. Schadler, and M. Niedrig. 2008. A survey on cases of tick-borne encephalitis in European countries. Euro Surveill. 13. [PubMed]
- 9.Duke, G. M., and A. C. Palmenberg. 1989. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J. Virol. 63:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumpis, U., D. Crook, and J. Oksi. 1999. Tick-borne encephalitis. Clin. Infect. Dis. 28:882-890. [DOI] [PubMed] [Google Scholar]
- 11.Ecker, M., S. L. Allison, T. Meixner, and F. X. Heinz. 1999. Sequence analysis and genetic classification of tick-borne encephalitis viruses from Europe and Asia. J. Gen. Virol. 80:179-185. [DOI] [PubMed] [Google Scholar]
- 12.Farrington, C. P. 1992. Estimating prevalence by group testing using generalized linear models. Stat. Med. 11:1591-1597. [DOI] [PubMed] [Google Scholar]
- 13.Federal Office for Public Health. 2009, posting date. Infectious diseases—information on tick-borne encephalitis. http://www.bag.admin.ch/themen/medizin/00682/00684/01114/index.html?lang=de.
- 14.Gresikova, M., and M. Kaluzova. 1997. Biology of tick-borne encephalitis virus. Acta Virol. 41:115-124. [PubMed] [Google Scholar]
- 15.Gresikova, M., M. Sekeyova, S. Stupalova, and S. Necas. 1975. Sheep milk-borne epidemic of tick-borne encephalitis in Slovakia. Intervirology 5:57-61. [DOI] [PubMed] [Google Scholar]
- 16.Gritsun, T. S., T. V. Frolova, A. I. Zhankov, M. Armesto, S. L. Turner, M. P. Frolova, V. V. Pogodina, V. A. Lashkevich, and E. A. Gould. 2003. Characterization of a Siberian virus isolated from a patient with progressive chronic tick-borne encephalitis. J. Virol. 77:25-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gritsun, T. S., V. A. Lashkevich, and E. A. Gould. 2003. Tick-borne encephalitis. Antiviral Res. 57:129-146. [DOI] [PubMed] [Google Scholar]
- 18.Heinz, F. X., M. S. Collett, R. H. Purcell, E. A. Gould, C. R. Howard, M. Houghton, R. J. M. Moormann, C. M. Rice, and H. J. Thiel. 2000. Family Flaviviridae, p. 859-878. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. Carstens, M. K. Estes, S. Lemon, J. Maniloff, M. A. Mayo, D. McGeogch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 19.International Scientific Working Group on TBE. 2009, posting date. Epidemiology of TBE. http://www.isw-tbe.info/tbe.aspx.
- 20.Korenberg, E. I., and Y. V. Kovalevskii. 1995. Variation in parameters affecting risk of human disease due to TBE virus. Folia Parasitol. (Praha) 42:307-312. [PubMed] [Google Scholar]
- 21.Labuda, M., O. Kozuch, E. Zuffova, E. Eleckova, R. S. Hails, and P. A. Nuttall. 1997. Tick-borne encephalitis virus transmission between ticks cofeeding on specific immune natural rodent hosts. Virology 235:138-143. [DOI] [PubMed] [Google Scholar]
- 22.Mahoney, W., N. M. J. Vermeulen, and I. Afonina. October 2008. Amplification methods. U.S. patent 20,080,248,474.
- 23.Martin, L. R., G. M. Duke, J. E. Osorio, D. J. Hall, and A. C. Palmenberg. 1996. Mutational analysis of the mengovirus poly(C) tract and surrounding heteropolymeric sequences. J. Virol. 70:2027-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moriarity, J. R., A. D. Loftis, and G. A. Dasch. 2005. High-throughput molecular testing of ticks using a liquid-handling robot. J. Med. Entomol. 42:1063-1067. [DOI] [PubMed] [Google Scholar]
- 25.Oehme, R., K. Hartelt, H. Backe, S. Brockmann, and P. Kimmig. 2002. Foci of tick-borne diseases in southwest Germany. Int. J. Med. Microbiol. 291(Suppl. 33):22-29. [DOI] [PubMed] [Google Scholar]
- 26.Padley, D. J., S. B. Lucas, and J. Saldanha. 2003. Elimination of false-negative hepatitis C virus RNA results by removal of inhibitors in cadaver-organ donor blood specimens. Transplantation 76:432-434. [DOI] [PubMed] [Google Scholar]
- 27.Pospisil, L., L. Jandasek, and J. Pesek. 1954. Isolation of new strains of meningoencephalitis virus in the Brno region during the summer of 1953. Lek List. 9:3-5. (In Czech.) [PubMed] [Google Scholar]
- 28.Puchhammer-Stockl, E., C. Kunz, C. W. Mandl, and F. X. Heinz. 1995. Identification of tick-borne encephalitis virus ribonucleic acid in tick suspensions and in clinical specimens by a reverse transcription-nested polymerase chain reaction assay. Clin. Diagn. Virol. 4:321-326. [DOI] [PubMed] [Google Scholar]
- 29.Ramakers, C., J. M. Ruijter, R. H. Deprez, and A. F. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 30.Rampas, J., and F. Gallia. 1949. Isolation of tick-borne encephalitis virus from ticks Ixodes ricinus. Cas. Lek. Cesk. 88:1179-1180. (In Czech.) [Google Scholar]
- 31.Randolph, S. E. 2001. The shifting landscape of tick-borne zoonoses: tick-borne encephalitis and Lyme borreliosis in Europe. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1045-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romanova, L., A. P. Gmyl, T. I. Dzhivanian, D. V. Bakhmutov, A. N. Lukashev, L. V. Gmyl, A. A. Rumyantsev, L. A. Burenkova, V. A. Lashkevich, and G. G. Karganova. 2007. Microevolution of tick-borne encephalitis virus in course of host alternation. Virology 362:75-84. [DOI] [PubMed] [Google Scholar]
- 33.Rudenko, N., M. Golovchenko, V. Cihlarova, and L. Grubhoffer. 2004. Tick-borne encephalitis virus-specific RT-PCR—a rapid test for detection of the pathogen without viral RNA purification. Acta Virol. 48:167-171. [PubMed] [Google Scholar]
- 34.Ruzek, D., H. Stastna, J. Kopecky, I. Golovljova, and L. Grubhoffer. 2007. Rapid subtyping of tick-borne encephalitis virus isolates using multiplex RT-PCR. J. Virol. Methods 144:133-137. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Schrader, C., and J. Suss. 1999. A nested RT-PCR for the detection of tick-borne encephalitis virus (TBEV) in ticks in natural foci. Zentralbl. Bakteriol. 289:319-328. [DOI] [PubMed] [Google Scholar]
- 37.Schwaiger, M., and P. Cassinotti. 2003. Development of a quantitative real-time RT-PCR assay with internal control for the laboratory detection of tick borne encephalitis virus (TBEV) RNA. J. Clin. Virol. 27:136-145. [DOI] [PubMed] [Google Scholar]
- 38.Sparagano, O. A., M. T. Allsopp, R. A. Mank, S. G. Rijpkema, J. V. Figueroa, and F. Jongejan. 1999. Molecular detection of pathogen DNA in ticks (Acari: Ixodidae): a review. Exp. Appl. Acarol. 23:929-960. [DOI] [PubMed] [Google Scholar]
- 39.Suss, J. 2003. Epidemiology and ecology of TBE relevant to the production of effective vaccines. Vaccine 21(Suppl. 1):S19-S35. [DOI] [PubMed] [Google Scholar]
- 40.Suss, J. 2008. Tick-borne encephalitis in Europe and beyond—the epidemiological situation as of 2007. Euro Surveill. 13:1-8. [PubMed] [Google Scholar]
- 41.Suss, J., and C. Schrader. 2004. Tick-borne human pathogenic microorganisms found in Europe and those considered nonpathogenic. I. Ticks and viruses. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 47:392-404. (In German.) [DOI] [PubMed] [Google Scholar]
- 42.Suss, J., C. Schrader, U. Falk, and N. Wohanka. 2004. Tick-borne encephalitis (TBE) in Germany—epidemiological data, development of risk areas and virus prevalence in field-collected ticks and in ticks removed from humans. Int. J. Med. Microbiol. 293(Suppl. 37):69-79. [DOI] [PubMed] [Google Scholar]
- 43.Wicki, R., P. Sauter, C. Mettler, A. Natsch, T. Enzler, N. Pusterla, P. Kuhnert, G. Egli, M. Bernasconi, R. Lienhard, H. Lutz, and C. M. Leutenegger. 2000. Swiss Army Survey in Switzerland to determine the prevalence of Francisella tularensis, members of the Ehrlichia phagocytophila genogroup, Borrelia burgdorferi sensu lato, and tick-borne encephalitis virus in ticks. Eur. J. Clin. Microbiol. Infect. Dis. 19:427-432. [DOI] [PubMed] [Google Scholar]
- 44.Zilber, L. A. 1939. Spring (spring-summer) epidemical tick-borne encephalitis. Arch. Biol. Nauk 56. (In Russian.)