Abstract
A microbial fuel cell (MFC) was inoculated with a random transposon insertion mutant library of Shewanella oneidensis MR-1 and operated with lactate as the sole fuel to select for mutants that preferentially grew in it. Agar plate cultivation of the resultant MFC enrichment culture detected an increased number of colonies exhibiting rough morphology. One such isolate, strain 4A, generated 50% more current in an MFC than wild-type MR-1. Determination of the transposon insertion site in strain 4A followed by deletion and complementation experiments revealed that the SO3177 gene, encoding a putative formyltransferase and situated in a cell surface polysaccharide biosynthesis gene cluster, was responsible for the increased current. Transmission electron microscopy showed that a layered structure at the cell surface, stainable with ruthenium red, was impaired in the SO3177 mutant (ΔSO3177), confirming that SO3177 is involved in the biosynthesis of cell surface polysaccharides. Compared to the wild type, ΔSO3177 cells preferentially attached to graphite felt anodes in MFCs, while physicochemical analyses revealed that the cell surface of ΔSO3177 was more hydrophobic. These results demonstrate that cell surface polysaccharides affect not only the cell adhesion to graphite anodes but also the current generation in MFCs.
Dissimilatory metal-reducing bacteria (DMRB) conserve energy for growth by coupling the oxidation of organic compounds to the reduction of metal compounds (29). DMRB are of great interest not only for their importance in the biogeochemical cycling of metals (25) but also for their utility in biotechnological processes, such as microbial fuel cells (MFCs) (24, 40). In recent years, the ability of many DMRB, including members of the genera Shewanella (5, 12, 20, 31), Geobacter (2), Aeromonas (34), Desulfobulbus (19), and Phodoferax (9), to generate current in MFCs has been described.
Among DMRB, Shewanella oneidensis MR-1 is one of the most extensively studied due to its metabolic versatility (28), annotated genome sequence (17), and genetic accessibility. In addition, since the first report in 1999 when this microorganism was shown to have the ability to transfer electrons to an anode without an exogenously added mediator (20), it has become a model organism for the study of microbial current generation in MFCs. Extensive studies have been performed to understand the mechanisms of extracellular electron transfer (EET) to solid materials, such as MFC anodes and metal oxides, in strain MR-1. Multiple mechanisms, including direct EET through the physical contact of bacterial cells via outer membrane (OM) cytochromes (42) and conductive nanowires (16) and mediated EET via self-produced electron shuttles such as quinones and flavins (27, 30, 39, 41), have been identified.
Although OM cytochromes and electron shuttles have been identified to play central roles in EET, it is reasonable to speculate that this complex catabolic process is also influenced by other (extra)cellular components. To date, only limited studies have been done to investigate other cellular components involved in EET (7). A useful approach for identifying unknown cellular components (and genes) associated with a particular phenotype involves the construction and screening of a random mutant library for obtaining mutants with altered phenotypes. In the present study, we constructed a random transposon (Tn) insertion mutant library of S. oneidensis MR-1 and obtained mutants with altered colony morphologies (rough morphotypes) after the selection of mutants in an MFC. Analyses of one of such mutants suggest that cell surface capsular polysaccharides affect not only the adhesion of cells to graphite anodes but also the current generation in MFCs.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. oneidensis MR-1 and Shewanella loihica PV-4 were obtained from American Type Culture Collection (ATCC). Strain PV-4 was used as a reference strain in some experiments, since we have found that MR-1 and PV-4 exhibit different behaviors in MFCs (31). Shewanella strains were cultured at 30°C in either Luria-Bertani (LB) medium or a modified lactate minimal medium (LMM) (28) comprised of 15 mM lactate, 9 mM (NH4)SO4, 5.7 mM K2HPO4, 3.3 mM KH2PO4, 2 mM NaHCO3, 30 mM HEPES (pH 7.4), 0.2 g liter−1 Casamino Acids, and 10 ml liter−1 each of an amino acid solution and a trace mineral solution. The amino acid solution contained 2 g liter−1 each of l-glutamic acid, l-arginine, and dl-serine, while the trace mineral solution contained 100 mM MgSO4·7H2O, 10 mM CaCl2·2H2O, 6.72 mM EDTA, 5.66 mM H3BO3, 1 mM NaCl, 5.4 mM FeSO4·7H2O, 0.5 mM CoCl2·6H2O, 0.5 mM NiCl2·6H2O, 0.39 mM Na2MoO4·4H2O, 0.15 mM Na2SeO3, 0.13 mM MnCl2·4H2O, 0.1 mM ZnSO4·7H2O, and 0.02 mM CuSO4·5H2O. Escherichia coli strains (31) were routinely cultured in LB medium at 37°C. The E. coli mating strain (WM6026) required 2,6-diaminopimelic acid (DAP) at 100 μg ml−1 for growth. When necessary, 15 μg ml−1 gentamicin (Gm) or 50 μg ml−1 kanamycin (Km) was added to culture media. Agar plates contained 1.6% Bacto agar (Difco).
MFC operation.
Current generation was evaluated using a single-chamber MFC equipped with a graphite felt anode (50 cm2; GF-80-3F [Sohgoh Carbon]) and an air cathode (approximately 20 cm2, 0.7 mg platinum cm−2, and four polytetrafluoroethylene layers) that was made as described elsewhere (11). Bacterial cells were inoculated at an initial optical density at 600 nm (OD600) of 0.005 into the MFC chamber containing 450 ml of LMM supplemented with 10 mM lactate. Upon depletion of lactate, a 4.5 M stock solution of lactate was injected into the reactor to raise the concentration of lactate back to 10 mM. Other MFC operational conditions were as described previously (31). The anode and cathode were connected with electric wires and an external resistor (100 Ω), and the voltage across the resistor was measured using a voltage data logger (HA-1510 [Graphtec]). Current (I [A]) was calculated using the equation, I = E/R (where E [V] is the cell voltage and R [Ω] is the resistance), while current density (A cm−2) was calculated using the anode projection area (50 cm2). A polarization curve was generated using a potentiostat (HSV-100; Hokuto Denko), from which the maximum power density (Pmax [W cm−2]) and short-circuit current (Isc [A cm−2]) were obtained as described elsewhere (40). Reproducibility was evaluated in at least three independent MFC operations.
Transposon mutagenesis and selection of mutants.
Transposon (Tn) mutagenesis of S. oneidensis MR-1 was carried out by filter mating with E. coli WM6026 harboring the suicide plasmid pBSL-T7-KAN2, containing mini-Tn10Kmr. Plasmid pBSL-T7-KAN2 was constructed as follows. The DNA fragment containing a Km resistance (Kmr) gene was amplified by PCR from Kan2-T7 transposon template DNA (Epicentre) using primers T7kan-2-SacI-F and T7kan-2-SacI-R (see Table S1 in the supplemental material) and was then ligated into vector pT7Blue (Novagen). After the nucleotide sequence of the resultant plasmid was confirmed, the insert was excised by SacI digestion and ligated with the large fragment of the SacI-digested plasmid pBSL180 (1). S. oneidensis MR-1 and E. coli WM6026(pBSL-T7-KAN2) were grown in LB medium until the stationary phase, and cells collected from 1 ml of each culture were suspended in 200 μl of 10 mM MgSO4 after being washed twice with the same solution. These cell suspensions were mixed, and cells were collected on filter membranes (0.2-μm pore size, mixed cellulose ester [Advantec]). After the cells on the membranes were grown on LB agar plates containing DAP at 30°C for 8 h, they were resuspended in 10 mM MgSO4, and the suspension was spread on LB plates to determine the CFU per ml.
LMM (1 liter) was inoculated with a cell suspension containing 105 Tn mutants, and they were cultivated aerobically until the OD600 reached 0.54 (5 × 108 CFU ml−1). Part of the culture, containing 4 × 108 CFU, was introduced into an MFC reactor, which was operated until a total of 40 mM lactate (four injections of the lactate solution) was completely consumed. Cells were collected from both the supernatant and the anode biofilm in the MFC reactor and spread onto LB plates for colony isolation.
Inverse PCR.
Total DNA of S. oneidensis mutant 4A was extracted using DNAzol reagent (Invitrogen) according to the manufacturer's instructions. After being digested with HincII, 100 ng of the DNA was self-ligated using the Ligation High kit (Toyobo) and used as a template for PCR using primers KAN-2 FP-1 and KAN-2 RP-1 (see Table S1 in the supplemental material) to amplify the DNA region flanking the Tn insert. The amplified DNA fragment was purified by agarose gel electrophoresis and subjected to sequence analysis.
Gene disruption and complementation.
The in-frame disruption of the SO3177 gene in strain MR-1 was performed using suicide plasmid pSMV-10 and a two-step homologous recombination method as described previously (31, 37). Briefly, a 1.6-kb fusion product, consisting of an upstream sequence of the SO3177 gene (766 bp), replacement sequence (82 bp), and downstream sequence (809 bp), was constructed by PCR and in vitro extension using primers listed in Table S1 in the supplemental material. This fusion product was ligated into pSMV10 at the SpeI site. The resultant plasmid, pSMV-3177, was introduced into MR-1 by filter mating with E. coli WM6026. Transconjugants were selected on LB plates containing Km, and these single-crossover clones were further cultivated for 20 h in LB medium lacking the antibiotics. The cultures were then spread onto LB plates containing 10% (wt/vol) sucrose to isolate Km-sensitive double-crossover mutants. The disruption of the SO3177 gene in these strains was confirmed by PCR. The resultant mutant strain in which the SO3177 gene was in-frame disrupted was designated ΔSO3177.
Plasmid pBBR1-3177 was constructed for the complementation of the SO3177 gene. The SO3177 gene was amplified using primers SO3177-F-HindIII and SO3177-R-XbaI (see Table S1 in the supplemental material), the PCR product obtained was digested using HindIII and XbaI, and the fragment was ligated into HindIII-XbaI-digested pBBR1MCS-5 (23). The resultant plasmid, pBBR1-3177, was introduced into ΔSO3177 cells by filter mating with E. coli WM6026.
TEM.
Shewanella cells were cultivated aerobically in LB medium. For preparing ruthenium red-stained samples (35), bacterial cells were first harvested by centrifugation, fixed with 1.25% glutaraldehyde, and stained with 0.05% ruthenium red in 0.1 M cacodylate buffer (pH 6.5) for 5 h at 4°C. Cells were then washed with the same buffer and postfixed in a 1% osmium tetroxide solution containing 0.05% ruthenium red for 2 h at 4°C. After the cells were rinsed in the cacodylate buffer, they were embedded in 1.5% agarose before dehydration using a graded ethanol series. For the preparation of samples without ruthenium red staining, bacterial pellets were fixed with 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2) at 4°C for 4 h and postfixed in 1% osmium tetroxide for 90 min at 4°C. Fixed cells were suspended in 1% aqueous uranyl acetate for 1 h at room temperature and embedded in 1.5% agarose before dehydration using a graded ethanol series. In both cases, the dehydrated blocks were embedded in Spurr resin. Ultrathin sections were prepared using an ultramicrotome (Ultracut-N; Leichert-Nissei), mounted on copper grids, and stained with uranyl acetate and lead citrate. Images of sections were obtained by transmission electron microscopy (TEM) at 75 kV (H-7000; Hitachi).
SDS-PAGE of LPS.
For analyzing lipopolysaccharides (LPS), cells grown overnight on an LB plate were subjected to proteinase K digestion followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining as described elsewhere (15, 18). For SDS-PAGE, 15% polyacrylamide gels were run in Tris-Tricine buffer (100 mM Tris, 50 mM Tricine, 0.1% SDS) for 60 to 90 min at 150 V until the loading dye (bromophenol blue) reached the bottom of the gel.
Protein assay of cells in MFC reactors.
To determine the protein content of planktonic cells, 1 ml of supernatant was collected from an MFC reactor. After centrifugation at 15,000 × g for 5 min, the pellet was resuspended in 100 μl of B-PER II bacterial protein extraction reagent (Pierce) to solubilize the proteins according to the manufacturer's instructions. The protein concentration was determined using the Micro bicinchoninic acid (BCA) protein assay kit (Pierce) according to the manufacturer's instructions, and the total protein content in the MFC reactor was calculated using the supernatant volume of 450 ml. To determine the protein content of cells attached to the MFC graphite felt anode, a round piece of anode with a diameter of 4.8 mm (0.181 cm2) was punched out from the anode and soaked in 300 μl of B-PER II. After vortexing for 2 min, the protein concentration was determined as described above and the total protein content on the anode was calculated based on an anode area of 50 cm2. To determine the protein content of cells attached to the air cathode, the entire biofilm was removed from the air cathode by scraping with a flat spurtle and uniformly suspended in 100 ml of phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4, pH 7.3). For the protein assay, 1 ml of this suspension was centrifuged at 15,000 × g for 5 min, and the pellet was resuspended in 300 μl of B-PER II. All measurements were performed in triplicate, and data were statistically analyzed by analysis of variance (ANOVA) in combination with the Ryan multiple-comparison test (http://www.hju.ac.jp/∼kiriki/anova4/) at a significance level of 0.05.
Adhesion to graphite felt.
S. oneidensis MR-1, the generated mutant strains, and S. loihica PV-4 were cultivated overnight aerobically in LB medium. These cells were washed with an LMM buffer (LMM without lactate, amino acids, Casamino Acids, and mineral solutions) and then resuspended in the same buffer at an OD600 of 0.3 in plastic test tubes. A round piece of graphite felt (0.181 cm2) was placed in each cell suspension, and the test tubes were incubated at 30°C with agitation (180 rpm). After a 2-h incubation, the protein contents in the cell suspensions and those of graphite-attached cells were determined as described above.
Zeta potentials and hydrophobicity assay.
For zeta potential measurements, Shewanella cells were cultured aerobically in LB medium and suspended in 0.01 M NaCl-0.05 M HEPES buffer (pH 7.4) at an OD600 of 0.1. Electrophoretic mobilities of the cells in these suspensions were measured using an ELSZ series zeta potential and particle size analyzer (Photal) and converted to zeta potentials by use of Smoluchowski's approximation (32). Hydrophobicity of cells was analyzed according to a method described elsewhere (6, 14) with slight modifications. Briefly, Shewanella cells were suspended in 2.4 ml of 0.15 M NaCl at an OD600 of 0.3 (approximately 3 × 108 CFU ml−1) and vortexed for 60 s in the presence of 0.4 ml hexadecane. The mixture was allowed to stand for 15 min to ensure that the two phases were completely separated before a sample (1 ml) was carefully removed from the aqueous phase for measuring the OD600. The percentage of cells transferred to the hexadecane phase was subsequently calculated using the equation affinity (%) = 100 × [1 − (A/A0)], where A0 is the OD600 of the bacterial suspension before mixing with hexadecane and A is the OD600 after mixing. Each measurement was performed in triplicate.
Autoaggregation assay.
An autoaggregation assay was performed as described elsewhere (14). Briefly, S. oneidensis MR-1 and the mutant strains were cultured overnight in LB medium, and cells were washed twice with LMM buffer. The cells were again suspended in LMM buffer at an OD600 of 0.7. Each cell suspension in a total volume of 2 ml was incubated in a test tube at 25°C without shaking, and the OD600 was measured using a spectrophotometer (DU800; Beckman).
RESULTS
Selection of mutants in MFC.
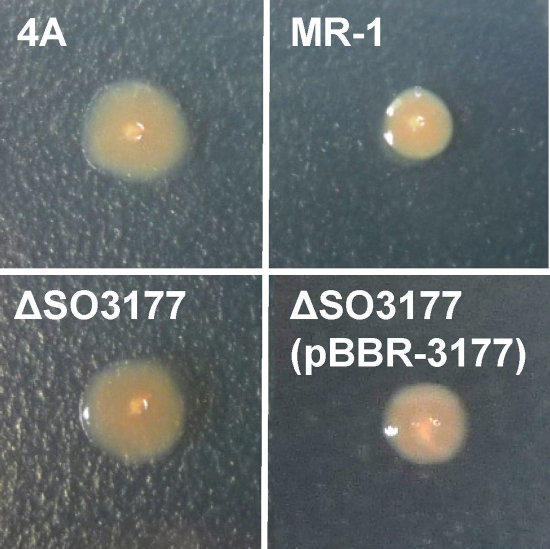
A library of S. oneidensis MR-1 random transposon mutants containing greater than 105 cells was introduced into an MFC reactor to cultivate mutants. Supplementation of the MFC with lactate (10 mM) resulted in generation of current (Fig. 1), and the trend and amount of current were similar to those for MFCs operated with wild-type MR-1 (data not shown). After the fourth addition of lactate was completely depleted, both planktonic and anode-attached cells were collected and spread onto LB plates for colony isolation. We found that substantial numbers of colonies (approximately 4% of total colonies from planktonic and anode-attached cell fractions) exhibited rough morphologies. One such mutant isolate was designated strain 4A, whose colony shape was distinct from the regular, round, and smooth morphologies exhibited by wild-type MR-1 colonies (Fig. 2). The number of rough colonies in the original mutant library was very small (less than 0.1%), indicating that rough morphotypes preferentially proliferated in the MFC. We were therefore interested in identifying disrupted genes in these rough colonies. Furthermore, since colony morphology is known to be influenced by cell surface structures (3, 14, 22), we also analyzed cell morphology and surface properties.
FIG. 1.
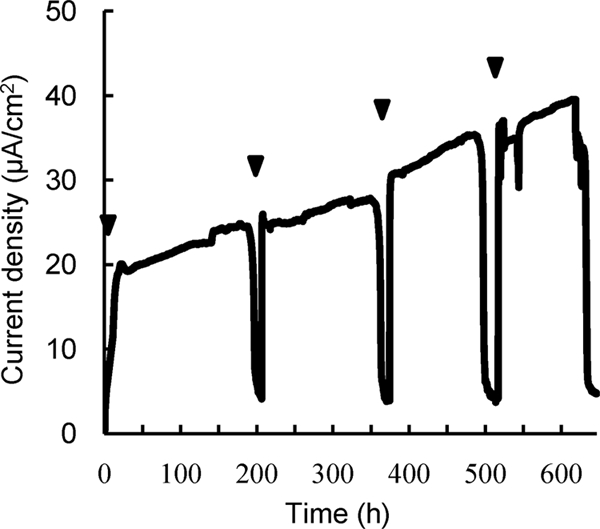
Current generation in an MFC inoculated with a random transposon mutant library of S. oneidensis MR-1 and used to select for mutants. Arrowheads indicate time points at which lactate (10 mM) was injected into the MFC.
FIG. 2.
Colony morphologies of S. oneidensis MR-1 and mutant strains on LB plates.
Strain 4A.
When an MFC was inoculated with strain 4A, it produced 1.4- to 1.5-fold more current than wild-type MR-1 (Fig. 3 A). To evaluate MFC performance, polarization and power curves were determined for the MFCs containing strain 4A or wild-type cells after the third addition of lactate (Fig. 3B and C). The Pmax and Isc for the 4A MFC (6.53 μW cm−2 and 44.2 μA cm−2, respectively) were approximately 50% higher than those for wild-type MR-1 (4.34 μW cm−2 and 27.5 μA cm−2, respectively).
FIG. 3.
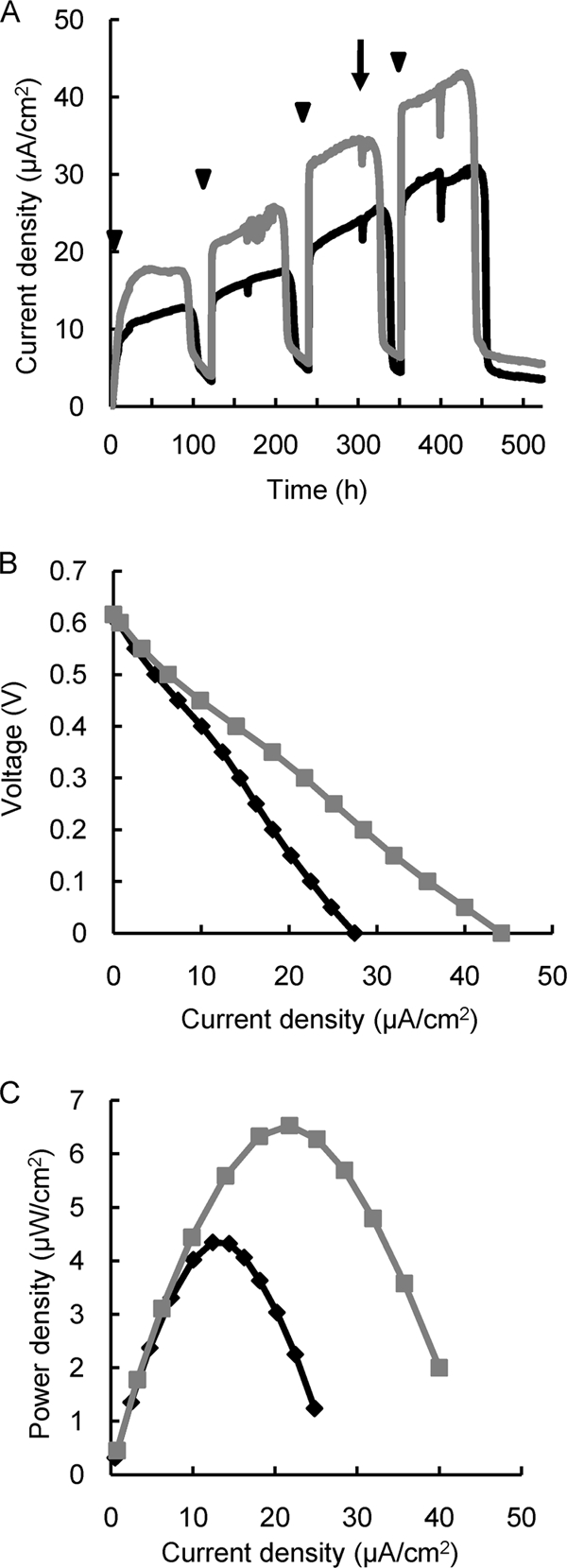
Current generation by mutant 4A and wild-type MR-1 in single-chamber MFCs. (A) Comparison of current density between MR-1 (black line) and 4A (gray line). (B and C) Polarization (B) and power (C) curves were measured at the time point indicated by the arrow. Arrowheads indicate time points at which lactate (10 mM) was injected into the MFCs. Reproducibility was examined in at least three independent operations, and typical data are shown.
To determine the transposon insertion site in strain 4A, we performed inverse PCR and sequence analysis of the resulting product. It was determined that the transposon had inserted into the SO3177 gene, which, based on the annotated genome sequence, encodes a protein of unknown function. The deduced amino acid sequence of SO3177 had the closest homology to a hypothetical protein (SUN1525) in Sulfurovum sp. strain NBC37-1 (50% identity), and a BLAST search suggested that it contained a formyltransferase domain. In the genome of strain MR-1, the SO3177 gene is located in a gene cluster, which codes for proteins possibly involved in the biosynthesis of cell surface polysaccharides (see Fig. S1 in the supplemental material).
Deletion and complementation of the SO3177 gene.
To confirm that the disruption of the SO3177 gene was responsible for the rough morphotype of and enhanced current generation by strain 4A, an in-frame deletion mutant of SO3177 (designated ΔSO3177) and a plasmid-complemented strain, ΔSO3177(pBBR-3177), were constructed. When cultivated on LB plates, ΔSO3177 displayed the identical rough morphotype as observed for strain 4A (Fig. 2), while ΔSO3177(pBBR-3177) formed smooth colonies similar to those of the wild-type strain (Fig. 2). The abilities of these mutants to generate current in MFCs were also analyzed and compared with that of wild-type MR-1. After the third addition of lactate, the ΔSO3177 and ΔSO3177(pBBR-3177) MFCs achieved, on average, Pmax values that were 148% and 92%, respectively, of that for a wild-type MFC. These results demonstrate that there is a relationship between the observed rough colony morphology in ΔSO3177 and the improved current generation in MFCs.
TEM and SDS-PAGE of cell surface polysaccharides.
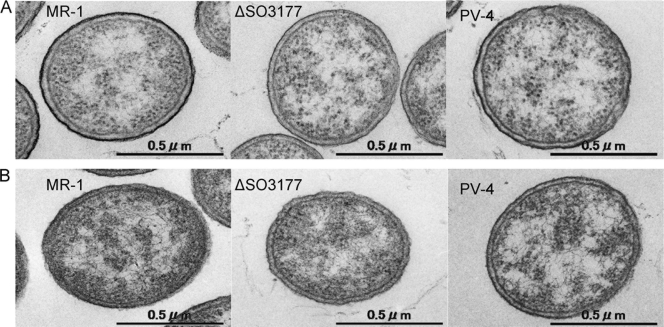
As colony morphology is known to be influenced by cell surface properties and the SO3177 gene was situated in a putative cell surface polysaccharide biosynthesis gene cluster, cell surface polysaccharides of the wild-type MR-1 and ΔSO3177 were examined by TEM and SDS-PAGE. Also included in these analyses for comparison was Shewanella loihica PV-4, since this strain exhibits unique behavior in an MFC that differs from that of strain MR-1 (31). When cells were stained with ruthenium red (a dye to stain polysaccharides) and visualized by TEM, bold lines appeared surrounding both MR-1 and PV-4 cells but not those of ΔSO3177 (Fig. 4). These data indicated that the disruption of the SO3177 gene impaired cell surface polysaccharides, such as capsular polysaccharides and lipopolysaccharides (LPS). We therefore compared the LPS structures of the wild type, 4A (SO3177::Tn), and ΔSO3177 by SDS-PAGE analysis (21, 38). However, no difference was detected among these strains (see Fig. S2 in the supplemental material), which indicated that the SO3177 gene is involved in the biosynthesis of capsular polysaccharides.
FIG. 4.
Transmission electron micrographs of S. oneidensis MR-1 and ΔSO3177 and S. loihica PV-4 with (A) and without (B) ruthenium red staining.
Cell distribution in MFCs.
In the single-chamber MFC used in this study, Shewanella cells could be localized to three areas of the reactors, i.e., anode biofilms, planktonic cells, and cathode biofilms. In the MFC containing ΔSO3177, the turbidity of the supernatant was much lower than that in the wild-type MFC (data not shown), suggesting that more mutant cells attached to the electrodes. In order to examine this hypothesis, wild-type MR-1, ΔSO3177, and ΔSO3177(pBBR-3177) cells from each area of the MFCs were collected after the third addition of lactate (after approximately 400 h of operation), and the protein contents in these samples were compared (Fig. 5). In the ΔSO3177 MFC, the amount of cells attaching to the anode graphite felt was 67% higher than that of wild-type cells, while the amount of ΔSO3177 cells in the supernatant was 36%. In contrast, the complemented strain ΔSO3177(pBBR-3177) showed a localization pattern similar to that of wild type. The total protein contents, which represented the sums of the protein contents in the three MFC areas, were not significantly different among the three strains. These results indicate that the disruption of SO3177 enhanced the ability of cells to adhere onto the anode graphite felt.
FIG. 5.
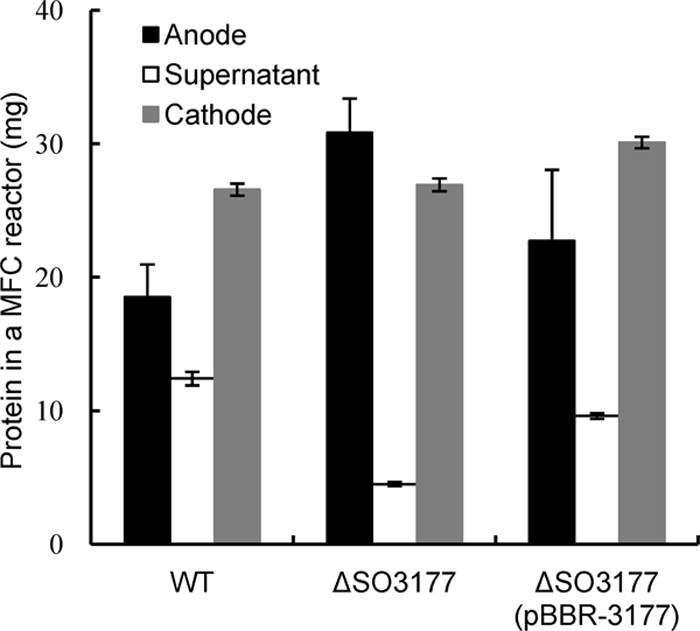
Protein contents of the anode biofilms, supernatant, and cathode biofilms in single-chamber MFCs, showing the distributions of the wild-type (WT), ΔSO3177, and ΔSO3177(pBBR-3177) cells. Error bars represent standard deviations of means that were calculated from assays performed in triplicate at a minimum.
Cell surface physicochemical characteristics.
The rough colony morphotype of the ΔSO3177 mutant suggested that cell surface physicochemical properties, such as charge and hydrophobicity, were affected by this mutation. In addition, such cell surface properties were assumed to be related to the cell adhesiveness to anode graphite felts. Thus, the cell surface charges, hydrophobicity, and adhesiveness to graphite felts of wild-type MR-1, 4A, ΔSO3177, and S. loihica PV-4 were measured and compared (Table 1). The SO3177 mutants (4A and ΔSO3177) were more electronegative and hydrophobic than wild-type MR-1. In addition, the two mutants were more adhesive to graphite felt, which was also observed in the MFC experiments. These results clearly demonstrate that capsular polysaccharides affect the physicochemical cell surface properties and adhesiveness to graphite felt. For S. loihica PV-4, the adhesiveness to graphite felt was nearly identical to that of wild-type MR-1. PV-4 cells were strongly electronegative but hydrophilic, suggesting that hydrophobic interaction is important for the adhesion of Shewanella cells to graphite felt.
TABLE 1.
Cell surface physicochemical properties of and adhesion to graphite felt by Shewanella cells
| Species and strain | Zeta potential (mV)a | Affinity to hexadecane (%)b | Adhesion to graphite felt (%)c |
|---|---|---|---|
| S. oneidensis | |||
| MR-1 | −3.1 ± 0.8 | 10.9 ± 0.1 | 26.6 ± 1.9 |
| 4A | −19.7 ± 1.2 | 31.9 ± 4.5 | NDd |
| ΔSO3177 | −16.4 ± 0.4 | 30.2 ± 4.5 | 55.2 ± 3.0 |
| ΔSO3177(pBBR-3177) | −5.1 ± 0.9 | ND | 23.2 ± 2.7 |
| S. loicha PV-4 | −23.0 ± 0.6 | 4.8 ± 1.9 | 22.7 ± 1.1 |
Measured in 0.01 M NaCl-0.05 M HEPES (pH 7.4). Values are means ± standard deviations (n = 6).
Percentage of cells partitioned into the hydrophobic solvent hexadecane. Values are means ± standard deviations (n = 3).
Percentage of cells adhered on a piece of graphite felt. Values are means ± standard deviations (n = 3).
ND, not determined.
The cell surface characteristics of these Shewanella strains were also assessed by the autoaggregation assay (Fig. 6 A and B). Figure 6A presents a photograph of cell suspensions in the autoaggregation assay (2 h after the incubation began), showing that the SO3177 disruptant (4A and ΔSO3177) cells were precipitated to larger extents than wild-type MR-1 and ΔSO3177(pBBR-3177) cells. Figure 6B shows time courses of OD600 in the liquid phases, indicating that the decreases in OD600 due to autoaggregation were much faster for the 4A and ΔSO3177 cells than for wild-type MR-1 and ΔSO3177(pBBR-3177) cells. These results suggest that the cell-cell adhesion is also enhanced by the disruption of the SO3177 gene.
FIG. 6.
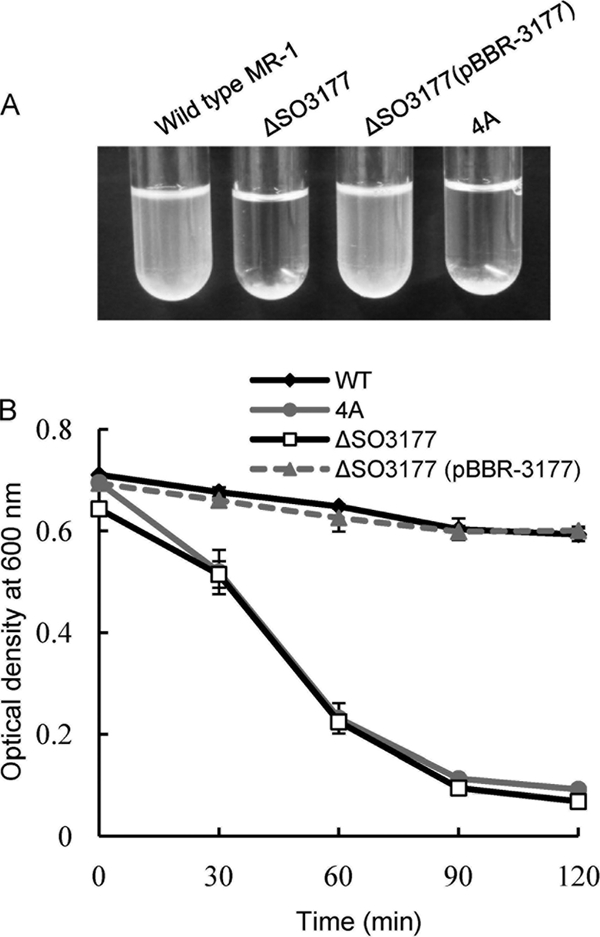
Autoaggregation of the wild-type MR-1 and two mutant strains. (A) Typical appearances of test tubes at 2 h after the incubation began. (B) Time courses of OD600. Error bars represent standard deviations that were calculated from triplicate assays.
DISCUSSION
In this study, we isolated and characterized a novel S. oneidensis MR-1 mutant, ΔSO3177, that generated approximately 50% more current in an MFC than the wild-type strain. The analyses of this mutant revealed that it is impaired in the biosynthesis of cell surface capsular polysaccharides, indicating that capsular polysaccharides of MR-1 exert a negative influence on current generation. Although capsular polysaccharides are known to aid bacterial survival under stressful environmental conditions (10, 26, 33), such components can exert negative effects on biotechnological applications.
In order to understand the mechanisms by which capsular polysaccharides affected the current generation in MFCs, the cell distribution in MFCs and cell surface physicochemical properties were examined. Compared to wild-type MR-1, the ΔSO3177 cells attached with higher frequency to the graphite felt anode (Fig. 5), cell autoaggregation was enhanced (Fig. 6), and the surface of the ΔSO3177 cells was more hydrophobic and electrochemically negative (Table 1). Taken together, these results support the idea that capsular polysaccharides modify cell surface physicochemical properties, resulting in the altered behaviors in MFCs and current generation. Although Shewanella cells have been reported to produce water-soluble electron-shuttling compounds, such as flavins and quinones (27, 30, 39), EET mediated by such electron-shuttling compounds can be limited by their diffusion when cells are physically distant from the solid electron acceptors. Moreover, electron-shuttling compounds are known to adsorb onto carbon electrodes and bacterial cells, especially in matured biofilms (27). It is therefore likely that cell adhesion to anodes could be an important event for efficient current generation in MFCs, even for bacteria such as Shewanella that can employ indirect EET mediated by electron shuttles.
Bacterial adhesion to solid surfaces and subsequent biofilm formation have been the focus of numerous investigations, particularly for those species involved in biofilm-mediated diseases (3, 14) and plant-microbe interactions (13, 36). These studies have demonstrated that cell surface (capsules and LPS) and/or extracellular polysaccharides strongly affect surface physicochemical properties and adhesiveness to solid surfaces (3, 14). In Shewanella, the physicochemical cell surface properties and abilities to adhere to hematite (α-Fe2O3) were compared among several different strains (22), demonstrating that strains expressing rough LPS with no capsule are more hydrophobic and electronegative than those possessing smooth LPS or capsules. Furthermore, cell adhesion to hematite was enhanced for strains possessing rough LPS without capsules (22). In another study, a mutant of Shewanella alga BrY deficient in the adhesion to amorphous Fe(III) oxide was isolated and characterized (8). Although this mutant was more hydrophilic than the wild type, the cell surface charge did not differ between them, suggesting that hydrophobic interactions are the major factor controlling the adhesion of bacteria cells to amorphous Fe(III) oxide. We also found that wild-type MR-1 was slow in reducing amorphous Fe(III) oxide compared to ΔSO3177 (our unpublished results).
The disruption of the SO3177 gene resulted in deficient biosynthesis of capsular polysaccharides. The SO3177 gene is located within a large gene cluster (approximately 38 kb) consisting of 35 annotated open reading frames (ORFs) from SO3160 to SO3194 (see Fig. S1 in the supplemental material). This gene cluster includes ORFs annotated as genes encoding polysaccharide-biosynthesis proteins, such as sugar epimerases (SO3189 and SO3173) and sugar transferases (SO3180, SO3176, SO3174, and SO3172). In addition, several ORFs exhibit relatively high homologies to genes in E. coli responsible for O-antigen biosynthesis, such as rfbB (75% amino acid identity to SO3188), rfbA (66% to SO3186), and rfbC (52% to SO3160), although MR-1 lacks O-antigen polysaccharides (38). Analyses of the deduced amino acid sequence of SO3177 revealed the presence of a conserved formyltransferase domain. In E. coli, it has been reported that the N-terminal domain of ArnA acts as a formyltransferase that is involved in the synthesis of a precursor of 4-amino-4-deoxy-l-arabinose required for the modification of LPS (4), suggesting that formyltransferases are involved in the biosynthesis of cell surface structures. However, since the homology between the N-terminal domain of ArnA and SO3177 is low (28% amino acid identity), it is difficult to conclusively annotate the SO3177 gene. Interestingly, there is no homologue of SO3177 in other Shewanella species, which may be related to the unique cell surface physicochemical properties displayed by MR-1, including very low surface charge, compared to other Shewanella strains (22).
In conclusion, the results of the present study suggest that capsular polysaccharides alter bacterial cell surface properties, resulting in an increased ability of the capsular polysaccharide-deficient mutant to adhere to a graphite anode and to generate current in an MFC. However, in addition to cell adhesiveness, there may also exist direct effects of the capsular polysaccharide deficiency on current generation; namely, electrically nonconductive capsular polysaccharides can interfere with the contact of outer membrane cytochromes to anodes and direct EET via them. Further studies are necessary to address this possibility. Previous studies have mainly focused on the electron transfer components directly involved in EET, such as OM cytochromes, to understand and engineer EET (and current generation in MFCs) by S. oneidensis MR-1 (5, 12, 31). Based on the results of this study, however, we suggest that cell surface engineering is also a possible scheme to improve bacterial current generation in MFCs.
Supplementary Material
Acknowledgments
This work was supported by the Exploratory Research for Advanced Technology (ERATO) program of the Japanese Science and Technology Agency (JST).
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene 160:59-62. [DOI] [PubMed] [Google Scholar]
- 2.Bond, D. R., and D. R. Lovley. 2003. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 69:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, A., and A. M. Chakrabarty. 1995. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J. Ind. Microbiol. 15:162-168. [DOI] [PubMed] [Google Scholar]
- 4.Breazeale, S. D., A. A. Ribeiro, A. L. McClerren, and C. R. Raetz. 2005. A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-l-arabinose. Identification and function of UDP-4-deoxy-4-formamido-l-arabinose. J. Biol. Chem. 280:14154-14167. [DOI] [PubMed] [Google Scholar]
- 5.Bretschger, O., A. Obraztsova, C. A. Sturm, I. S. Chang, Y. A. Gorby, S. B. Reed, D. E. Culley, C. L. Reardon, S. Barua, M. F. Romine, J. Zhou, A. S. Beliaev, R. Bouhenni, D. Saffarini, F. Mansfeld, B. H. Kim, J. K. Fredrickson, and H. Nealson. 2007. Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl. Environ. Microbiol. 73:7003-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briandet, R., T. Meylheuc, C. Maher, and M. N. Bellon-Fontaine. 1999. Listeria monocytogenes Scott A: cell surface charge, hydrophobicity, and electron donor and acceptor characteristics under different environmental growth conditions. Appl. Environ. Microbiol. 65:5328-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns, J. L., B. R. Ginn, D. J. Bates, S. N. Dublin, J. V. Taylor, R. P. Apkarian, S. Amaro-Garcia, A. L. Neal, and T. J. DiChristina. 2010. Outer membrane-associated serine protease involved in adhesion of Shewanella oneidensis to Fe(III) oxides. Environ. Sci. Technol. 44:68-73. [DOI] [PubMed] [Google Scholar]
- 8.Caccavo, F., P. C. Schamberger, K. Keiding, and P. H. Nielsen. 1997. Role of hydrophobicity in adhesion of the dissimilatory Fe(III)-reducing bacterium Shewanella alga to amorphous Fe(III) oxide. Appl. Environ. Microbiol. 63:3837-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri, S. K., and D. R. Lovley. 2003. Electricity generation by direct oxidation of glucose in mediatorless microbial fuel cells. Nat. Biotechnol. 21:1229-1232. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J., S. M. Lee, and Y. Mao. 2004. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157:H7 to osmotic and oxidative stress. Int. J. Food Microbiol. 93:281-286. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, S., H. Liu, and B. E. Logan. 2006. Increased power generation in a continuous flow MFC with advective flow through the porous anode and reduced electrode spacing. Environ. Sci. Technol. 40:2426-2432. [DOI] [PubMed] [Google Scholar]
- 12.Coursolle, D., D. B. Baron, D. R. Bond, and J. A. Gralnick. 2009. The Mtr respiratory pathway is essential for reducing flavins and electrodes in Shewanella oneidensis. J. Bacteriol. 192:467-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danhorn, T., and C. Fuqua. 2007. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61:401-422. [DOI] [PubMed] [Google Scholar]
- 14.Davey, M. E., and M. J. Duncan. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J. Bacteriol. 188:5510-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fomsgaard, A., M. A. Freudenberg, and C. Galanos. 1990. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J. Clin. Microbiol. 28:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorby, Y. A., S. Yanina, J. S. McLean, K. M. Rosso, D. Moyles, A. Dohnalkova, T. J. Beveridge, I. S. Chang, B. H. Kim, K. S. Kim, D. E. Culley, S. B. Reed, M. F. Romine, D. A. Saffarini, E. A. Hill, L. Shi, D. A. Elias, D. W. Kennedy, G. Pinchuk, K. Watanabe, S. Ishii, B. Logan, K. H. Nealson, and J. K. Fredrickson. 2006. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. U. S. A. 103:11358-11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 18.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes, D. E., D. R. Bond, and D. R. Lovley. 2004. Electron transfer by Desulfobulbus propionicus to Fe(III) and graphite electrodes. Appl. Environ. Microbiol. 70:1234-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, B. H., H. J. Kim, M. S. Moon, and D. H. Park. 1999. Direct electrode reaction of Fe(III)-reducing bacterium Shewanella putrefaciens. J. Microbiol. Biotechnol. 9:127-131. [Google Scholar]
- 21.Korenevsky, A. A., E. Vinogradov, Y. Gorby, and T. J. Beveridge. 2002. Characterization of the lipopolysaccharides and capsules of Shewanella spp. Appl. Environ. Microbiol. 68:4653-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korenevsky, A., and T. J. Beveridge. 2007. The surface physicochemistry and adhesiveness of Shewanella are affected by their surface polysaccharides. Microbiology 153:1872-1883. [DOI] [PubMed] [Google Scholar]
- 23.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, 2nd, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 24.Logan, B. E., B. Hamelers, R. Rozendal, U. Schröder, J. Keller, S. Freguia, P. Aelterman, W. Verstraete, and K. Rabaey. 2006. Microbial fuel cells: methodology and technology. Environ. Sci. Technol. 40:5181-5192. [DOI] [PubMed] [Google Scholar]
- 25.Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219-286. [DOI] [PubMed] [Google Scholar]
- 26.Mao, Y., M. P. Doyle, and J. Chen. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:3811-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsili, E., D. B. Baron, I. D. Shikhare, D. Coursolle, J. A. Gralnick, and D. R. Bond. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U. S. A. 105:3968-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers, C. R., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 29.Nealson, K. H., and B. L. Cox. 2002. Microbial metal-ion reduction and Mars: extraterrestrial expectations? Curr. Opin. Microbiol. 5:296-300. [DOI] [PubMed] [Google Scholar]
- 30.Newman, D. K., and R. Kolter. 2000. A role for excreted quinones in extracellular electron transfer. Nature 405:94-97. [DOI] [PubMed] [Google Scholar]
- 31.Newton, G. J., S. Mori, R. Nakamura, K. Hashimoto, and K. Watanabe. 2009. Analyses of current-generation mechanisms of Shewanella loihica PV-4 and Shewanella oneidensis MR-1 in microbial fuel cells. Appl. Environ. Microbiol. 75:7674-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noordmans, J., J. Kempen, and H. J. Busscher. 1993. Automated image analysis to determine zeta potential distributions in particulate microelectrophoresis. J. Colloid Interface Sci. 156:394-399. [Google Scholar]
- 33.Ophir, T., and D. L. Gutnick. 1994. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl. Environ. Microbiol. 60:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pham, C. A., S. J. Jung, N. T. Phung, J. Lee, I. S. Chang, B. H. Kim, H. Yi, and J. Chun. 2003. A novel electrochemically active and Fe(III)-reducing bacterium phylogenetically related to Aeromonas hydrophila, isolated from a microbial fuel cell. FEMS. Microbiol. Lett. 223:129-134. [DOI] [PubMed] [Google Scholar]
- 35.Puech, V., M. Chami, A. Lemassu, M. A. Lanéelle, B. Schiffler, P. Gounon, N. Bayan, R. Benz, and M. Daffé. 2001. Structure of the cell envelope of corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology 147:1365-1382. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez-Navarro, D. N., M. S. Dardanelli, and J. E. Ruíz-Saínz. 2007. Attachment of bacteria to the roots of higher plants. FEMS Microbiol. Lett. 272:127-136. [DOI] [PubMed] [Google Scholar]
- 37.Saltikov, C. W., and D. K. Newman. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U. S. A. 100:10983-10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vinogradov, E., A. Korenevsky, and T. J. Beveridge. 2003. The structure of the rough-type lipopolysaccharide from Shewanella oneidensis MR-1, containing 8-amino-8-deoxy-Kdo and an open-chain form of 2-acetamido-2-deoxy-d-galactose. Carbohydr. Res. 338:1991-1997. [DOI] [PubMed] [Google Scholar]
- 39.Von Canstein, H., J. Ogawa, S. Shimizu, and J. R. Lloyd. 2008. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74:615-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe, K. 2008. Recent developments in microbial fuel cell technologies for sustainable bioenergy. J. Biosci. Bioeng. 106:528-536. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe, K., M. Manefield, M. Lee, and A. Kouzuma. 2009. Electron shuttles in biotechnology. Curr. Opin. Biotechnol. 20:633-641. [DOI] [PubMed] [Google Scholar]
- 42.Xiong, Y., L. Shi, B. Chen, M. U. Mayer, B. H. Lower, Y. Londer, S. Bose, M. F. Hochella, J. K. Fredrickson, and T. C. Squier. 2006. High-affinity binding and direct electron transfer to solid metals by the Shewanella oneidensis MR-1 outer membrane c-type cytochrome OmcA. J. Am. Chem. Soc. 128:13978-13979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.