Abstract
The symbiosis between Sinorhizobium fredii USDA257 and soybean [Glycine max (L.) Merr.] exhibits a high degree of cultivar specificity. USDA257 nodulates primitive soybean cultivars but fails to nodulate agronomically improved cultivars such as McCall. In this study we provide evidence for the involvement of a new genetic locus that controls soybean cultivar specificity. This locus was identified in USDA257 by Tn5 transposon mutagenesis followed by nodulation screening on McCall soybean. We have cloned the region corresponding to the site of Tn5 insertion and found that it lies within a 1.5-kb EcoRI fragment. DNA sequence analysis of this fragment and an adjacent 4.4-kb region identified an operon made up of three open reading frames encoding proteins of deduced molecular masses of 41, 13, and 104 kDa, respectively. These proteins revealed significant amino acid homology to glycine cleavage (gcv) system T, H, and P proteins of Escherichia coli and other organisms. Southern blot analysis revealed the presence of similar sequences in diverse rhizobia. Measurement of β-galactosidase activity of a USDA257 strain containing a transcriptional fusion of gcvT promoter sequences to the lacZ gene revealed that the USDA257 gcvTHP operon was inducible by glycine. Inactivation of either gcvT or gcvP of USDA257 enabled the mutant to nodulate several agronomically improved North American soybean cultivars. These nodules revealed anatomical features typical of determinate nodules, with numerous bacteroids within the infected cells. Unlike for the previously characterized soybean cultivar specificity locus nolBTUVW, inactivation of the gcv locus had no discernible effect on the secretion of nodulation outer proteins of USDA257.
Sinorhizobium fredii USDA257 is a fast-growing rhizobium that forms nitrogen-fixing nodules on Glycine max, Glycine soja, Neonotonia wightii, Phaseolus vulgaris, and several other legumes (24, 36). The symbiotic relationship between S. fredii USDA257 (here called USDA257) and soybean is of particular scientific and economic interest because this bacterium nodulates soybean in a cultivar-specific manner (13, 19). USDA257 effectively nodulates the primitive soybean cultivar Beijing but fails to nodulate agronomically improved cultivars such as McCall (2, 13). Histological studies have shown that USDA257 is able to infect McCall root hairs and induce cortical cell divisions but fails to form infection threads (5).
Previously, we have demonstrated the presence of negatively acting nodulation genes by creating Tn5 mutants (14). In one of the mutants (DH4) the Tn5 insertion was located in sym plasmid, while in the second mutant (DH5), the mutation was located in the bacterial chromosome. In DH4, the Tn5 insertion was located in an 8.0-kb EcoRI fragment which harbors the nolXWBTUV locus (34). Disruption of any these genes expands the host range of S. fredii USDA257. Subsequent studies have demonstrated that this locus is part of a USDA257 type III secretion system (T3SS) (27, 34). Both DH4 and DH5 were able to form nodules on agronomically improved soybean cultivars. In contrast to DH4, which formed nitrogen-fixing nodules, the DH5 formed only ineffective nodules (14). The gene nolC, which is inactivated by Tn5 insertion in DH5, shows homology to dnaJ of Escherichia coli, a heat-shock gene (22).
In the present work, we report the identification and characterization of another genetic locus in USDA257 that regulates soybean cultivar specificity. This chromosomal locus was identified by random mutagenesis with a Tn5 transposon, followed by nodulation screening on different soybean hosts. Sequence analysis showed that a gene that codes for the aminomethyltransferase precursor (glycine cleavage system T protein) was disrupted at the N terminus by the Tn5 transposon. The gcv mutants of USDA257, unlike the wild type, were able to form nitrogen-fixing nodules on several agronomically improved North American soybean cultivars.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Rhizobia were grown on a reciprocal shaker at 30°C in yeast extract mannitol (YEM) medium (48), and E. coli was cultured in Luria-Bertani broth at 37°C (42). When appropriate, antibiotics were added at the following concentrations: tetracycline, 10 μg/ml; kanamycin, 50 μg/ml; spectinomycin, 50 μg/ml; ampicillin, 100 μg/ml; gentamicin, 10 μg/ml; and trimethoprim, 10 μg/ml (for counter-selection against E. coli donor strains).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. fredii USDA257 | Nod+ on soybean cultivar Beijing | USDA-ARS |
| S. fredii USDA205 | Nod+ on soybean cultivar Beijing | USDA-ARS |
| S. meliloti USDA1002 | Nod+ on alfalfa | USDA-ARS |
| S. terangae USDA4894 | Nod+ on acacia | USDA-ARS |
| B. liaoningense USDA3622 | Nod+ on soybean | USDA-ARS |
| R. galegae USDA4128 | Nod+ on Galega spp. | USDA-ARS |
| R. hainanense USDA3588 | Nod+ on Desmodiumspp. | USDA-ARS |
| R. huautlaense USDA4900 | Nod+ on Sesbania herbacea | USDA-ARS |
| R. tropici USDA9030 | Nod+ on Phaseolus vulgaris | USDA-ARS |
| R. leguminosarum USDA2370 | Nod+ on pea | USDA-ARS |
| M. amorphae USDA1001 | Nod+ on Amorpha fruticosa | USDA-ARS |
| S. fredii USDA191 | Nod+ on soybean | USDA-ARS |
| S. fredii HBK257-15A | USDA257 derivative containing a Tn5 insertion in gcvT; Kmr | This work |
| S. fredii HBKgcvT-Ω | USDA257 derivative containing an Ω insertion in gcvT; Specr | This work |
| S. fredii HBKgcvP-Ω | USDA257 derivative containing an Ω insertion in gcvP; Specr | This work |
| E. coli HB101 | Restriction-minus, recA background | Takara Bio Inc. |
| E. coli DH5α | λ− φ80dlacZDM15 D(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Gibco BRL |
| Plasmids | ||
| pGEM-7zf(+) | Apr | Promega |
| pBluescript II SK(+) | Apr | Stratagene |
| pGEM-T Easy | Apr | Promega |
| pMP220 | Tcr | 45 |
| pJQ200sk | Gmr | 38 |
| pGCV-1 | Kmr Tcr; USDA257 gcv-Tn5 in pLAFR1 | This work |
| pGCV-2 | Tcr; USDA257 gcv in pLAFR1 | This work |
| pGCV-3 | Apr; 1.5-kb USDA257 gcvTEcoRI fragment in pGEM-7zf(+) | This work |
| pGCV-4 | Apr; 5.7-kb EcoRII fragment containing part of gcvT and gcvHP in pBluescript II SK(+) | This work |
| pGCV-5 | Tcr; 1.5-kb EcoRI fragment from pGVC-3 cloned in pMP220 | This work |
| pGCV-6 | Apr; 3.4-kb PstI fragment from pHKCS-4 in pBluescript II SK(+) | This work |
| pGCV-6Ω | Apr Spcr; 2.0-kb Ω fragment cloned into EcoRI site of pHKCS-6 | This work |
| pGCV-7Ω | Gmr Specr; 5.4-kb PstI fragment from pHKCS-6Ω cloned in pJQ200sk | This work |
Molecular techniques.
Recombinant DNA techniques were performed by using standard methods (42). Rhizobial genomic DNA was isolated by the method of Jagadish and Szalay (15), and DNA probes were labeled with [32P]dCTP by using a Multiprime DNA labeling system (Amersham LifeScience, Cleveland, OH). Restriction mapping, cloning of restriction fragments, and Southern blot analysis were performed by using standard protocols (42).
Tn5 mutagenesis and screening.
USDA257 Tn5 mutants were created with the help of suicide vector pSUP1011 as described by Heron et al. (14). Mutants capable of nodulating McCall soybean, which is not nodulated by parental strain USDA257, were assayed by pooling kanamycin-resistant colonies that were adjusted turbidimetrically to 1 × 108 cells/ml. Roots of 3-day-old McCall seedlings were inoculated with 200 μl of bacterial suspension and transferred to aseptically prepared Leonard jars that had been filled with vermiculite (21). The jars were kept in a growth chamber at a light intensity of 400 μmol of photons/m2/s with a 12-h photoperiod. Nodulation was evaluated 20 days after inoculation. USDA257 Tn5 mutants that produced nodules on McCall soybean were recovered by squeezing the nodule contents on a YEM agar plate containing kanamycin.
Cloning and nucleotide sequence analysis of the genes encoding the glycine cleavage system of USDA257.
A cosmid library of the parental strain USDA257 and the Tn5 mutant strain USDA257-15A in pLAFR1 (8) was constructed as described previously (14). To identify cosmid clones containing the Tn5 insertion, the library was streaked on YEM plates containing tetracycline and kanamycin. DNA from four cosmid clones that were resistant to both of the antibiotics were digested individually with EcoRI and BamHI and subjected to Southern blot analysis. An internal HindIII fragment of Tn5 was used as a probe to identify DNA fragments harboring the Tn5. A 7.5-kb EcoRI fragment, which contained USDA257 DNA sequences flanking the Tn5 insertion, was used to probe a cosmid library of the parental strain USDA257. Three cosmid clones that yielded positive hybridization signals were identified. Cosmid DNA was isolated from these clones, restricted individually with EcoRI, and fractionated by agarose gel electrophoresis. The DNA was transferred to a nitrocellulose membrane and hybridized with the 32P-labeled 7.5-kb EcoRI fragment containing DNA sequences flanking the Tn5 insertion. All the positive cosmid clones exhibited strong hybridization with a 1.5-kb EcoRI fragment. This EcoRI fragment from one of the cosmid clones (pGCV-2) was cloned into pGEM 7zf(+) to obtain pGCV-3 (Table 1). DNA sequencing was performed at the University of Missouri DNA Core Facility by utilizing appropriate primers synthesized by Integrated DNA Technologies (Coralville, IA).
Construction of gcvT-lacZ fusion constructs.
A 1.5-kb EcoRI DNA fragment which includes the putative promoter sequences of the gcvT was cloned in both orientations in the promoter probe plasmid pMP220 (45). Each of these constructs was introduced into S. fredii USDA257 by triparental mating using the helper plasmid pRK2013 (7). Transconjugants were grown for 2 days in YEM medium, followed by centrifugation to pellet the cells and suspension in minimal medium. Afterwards, cells were grown in the absence and in the presence of 200 mM glycine for 4 to 6 h. β-Galactosidase activity (35) was measured in each of the induced and noninduced samples.
Mutagenesis and marker exchange of USDA257 gcvT and gcvP genes.
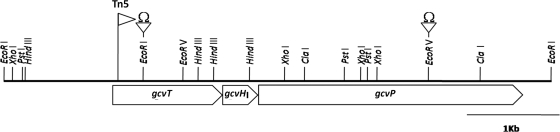
A 3.4-kb PstI fragment (Fig. 1) from pGCV-4 was subcloned into modified pBluescript II SK(+) in which the EcoRI site was removed to produce pGCV-6. For construction of the gcvT mutant, a 2.0-kb Ω fragment was cloned into an EcoRI site of pGCV-6 to produce pGCV-6Ω. The 5.4-kb PstI fragment from pGCV-6Ω was cloned into the suicide plasmid pJQ200sk to produce pGCV-7Ω. For construction of a gcvP mutant, a similar strategy was used, resulting in the insertion of the Ω fragment into an EcoRV site (Fig. 1). The gcvTΩ and gcvPΩ constructs were mobilized into USDA257 by triparental mating with helper plasmid pRK2013 (7). Marker exchange was achieved by selection on YEM medium plates containing 5% (wt/vol) sucrose. Mutants were confirmed by Southern blot hybridization with the wild-type region.
FIG. 1.
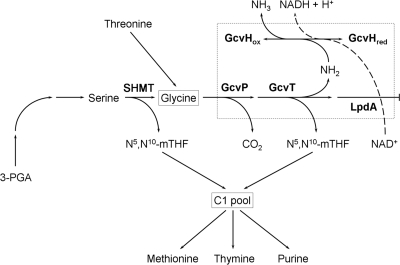
Metabolic pathway of the glycine cleavage enzyme system. This system is a multimeric assembly of four loosely associated proteins known as P protein (GcvP pyridoxal phosphate-containing glycine decarboxylase), H protein (GcvH, lipoic acid-containing carrier), T protein (GcvT, tetrahydrofolate requiring aminomethyltransferase or glycine synthase), and L protein (GcvL, lipoamide dehydrogenase). 3-PGA, 3-phosphoglycerate; N5,N10-mTHF, N5,N10-methylene tetrahydrofolate; C1 pool, pool of compounds containing only one carbon.
Isolation of extracellular proteins and Western blot analysis.
USDA257 and the gcvT mutant were grown in YEM medium in either the presence or absence of 1 μM apigenin for 48 h at 30°C. Bacterial cells were pelleted by centrifugation at 12,000 × g for 15 min. To the resulting supernatant 3 volumes of ice-cold acetone was added and left at −20°C overnight. Precipitated proteins were recovered by centrifugation as before and resuspended in SDS sample buffer. Protein samples were boiled for 3 min, and aliquots were resolved by 15% SDS-PAGE (28) using a Hoefer SE260 minigel electrophoresis apparatus (GE Healthcare, Piscataway, NJ). Resolved proteins were visualized by silver staining. Western blot analysis was performed using antibodies raised against the entire Nop (for nodulation outer protein) protein SR (also known as signal response [SR] protein) at a final dilution of 1:10,000. Immunoreactive proteins were detected with an enhanced chemiluminescent substrate (Super Signal West Pico kit; Pierce Biotechnology, Rockford, IL) according to the manufacturer's recommendations.
Nodulation tests.
Soybean seeds were surface sterilized and germinated on water agar plates as described previously (21). Cells of USDA257 and gcv mutants were harvested from liquid YEM medium cultures by centrifugation at 7,700 × g for 15 min. The cell concentrations were adjusted turbidimetrically to 1 × 108 cells/ml. Three-day-old seedlings were dip inoculated with bacterial cells and transferred immediately to autoclaved Leonard jars containing vermiculite and Jensen's nitrogen-free solution (21). Plants were placed in a growth chamber that was set at a constant temperature of 28°C with a light intensity of 500 μmol of photons/m2/s with 12 h of light. Nodules were counted at 20 days postinoculation. Nodulation experiments were repeated two times with five plants per treatment in each experiment. Acetylene reduction rates were determined by the method of Schwinghamer et al. (44).
Light and transmission electron microscopy.
Embedment of soybean nodules in paraffin was carried out as described earlier (26). Paraffin-embedded nodules were sectioned (10 μm thick) with a microtome and stained with hematoxylin and eosin. For ultrastructural studies, soybean nodules harvested at 20 days after inoculation were dissected into small pieces (2 to 4 mm) with a razor blade and fixed at room temperature for 4 h in buffered 2.5% glutaraldehyde (pH 7.2; 50 mM sodium phosphate). The tissue samples were washed four times at 15-min intervals with 50 mM phosphate buffer (pH 7.2) and postfixed with 2% aqueous osmium tetroxide for 1 h at room temperature. After extensive rinses in distilled water, the samples were dehydrated in a graded acetone series and infiltrated with Spurr's resin. Thin sections were cut with a diamond knife and collected on uncoated copper grids, stained with 0.5% aqueous uranyl acetate and 0.4% aqueous lead citrate, and viewed with a JEOL JEM 100B (Tokyo, Japan) electron microscope at 100 kV.
Nucleotide sequence accession number.
USDA257 gcvTHP sequences have been deposited in the GenBank database under accession number GQ214396.
RESULTS AND DISCUSSION
Isolation of a Tn5 mutant regulating soybean cultivar specificity.
Previously, two Tn5 mutants of USDA257, DH4 and DH5, that are involved in regulating soybean cultivar specificity were identified and characterized (4, 14, 20, 22, 34). The genes interrupted by the Tn5 insertion in DH4 was found to code for a type III secretion system (27), while in the case of DH5 the Tn5 inactivated a gene having homology to dnaJ of E. coli (22). In order to identify additional genes involved in soybean cultivar specificity, we extended our search by Tn5 transposon mutagenesis. About 12,000 presumptive Tn5-containing mutants of USDA257 were tested on McCall soybeans, which is not nodulated by the parental strain USDA257. A single mutant, HB15A, capable of nodulating McCall soybeans was selected. Rhizobia were isolated from the nodules and confirmed to be kanamycin resistant and a derivative of USDA257.
As a first step in identifying and characterizing the gene inactivated by Tn5 insertion, total DNA from the mutant was isolated, digested with either EcoRI or BamHI, and subjected to Southern blot analysis utilizing the internal HindIII fragment of Tn5 as a probe. The mutant revealed a single hybridizing EcoRI fragment and two BamHI fragments (data not shown). Since Tn5 has no EcoRI site and contains a single BamHI site, the results of our Southern blot analysis indicate that the mutant contains a single copy of the transposon. To isolate the gene inactivated by Tn5, we constructed a cosmid library of the mutant in pLAFR1 and screened the genomic library with a radiolabeled internal HindIII fragment of Tn5 as a probe. Cosmid DNA from several positive clones was isolated, digested with EcoRI, and subjected to Southern hybridization. All of these cosmid clones showed a single 7.5-kb hybridizing EcoRI fragment. One such cosmid clone (pGCV-1) was selected for further characterization. The Tn5-containing 7.5-kb EcoRI fragment was subcloned into the vector pBSKS. In order to isolate the wild-type fragment, we constructed a genomic DNA library of the parental strain S. fredii USDA257. This library was screened with a 32P-labeled Tn5-containing 7.5-kb EcoRI fragment; six positive clones were identified, and one was studied in detail. Southern blot analysis indicated that the Tn5 transposon was located within a 1.5-kb EcoRI fragment. We also performed Southern blot analysis using genomic DNA isolated from S. fredii USDA192 and its sym plasmid-cured derivative. In both cases strong hybridization was observed to a 5.7-kb EcoRI fragment, suggesting that this newly identified gene is not localized in the sym plasmid.
Since the Tn5 insertion was identified to a 1.5-kb EcoRI DNA fragment, we subcloned this fragment and sequenced it in its entirety. Nucleotide sequence analysis identified an incomplete open reading frame (ORF) at the 3′ region of the 1.5-kb DNA fragment. This partial ORF showed extensive homology to the glycine cleavage (gcv) system T protein of E. coli and several other organisms. In E. coli and in other organisms, the T protein is a subunit of the glycine decarboxylase complex (47). This Gcv complex is composed of four proteins: GcvP, GcvH, GcvT, and GcvL (Fig. 1). The GcvP protein (glycine dehydrogenase) catalyzes the pyridoxal-phosphate (pyridoxal-P)-dependent decarboxylation of glycine and transfer of the remaining aminomethyl moiety to the lipoyl prosthetic group of the GcvH protein. The GcvT protein (aminomethyltransferase) catalyzes the release of ammonia from the intermediate attached to the H protein and the synthesis of methylenetetrahydrofolate in the presence of tetrahydrofolate (Fig. 1). To ascertain if a similar operon structure is also present in USDA257, we subcloned the adjacent 5.7-kb EcoRI fragment and determined its nucleotide sequence. The DNA sequence was subjected to computer analysis, and three potential ORFs were identified. All three ORFs were of the same polarity, and they appeared to be part of an operon (Fig. 2). The first ORF was comprised of 387 amino acids encoding a protein with a molecular mass of 41 kDa. The second ORF, which initiated 15 bp after the termination of the first ORF, contained 120 amino acids and encoded a protein with a molecular mass of 12.7 kDa. The third ORF was the largest among the three ORFs, initiated immediately after the second ORF, and was made up of 954 amino acids with a molecular mass of 104 kDa.
FIG. 2.
Coordinated physical and genetic maps of gcv region of S. fredii USDA257. The orientation of the three ORFs and the location of the Tn5 and omega cassette insertions are also shown.
Sequence homology among the glycine cleavage system from different organisms.
The deduced amino acid sequences of the three open reading frames were subjected to BLAST analysis, which revealed high sequence identities with proteins encoding the glycine cleavage system from other rhizobia. On the basis of extensive sequence homology, we have named the three ORFs gcvT, gcvH and gcvP. The amino acid sequence of USDA257 gcvT shows 80 to 87% overall sequence similarity with gcvT proteins from Sinorhizobium medicae, Sinorhizobium meliloti, Rhizobium leguminosarum bv. trifolii, and Rhizobium etli. Similarly, a high degree of sequence similarity with gcvH (85 to 93%) and gcvP (84 to 91%) proteins among these rhizobia were also detected. The glycine cleavage system is widely distributed in both prokaryotes and eukaryotes. In order to determine the percent sequence similarity among the GcvT proteins from different organisms, the deduced amino acid sequence of the GcvT protein from S. fredii USDA257 was aligned with the corresponding protein sequences from S. meliloti, Agrobacterium, E. coli, Arabidopsis, and human and bovine GcvT proteins (Fig. 3). USDA257 revealed 84.5% and 72.3% sequence similarity to the GcvT protein from S. meliloti and Agrobacterium, respectively. USDA257 also showed 35 to 38% amino acid sequence similarity to GcvT from eukaryotic organisms like Arabidopsis and bovine and human proteins (Fig. 3). In contrast, USDA257 and E. coli showed only 23.8% amino acid sequence similarity.
FIG. 3.
Multiple amino acid sequence alignment of GcvT proteins from different organisms. (A) The accession numbers of the sequences are as follows: S. meliloti, AL591688; Agrobacterium, AE007869; E. coli, AP009048.1; Arabidopsis, AT1G11860; human, NM_000481; bovine, NM_177485. Sequences are aligned with the USDA257 GcvT sequence (GQ214396). Positions of amino acid identity are boxed. Multiple sequence alignment analysis was performed using the CLUSTAL W program from UniProt Knowledgebase (http://www.uniprot.org/). (B) Phylogenetic tree of GcvT. The scale shown in the phylogenetic tree represents the branch distance as the number of changes in character state between GcvT proteins from different organisms.
Computerized comparisons of the USDA257 gcv operon revealed the presence of similar sequences in S. medicae, S. meliloti, R. leguminosarum bv. trifolii, and R. etli. This prompted us to search for homologous sequences in several other rhizobia. The results of Southern blotting in which EcoRI-digested genomic DNAs of 11 rhizobial strains were probed with a 430-bp EcoRI and EcoRV DNA fragment corresponding to gcvT demonstrated that a single strong hybridizing fragment was detected in Bradyrhizobium liaoningense USDA3622, Mesorhizobium amorphae USDA1001, Rhizobium galegae USDA4128, Rhizobium hainanense USDA3588, Rhizobium huautlaense USDA4900, Rhizobium tropici USDA9030, R. leguminosarum USDA2370, S. meliloti USDA1002, and Sinorhizobium terangae USDA4894 (Fig. 4). The sizes of the hybridizing fragments showed marked differences. S. fredii USDA205 and Sinorhizobium saheli USDA4894, however, revealed two strongly hybridizing EcoRI fragments (Fig. 4), indicating either the presence of two copies of gcvT or an additional EcoRI site in the gcvT coding region.
FIG. 4.
Southern blot analysis of gcvT in rhizobia. Genomic DNAs from B. liaoningense USDA3622 (lane 1), R. galegae USDA4128 (lane 2), R. hainanense USDA3588 (lane 3), R. huautlaense USDA4900 (lane 4), R. tropici USDA9030 (lane 5), M. amorphae USDA1001 (lane 6), S. fredii USDA205 (lane 7), S. meliloti USDA1002 (lane 8), S. terangae USDA4894 (lane 9), R. leguminosarum USDA2370 (lane 10), and S. saheli USDA4893 (lane 11) were restriction enzyme digested with EcoRI and separated electrophoretically in 0.8% agarose. The gel was blotted onto nitrocellulose and probed with the 32P-labeled S. fredii USDA257 gcvT gene. Molecular weight markers in kilobases are shown on the left side of the figure.
Recently, a number of genome projects for rhizobia have been completed, facilitating insights into the genome architecture of these symbiotic bacteria (31). An examination of the available complete rhizobial genome sequences revealed the presence of a gcv locus in Rhizobium sp. NGR234 (43), S. meliloti 1021 (9), R. leguminosarum bv. viciae (50), R. leguminosarum bv. trifolii (39), R. etli (11), Bradyrhizobium japonicum USDA110 (17), Bradyrhizobium sp. ORS278 (10), Azorhizobium caulinodans ORS571 (29), and Mesorhizobium loti MAFF303099 (16). Additionally, a gcv locus was also identified in the genome of the betaproteobacteria Burkholderia phymatum STM815 and Cupriavidus taiwanensis LMG19424 (1), both of which form root nodules on legumes. The organization of the gcv operon in these symbiotic nitrogen-fixing bacteria was similar to that of S. fredii USDA257 with the exception of R. leguminosarum bv. viciae and A. caulinodans ORS 571. In the case of R. leguminosarum bv. viciae, the gcvH was not identified in between gcvT and gcvP (Fig. 2). In contrast, the gcvP was not a part of the gcv operon in A. caulinodans ORS571. Proteins encoded by the gcv loci from these diverse symbiotic bacteria had similar molecular weights and exhibited significant amino acid sequence similarities between them. The amino acid sequence similarities of GcvT, GcvH, and GcvP among USDA257, Rhizobium sp. NGR234, and S. meliloti ranged from 85 to 95%. A similar comparison of the amino acid sequences between these proteins among USDA257 and the two betaproteobacteria B. phymatum STM815 and C. taiwanensis LMG19424 revealed similarities ranging from 40 to 58%, indicating a distinct phylogenetic relationship among these diverse symbiotic bacteria.
USDA257 gcvTHP operon is inducible by glycine.
In E. coli, the expression of the glycine cleavage enzyme system is induced by glycine (33, 46), and the transcriptional regulation of the gcvTHP operon has been studied extensively. To test if the USDA257 gcvTHP operon is also inducible by glycine, we cloned a 1.5-kb EcoRI fragment (Fig. 2), which contains about 1,173 bp of DNA sequence upstream of the gcvT start codon, in both orientations into the promoter probe plasmid pMP220. Each of these constructs was then introduced into S. fredii USDA257, and their responsiveness to the addition of 200 mM glycine was evaluated by measuring the β-galactosidase activity of the transconjugants. USDA257 carrying the pMP220 or 1.5-kb EcoRI fragment cloned in the wrong orientation in either the presence or absence of glycine revealed a basal level of β-galactosidase activity (less than 200 Miller units). USDA257 carrying the 1.5-kb EcoRI fragment cloned in the correct orientation and grown in the absence of glycine showed 224 ± 22 Miller units of β-galactosidase activity. However, when this transconjugant was grown in the presence of glycine, the activity of β-galactosidase was about seven times greater (1,646 ± 90 Miller units) than the values obtained from the uninduced cultures, indicating that the USDA257 gcvTHP operon is inducible by glycine.
Inactivation of gcvTHP operon of USDA257 alters symbiotic behavior.
The effect of inactivation of the glycine cleavage system was examined on several agronomically improved North American soybean cultivars (Table 2). Both USDA257 and USDA257-15A were able to form nitrogen-fixing nodules on Beijing soybean. In contrast, when agronomically improved soybean cultivars were inoculated with USDA257, no nodules were produced while the gcvT mutant (USDA257-15A) formed large nodules whose interiors were pink, presumably due the accumulation of leghemoglobin. Nodules harvested from agronomically improved cultivars 20 days after inoculation with USDA257-15A showed significant acetylene reduction, ranging from 18.8 to 32.6 μmol/h/g of fresh nodule weight. We also complemented the gcvT mutant by introducing a cosmid clone carrying the wild-type gcv operon of USDA257 and tested its nodulation phenotype on McCall soybean. Interestingly, even the complemented strain produced nodules on McCall soybean. However, the rhizobia recovered from those nodules were resistant to kanamycin but not to tetracycline. This indicates that the bacteria forming these nodules had lost the cosmid containing the wild-type gcv operon. This result is to be expected because selection generally favors the Nod+ phenotype (14). To obtain further evidence that the glycine cleavage system regulates the host range of USDA257, we created two independent mutants (gcvTΩ and gcvPΩ) and tested their ability to form nodules on several agronomically improved North American soybean cultivars (Table 2). Both of these mutants were able to form nitrogen-fixing nodules with all the soybean hosts. Thus, inactivation of the glycine cleavage system clearly enables USDA257 to initiate nitrogen-fixing nodules on agronomically improved North American soybean cultivars.
TABLE 2.
Symbiotic phenotypes of USDA257 and gcv mutants
| Legume species | Nodulation response witha: |
|||
|---|---|---|---|---|
| USDA257 | HBK257-15A | HBKgcvT-Ω | HBKgcvP-Ω | |
| Glycine max cultivar or strain | ||||
| Beijing | + | + | + | + |
| McCall | − | + | + | + |
| Jack | − | + | + | + |
| Clark | − | + | + | + |
| Maverick | − | + | + | + |
| Pio94Y20 | − | + | + | + |
| MFA4249 | − | + | + | + |
| Vigna unguiculata | + | + | + | + |
+, nitrogen-fixing nodules; −, no nodules.
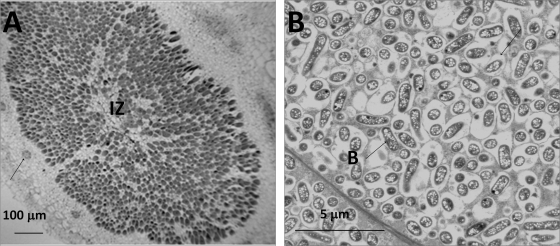
S. fredii USDA191, a close relative of USDA257, forms nitrogen-fixing nodules on agronomically improved soybean cultivars (2, 4, 13). To investigate if the nodules initiated by USDA257 gcv mutants were similar to those of USDA191, we examined the anatomical structure of the nodules with both light and transmission electron microscopy. Light microscopic examination of nodules initiated by gcv mutants on agronomically improved soybean cultivars exhibited a structure that was typical of determinate nodules (Fig. 5 A). The nodule, which was differentiated into a central bacteria-filled region, was separated from the outer cortex region by a layer of sclerenchyma cells. In addition, several vascular bundles were seen in the outer cortex. Transmission electron microscopy of thin sections of the central region revealed the presence of cells that were filled with bacteria that were enclosed by symbiosomes (Fig. 5B). These bacteroids contained numerous prominent polyhydroxybutyrate inclusions. The anatomical features of nodules induced by gcv mutants and those from USDA191 were indistinguishable.
FIG. 5.
Anatomy of soybean (G. max cv. McCall) nodules formed by the USDA257 gcvT mutant. (A) Light micrograph of a paraffin section of soybean nodule revealing a central infected zone (IZ). The arrows point to vascular bundles in the cortex. (B) Transmission electron micrograph of soybean nodule showing the presence of numerous bacteroids (B) which are surrounded by symbiosomes. Note the presence of numerous poly-β-hydroxybutyrate inclusions inside the bacteroids.
Inactivation of the gcvTHP operon does not affect the secretion of nodulation outer proteins.
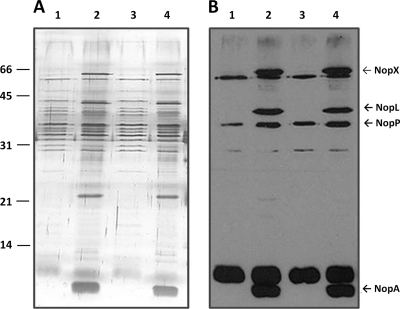
Previously, we have demonstrated that USDA257, when grown in the presence of the nod gene inducing flavonoids, can secrete nod factors and several extracellular proteins into the rhizosphere (3, 23). The secreted proteins were termed nodulation outer proteins (Nops), and their secretion was regulated by both nodD1 and nodD2 and was dependent on the soybean cultivar specificity locus nolBTUVW (25). This locus was found to code for components of the type III secretion system (T3SS) (20, 27). Mutants in this locus are defective in T3SS and consequently secrete only some or none of the nodulation outer proteins that have been demonstrated to have profound effects on nodulation (6, 12, 32, 49). For example, USDA257DH4, a Tn5 mutant of USDA257, is unable to secrete several Nops. Consequently, this mutant, unlike the wild-type strain, was able to nodulate agronomically improved soybean cultivars, suggesting an important role for the Nops in regulating soybean cultivar specificity (27, 30). To determine if the mutation in the gcv operon of USDA257 has any effect on Nops, we isolated extracellular proteins from USDA257 and the gcv mutant that were grown in either the presence or absence of apigenin, a potent inducer of the nod genes of USDA257 (21, 37). SDS-PAGE analysis followed by silver staining of the gels revealed no major changes in the protein profiles between the parental strain and the gcv mutant (Fig. 6 A). Western blot analysis utilizing antibodies raised against flavonoid-induced extracellular proteins of USDA257 clearly demonstrates that USDA257 secretes several proteins into the extracellular milieu when it is grown in the presence of flavonoids (Fig. 6B). This includes NopA, NopB, NopL, NopP, and NopX. The same set of Nops was also elaborated by the gcv mutant (Fig. 6). It is clear from this observation that gcv mutation has no obvious effect on the Nop production of USDA257. This observation is in drastic contrast with USDA257DH4, another Tn5 mutant extensively studied in our laboratory (3, 4, 14, 20, 25, 27). Thus, it is evident that in addition to Nops, there are other yet unidentified factors that could influence soybean cultivar specificity.
FIG. 6.
Extracellular protein profile and immunoblot analysis of USDA257 and the USDA257 gcvT mutant. Extracellular proteins were prepared from cells grown in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 1 μM apigenin. The proteins were resolved by 15% SDS-PAGE and silver stained (A) or transferred to a nitrocellulose membrane for immunological analysis with antibodies raised against Nops (B). Lanes 1 and 2, extracellular proteins from USDA257; lanes 3 and 4, extracellular proteins from the USDA257 gcvT mutant. The immunoreactive proteins are identified on the right side of the figure.
The glycine cleavage system is the most important pathway in serine and glycine catabolism in various vertebrates including humans. Nonketotic hyperglycinemia, a genetic disorder characterized by abnormally high levels of glycine in human infants, results from defective glycine cleavage activity (18). It is not clear why inactivation of the glycine cleavage system of USDA257 enables this strain to nodulate soybean cultivar McCall. Previously, a Tn5 mutant of B. japonicum was reported to elicit pseudonodule-like structures on soybean (41). Subsequent studies have shown that the Tn5 mutation was located in the glyA gene which encodes serine hydroxymethyltransferase (SHMT) (Fig. 1) (40). SHMT catalyzes the biosynthesis of glycine from serine and the transfer of a one-carbon unit to tetrahydrofolate (40). Even though the specific role of SHMT and its reaction products in causing defects in early nodule and bacteroid development is not fully understood, it is speculated that an adequate supply of glycine and/or a functioning C1 metabolism are essential for effective nodulation of soybean by B. japonicum (40). In contrast to B. japonicum, where inactivation of glyA leads to ineffective nodulation, the inactivation of the gcv locus of USDA257 leads to effective nodulation of soybean cultivars that are not originally nodulated by the parental strain. It is believed that both the gly and gcv loci are involved in the generation of C1 units and that they serve as the primary and secondary sources of C1 units, respectively (33). C1 units are used in a variety of biochemical reactions, including the synthesis of purines, histidine, thymine, and methionine and the formylation of aminoacylated initiator tRNA (46). We speculate that inactivation of the gcv locus could have altered some essential function of the cell, resulting in modification or synthesis of signal molecule(s) that may regulate soybean cultivar specificity. Further studies are required to elucidate the precise role of the gcv locus in regulating soybean cultivar specificity.
Acknowledgments
We thank Nathan Oehrle for critically reviewing the manuscript. Microscopy was carried out at the Electron Microscopy Facility of the University of Missouri School of Veterinary Medicine.
The use of product names is necessary to report factually on available data; however, neither the University of Missouri nor USDA either guarantees or warrants any product mentioned, and the use of the name by the University of Missouri or USDA implies no approval of the product to the exclusion of others that may be suitable.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Amadou, C., G. Pascal, S. Mangenot, M. Glew, C. Bontemps, D. Capela, S. Carrère, S. Cruveiller, C. Dossat, A. Lajus, M. Marchetti, V. Poinsot, Z. Rouy, B. Servin, M. Saad, C. Schenowitz, V. Barbe, J. Batut, C. Médigue, and C. Masson-Boivin. 2008. Genome sequence of the β-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res. 18:1472-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balatti, P. A., and S. G. Pueppke. 1992. Identification of North American soybean lines that form nitrogen-fixing nodules with Rhizobium fredii USDA257. Can. J. Plant Sci. 72:49-55. [Google Scholar]
- 3.Bec-Ferte, M.-P., H. B. Krishnan, D. Prome, A. Savagnac, S. G. Pueppke, and J.-C. Prome. 1994. Structures of nodulation factors from the nitrogen-fixing soybean symbiont Rhizobium fredii USDA257. Biochemistry 33:11782-11788. [DOI] [PubMed] [Google Scholar]
- 4.Bellato, C., H. B. Krishnan, T. Cubo, F. Temprano, and S. G. Pueppke. 1997. The soybean cultivar specificity gene nolX is present, expressed in a nodD-dependent manner, and of symbiotic significance in cultivar-nonspecific strains of Rhizobium (Sinorhizobium) fredii. Microbiology 143:1381-1388. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee, A., P. A. Balatti, W. Gibbons, and S. G. Pueppke. 1990. Interaction of Rhizobium fredii USDA257 and nodulation mutants derived from it with the agronomically improved soybean cultivar McCall. Planta 180:301-311. [DOI] [PubMed] [Google Scholar]
- 6.Deakin, W. J., and W. J. Broughton. 2009. Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat. Rev. Microbiol. 7:312-320. [DOI] [PubMed] [Google Scholar]
- 7.Figurski, D., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 9.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 10.Giraud, E., L. Moulin, D. Vallenet, V. Barbe, E. Cytryn, J.-C. Avarre, M. Jaubert, D. Simon, F. Cartieaux, Y. Prin, G. Bena, L. Hannibal, J. Fardoux, M. Kojadinovic, L. Vuillet, A. Lajus, S. Cruveiller, Z. Rouy, S. Mangenot, B. Segurens, C. Dossat, W. L. Franck, W.-S. Chang, E. Saunders, D. Bruce, P. Richardson, P. Normand, B. Dreyfus, D. Pignol, G. Stacey, D. Emerich, A. Vermeglio, C. Medigue, and M. Sadowsky. 2007. Legumes symbioses: absence of Nod genes in photosynthetic Bradyrhizobia. Science 316:1307-1312. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez, V., R. I. Santamaria, P. Bustos, I. Hernandez-Gonzalez, A. Medrano-Soto, G. Moreno-Hagelsieb, S. C. Janga, M. A. Ramirez, V. Jimenez-Jacinto, J. Collado-Vides, and G. Davila. 2006. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. U. S. A. 103:3834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Göttfert, M., S. Röthlisberger, C. Kündig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heron, D. S., and S. G. Pueppke. 1984. Mode of infection, nodulation specificity, and indigenous plasmids of 11 fast-growing Rhizobium japonicum strains. J. Bacteriol. 160:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heron, D. S., T. Érsek, H. B. Krishnan, and S. G. Pueppke. 1989. Nodulation mutants of Rhizobium fredii USDA257. Mol. Plant Microbe Interact. 2:4-10. [Google Scholar]
- 15.Jagadish, M. N., and A. A. Szalay. 1984. Directed transposon Tn5 mutagenesis and complementation in slow-growing, broad host range cowpea Rhizobium. Mol. Gen. Genet. 196:290-300. [Google Scholar]
- 16.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 18.Kanno, J., T. Hutchin, F. Kamada, A. Narisawa, Y. Aoki, Y. Matsubara, and S. Kure. 2007. Genomic deletion within GLDC is a major cause of non-ketotic hyperglycinaemia. J. Med. Genet. 44:e69. doi: 10.1136/jmg.2006.043448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keyser, H. H., B. B. Bohlool, T. S. Hu, and D. F. Weber. 1982. Fast-growing rhizobia isolated from root nodules of soybean. Science 215:1631-1632. [DOI] [PubMed] [Google Scholar]
- 20.Kovacs, L. G., P. A. Balatti, H. B. Krishnan, and S. G. Pueppke. 1995. Transcriptional organization and expression of nolXWBTUV, a locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 17:923-933. [DOI] [PubMed] [Google Scholar]
- 21.Krishnan, H. B., and S. G. Pueppke. 1991. Sequence and analysis of the nodABC region of Rhizobium fredii USDA257, a nitrogen-fixing symbiont of soybean and other legumes. Mol. Plant Microbe Interact. 4:512-520. [DOI] [PubMed] [Google Scholar]
- 22.Krishnan, H. B., and S. G. Pueppke. 1991. nolC, a Rhizobium fredii gene involved in cultivar-specific nodulation of soybean, shares homology with a heat-shock gene. Mol. Micriobiol. 5:737-745. [DOI] [PubMed] [Google Scholar]
- 23.Krishnan, H. B., and S. G. Pueppke. 1993. Flavonoid inducers of nodulation genes stimulate Rhizobium fredii USDA257 to export proteins into the environment. Mol. Plant Microbe Interact. 6:107-113. [DOI] [PubMed] [Google Scholar]
- 24.Krishnan, H. B., and S. G. Pueppke. 1994. Host range, RFLP, and antigenic relationships between Rhizobium fredii strains and Rhizobium sp. NGR234. Plant Soil 161:21-29. [Google Scholar]
- 25.Krishnan, H. B., C.-L. Kuo, and S. G. Pueppke. 1995. Elaboration of flavonoid-induced proteins by the nitrogen-fixing soybean symbiont Rhizobium fredii is regulated by both nodD1 and nodD2, and is dependent on the cultivar-specificity locus, nolXWBTUV. Microbiology 141:2245-2251. [Google Scholar]
- 26.Krishnan, H. B., W.-S. Kim, J. S. Hyung, K. Y. Kim, and J. Guoqiao. 2003. Citrate synthase mutants of Sinorhizobium fredii USDA257 form ineffective nodules with aberrant ultrastructure. Appl. Environ. Microbiol. 69:3561-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishnan, H. B., J. Lorio, W.-S. Kim, G. Jiang, K. Y. Kim, M. DeBoer, and S. G. Pueppke. 2003. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant Microbe Interact. 16:617-625. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lee, K. B., P. De Backer, T. Aono, C. T. Liu, S. Suzuki, T. Suzuki, T. Kaneko, M. Yamada, S. Tabata, D. M. Kupfer, F. Z. Najar, G. B. Wiley, B. Roe, T. T. Binnewies, D. W. Ussery, W. D'Haeze, J. D. Herder, D. Gevers, D. Vereecke, M. Holsters, and H. Oyaizu. 2008. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorio, J. C., W.-S. Kim, and H. B. Krishnan. 2004. NopB, a soybean cultivar-specificity protein from Sinorhizobium fredii USDA257, is a type III secreted protein. Mol. Plant Microbe Interact. 17:1259-1268. [DOI] [PubMed] [Google Scholar]
- 31.MacLean, A. M., T. M. Finan, and M. J. Sadowsky. 2007. Genomes of the symbiotic nitrogen-fixing bacteria of legumes. Plant Physiol. 144:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marie, C., W. J. Broughton, and W. J. Deakin. 2001. Rhizobium type III secretion systems: legume charmers or alarmers? Curr. Opin. Plant Biol. 4:336-342. [DOI] [PubMed] [Google Scholar]
- 33.Meedel, T. H., and L. I. Pizer. 1974. Regulation of one-carbon biosynthesis and utilization in Escherichia coli. J. Bacteriol. 118:905-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meinhardt, L. W., H. B. Krishnan, P. A. Balatti, and S. G. Pueppke. 1993. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 9:17-29. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Pueppke, S. G., and W. J. Broughton. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant Microbe Interact. 12:293-318. [DOI] [PubMed] [Google Scholar]
- 37.Pueppke, S. G., M. C. Bolaños-Vásquez, D. Werner, M.-P. Bec-Ferté, J. C. Promé, and H. B. Krishnan. 1998. Release of flavonoids by the soybean cultivars McCall and Peking and their perception as signals by the nitrogen-fixing symbiont Sinorhizobium fredii. Plant Physiol. 117:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 39.Reeve, W., G. O'Hara, P. Chain, J. Ardley, L. Bräu, K. Nandesena, R. Tiwari, S. Malfatti, H. Kiss, A. Lapidus, A. Copeland, M. Nolan, M. Land, N. Ivanova, K. Mavromatis, V. Markowitz, N. Kyrpides, V. Melino, M. Denton, R. Yates, and J. Howieson. 2010. Complete genome sequence of Rhizobium leguminosarum bv trifolii strain WSM2304, an effective microsymbiont of the South American clover Trifolium polymorphum. Stand. Genomic Sci. 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossbach, S., and H. Hennecke. 1991. Identification of glyA as a symbiotically essential gene in Bradyrhizobium japonicum. Mol. Microbiol. 5:39-47. [DOI] [PubMed] [Google Scholar]
- 41.Rossbach, S., T. Gloudemans, T. Bisseling, D. Studer, B. Kaluza, S. Ebeling, and H. Hennecke. 1989. Genetic and physiologic characterization of a Bradyrhizobium japonicum mutant defective in early bacteroid development. Mol. Plant Microbe Interact. 2:233-240. [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Schmeisser, C., H. Liesegang, D. Krysciak, N. Bakkou, A. Le Quéré, A. Wollherr, I. Heinemeyer, B. Morgenstern, A. Pommerening-Röser, M. Flores, R. Palacios, S. Brenner, G. Gottschalk, R. A. Schmitz, W. J. Broughton, X. Perret, A. W. Strittmatter, and W. R. Streit. 2009. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl. Environ. Microbiol. 75:4035-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwinghamer, E. A., H. J. Evans, and M. D. Dawson. 1970. Evaluation of effectiveness in mutant strains of Rhizobium by acetylene reduction relative to other criteria of N2 fixation. Plant Soil 33:192-212. [Google Scholar]
- 45.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 46.Stauffer, G. V. 1987. Biosynthesis of serine and glycine, p. 412-418. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 47.Stauffer, L. T., S. J. Fogarty, and G. V. Stauffer. 1994. Characterization of the Escherichia coli gcv operon. Gene 142:17-22. [DOI] [PubMed] [Google Scholar]
- 48.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Oxford, England.
- 49.Viprey, V., A. Del Greco, W. Golinowski, W. J. Broughton, and X. Perret. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381-1389. [DOI] [PubMed] [Google Scholar]
- 50.Young, J. P., L. Crossman, A. Johnston, N. Thomson, Z. Ghazoui, K. Hull, M. Wexler, A. Curson, J. Todd, P. Poole, T. Mauchline, A. East, M. Quail, C. Churcher, C. Arrowsmith, I. Cherevach, T. Chillingworth, K. Clarke, A. Cronin, P. Davis, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, S. Whitehead, and J. Parkhill. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]