Abstract
Phylogenetic analysis of 16S rRNA, nodC, and nifH genes of four bacterial strains isolated from root nodules of Phaseolus vulgaris grown in Morocco soils were identified as Burkholderia phymatum. All four strains formed N2-fixing nodules on P. vulgaris and Mimosa, Acacia, and Prosopis species and reduced acetylene to ethylene when cultured ex planta.
Until 2001 all known bacteria involved in root nodule symbioses with leguminous plants were classified as members of the order Rhizobiales of the Alphaproteobacteria, including Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium (28, 36, 38). Moulin et al. (21), however, described two Burkholderia nodule-forming strains isolated from Machaerium lunatum in French Guiana and Aspalathus carnosa in South Africa, respectively, this being the first report on the presence of a betaproteobacterium within root nodules of legumes. Later, the strains were formally classified as Burkholderia phymatum STM815T and Burkholderia tuberum STM678T, respectively (33). Burkholderia species are the predominant isolates from nodules of mimosoid legumes from Panama (2), Costa Rica (3), Taiwan (4, 6), Brazil (5, 7), Venezuela (5), and Madagascar (24), which indicates a high affinity of Burkholderia for forming effective symbioses with Mimosa. Diazotrophy is well represented in Burkholderia; among the more than 55 species presently classified as Burkholderia, 9 have been shown to fix N2 ex planta by using either the acetylene reduction activity (ARA) assay or the presence of nifH genes encoding nitrogenase reductase (3, 5, 11, 24) and more recently by 15N2 isotopic dilution experiments (17).
Common bean (Phaseolus vulgaris) is an herbaceous leguminous plant which establishes N2-fixing symbiosis with at least 5 species of the genus Rhizobium. Rhizobium etli is the predominant species in America (29) and is also detected in Europe and Africa (13, 20). Rhizobium leguminosarum bv. phaseoli is commonly found in Europe (13) and has also been reported to be present in Tunisia (20) and Colombia (10). Rhizobium tropici is found in acid soils of South America and is also present in Europe and several African countries (18). Rhizobium giardinii has been detected only in European and Tunisian soils (1, 20), and Rhizobium gallicum has been found nodulating beans in Europe, North Africa, and Mexico (1, 13). In this study we report on the isolation and characterization of B. phymatum from root nodules of P. vulgaris grown in alkaline soils from Morocco. Our results show that strains formed effective nodules on species of Mimosa, Acacia, and Prosopis and fixed atmospheric N2 under free-living conditions.
Soil was taken from a field near Oulade Mansour (34°47′N, 2°15′W, Oujda province, Morocco) where maize and common bean have traditionally been grown as rotational crops without N fertilization. Soil had a sandy-clay texture and the following characteristics: pH (in water), 8.1; 55.18% sand; 17.17% silt; 27.65% clay; 6.1% carbonates; 7.69% organic carbon; 0.069 total nitrogen. Seeds of P. vulgaris cv. Flamingo were surface sterilized (96% ethanol for 30 s followed by immersion in 15% [vol/vol] H2O2 for 8 min), washed several times with sterile water, germinated in the dark, and planted in 1-kg pots containing soil and sterile sand (1:1, vol/vol). Plants were grown for 30 days in controlled environmental chambers under conditions previously described (9). Nodules were collected, pooled together, surface sterilized with 2.5% HgCl2 for 5 min, and rinsed thoroughly with sterile distilled water. Then, 12 nodules were placed independently on petri dishes and crushed in a drop of sterile water with a sterile glass rod. The resulting suspension was streaked onto petri dishes containing either yeast extract-mannitol (YEM) medium (35) or peptone-mineral salts-yeast extract (PSY) medium (25). After incubation of the plates at 30°C for 7 days, CFU which represented all of the colony types that could be distinguished by microscopic observation of living cells were chosen. After identification, Burkholderia strains were routinely grown in BAc medium (12).
For DNA extraction and PCR amplifications, genomic DNA was isolated from bacterial cells using the RealPure genomic DNA extraction kit (Durviz, Spain) according to the manufacturer's instructions. Repetitive extragenic palindromic (REP) fingerprinting was performed using primers REPIR-I and REP2-I according to the method of de Bruijn (8). PCR amplifications of the 16S rRNA gene fragments were done with the Bphym-F and Bphym-R species-specific primer pair (37).
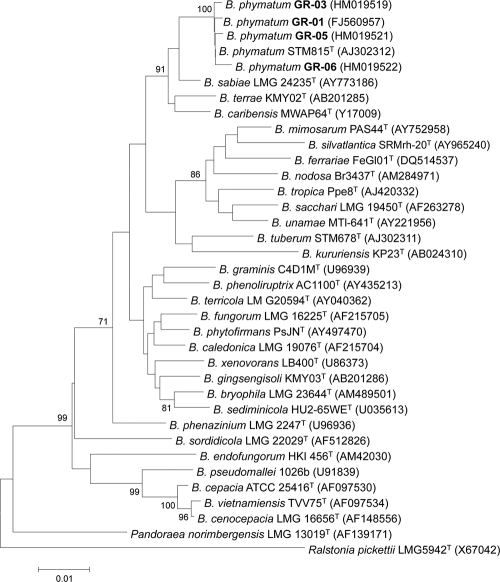
After isolation from root nodules, 52 strains forming morphologically different colonies were obtained and grouped in two main clusters after REP-PCR fingerprinting (data not shown), a technique that is extensively used to group bacteria at subspecies or strain level (8, 34) and has proven to be a powerful tool for studies of microbial ecology and evolution (14). The nearly complete sequence of the 16S rRNA gene from a representative strain of each REP-PCR group was obtained and compared with those held in GenBank. Forty-six strains in cluster I were members of the family Rhizobiaceae from the Alphaproteobacteria. Another 4 strains, here referred to as GR strains (GR01 to GR04), grouped in cluster II and were classified into the family Burkholderiaceae within the Betaproteobacteria. The remaining two strains have not been clearly classified as yet. The four GR strains have almost identical 16 rRNA gene sequences, and BLAST searches showed that they were phylogenetically close (99% identity) to B. phymatum STM815T, a strain originally isolated from the papilionoid legume Machaerium lunatum (21, 33). A phylogenetic analysis including 30 Burkholderia reference strains showed that strains from root nodules of P. vulgaris form a tight cluster with B. phymatum STM815T (Fig. 1).
FIG. 1.
Phylogenetic tree showing the positions of four P. vulgaris-isolated strains, GR01, GR03, GR05, and GR06, within the genus Burkholderia based on 16S rRNA gene sequence comparisons. One thousand bootstrap samplings were performed. The NCBI GenBank accession number for each strain is shown in parentheses. The bar represents one nucleotide substitution per 1,000 nucleotides. The multiple alignments of the sequences were performed with CLUSTAL W software (30). The tree topology was inferred by the neighbor-joining method (27), based on 1,310 DNA sites, and the distance matrix method was performed according to the method of Jukes and Cantor (15) using the program MEGA version 2.1 (16).
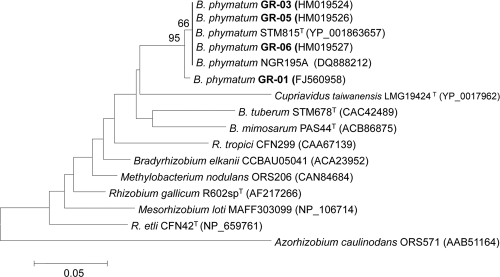
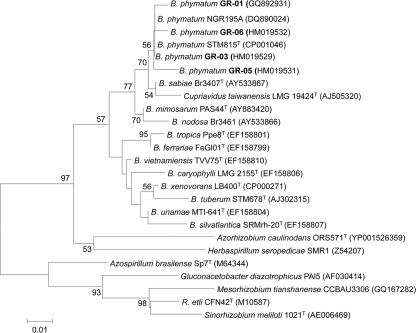
The nodC gene was amplified with the primer pairs and conditions previously described (11). Amplification of the nodC gene from each GR strain yielded a DNA fragment of about 0.4 kb (data not shown) whose nucleotide sequences were identical for all four strains and showed 99% identity to those of B. phymatum strains STM815T and NGR195A (11). A phylogenetic tree inferred from NodC sequences from B. phymatum strains and members of the order Rhizobiales is shown in Fig. 2. Primers IGK (23) and NDR-1 (31) were used for amplification of the nifH genes as indicated earlier (22). PCR amplifications of the nifH gene and further sequencing from each GR strain revealed that they all had almost identical DNA sequences, which were 99% identical to those of B. phymatum STM815T and NGR195A (11). A phylogenetic tree based on NifH sequences showing the relationships between B. phymatum and other Burkholderia and rhizobial species is shown in Fig. 3.
FIG. 2.
Phylogenetic tree inferred from NodC sequences shows the positions of four P. vulgaris-isolated strains, GR01, GR03, GR05, and GR06, within the genus Burkholderia. The tree topology was inferred by the neighbor-joining method (27) based on 195 sites. The bar represents the number of amino acid substitutions per site. One thousand bootstrap samplings were performed. The NCBI GenBank accession number for each strain is shown in parentheses.
FIG. 3.
Phylogenetic tree inferred from NifH sequences shows the positions of four P. vulgaris-isolated strains, GR01, GR03, GR05, and GR06, within the genus Burkholderia. The tree topology was inferred by the neighbor-joining method (27) based on 195 sites. The bar represents the number of amino acid substitutions per site. One thousand bootstrap samplings were performed. The NCBI GenBank accession number for each strain is shown in parentheses.
For nodulation tests, seeds of Glycine max, Cicer arietinum, Pisum sativum, Lens culinaris, Lotus corniculatus, and Medicago sativa were surface sterilized as described above for common beans. Seeds of Mimosa, Leucaena, Prosopis, and Acacia were surface sterilized with concentrated sulfuric acid for 10 min followed by 3% sodium hypochlorite for 10 min and then washed thoroughly with sterile water. Plant cultivation was carried out as indicated above. Acetylene reduction activity (ARA) by nodulated plants was assayed on detached root systems excised at the cotyledonary node as previously described (19). The GR strains are true symbionts of P. vulgaris as, after nodule isolation, they were able to establish new effective symbiosis with common beans, with values of ARA ranging from 492 to 525 μmol ethylene/plant/h. B. phymatum STM815T also infected P. vulgaris, but the efficiency of the symbiosis, determined as plant dry weight (1.06 ± 0.18 g/plant/h), was half of that found in plants nodulated by the GR strains. These strains also nodulated Mimosa pigra, Acacia cochliacantha, Acacia bilimeki, Leucaena glauca, and Prosopis laevigata but were unable to form nodules on P. sativum, L. culinaris, L. corniculatus, M. sativa, G. max, and C. arietinum.
Diazotrophy is common among Burkholderia species, as shown recently by N2 isotopic dilution studies (17). Under free-living conditions, ARA by the GR strains was tested in semisolid JMV medium as indicated earlier (26). At the end of the experiments, the culture purity was routinely checked by plating to verify uniform colony morphology. Like strain STM815T, strains isolated from P. vulgaris also had nitrogenase activity when grown ex planta. Values of activity, however, were about half of that detected in strain STM815T (83 ± 15 nmol C2H4/h).
Preparation of whole-cell proteins and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) assays were performed as described previously (12). Protein profiles were compared with type and reference strains of legume-nodulating Burkholderia species. P. vulgaris-isolated strains showed SDS-PAGE protein profiles (evaluated by visual comparison) almost identical to those from the STM815T type strain of B. phymatum but clearly different from those of other legume-nodulating Burkholderia species (Fig. 4). No differences were found when API 20 NE and API 50 CH strips were used to check for differences in nitrogen and carbon sources between B. phymatum STM815T and the GR strains (data not shown).
FIG. 4.
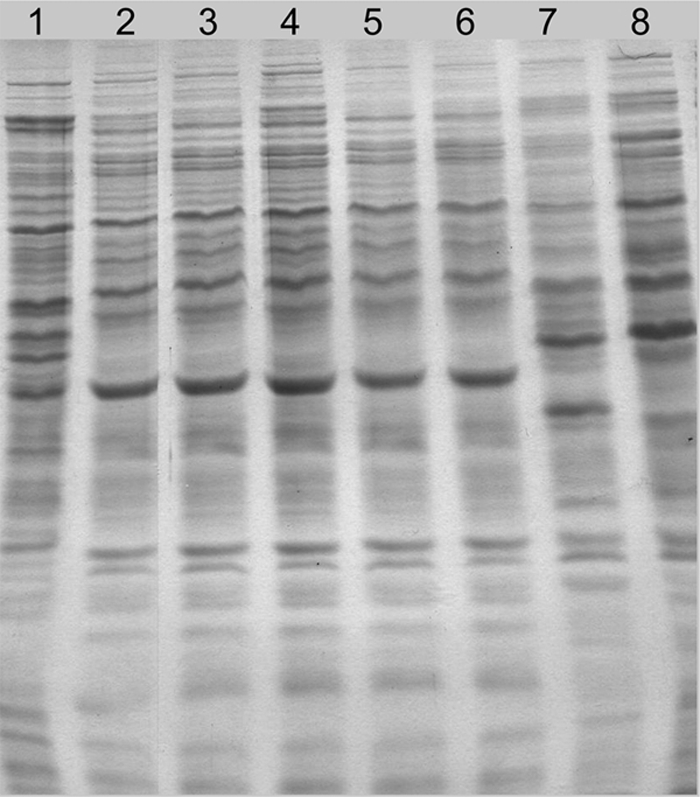
Protein electrophoregrams (SDS-PAGE) of P. vulgaris-isolated strains and type strains of known legume-nodulating Burkholderia species. Lane 1, B. tuberum STM678T; lane 2, B. phymatum STM815T; lanes 3 to 6, B. phymatum GR01, GR03, GR04, and GR06, respectively; lane 7, B. mimosarum PAS44T; lane 8, B. nodosa Br3437T.
Based on 16S rRNA gene sequences and protein profiles, which provide strong evidence for the delineation of bacterial species (32), the Phaseolus-isolated strains could be assigned to the species B. phymatum. Moreover, analysis of the phylogenetic relationships of such sequences and other Burkholderia species showed that they formed a robust clade with B. phymatum STM815T. In addition, sequencing of the nodC and nifH genes revealed that the phylogenetically closest bacterial species was B. phymatum STM815T. All these results support the affiliation of Phaseolus-isolated strains as B. phymatum. Since genomic DNAs from the GR strains had the same DNA band pattern after REP-PCR fingerprinting and extremely similar 16S rRNA and nifH gene sequences, as well as identical nodC sequences, the four strains could be derived from a single clone. Our results also suggest that strains of B. phymatum isolated from Mimosa and Phaseolus have acquired their symbiosis genes either from a common ancestor or by lateral transfer between them, the direction of transfer being unknown. Although limited to three isolates, strains NGR114 and NGR195A from Mimosa invisa and Mimosa pudica in Papua New Guinea, respectively (11), and STM815T from M. lunatum in French Guiana (21, 33), and four strains from P. vulgaris in Morocco, our results raise questions concerning the biogeographical, environmental, and host taxon distribution of B. phymatum nodule symbionts. B. phymatum was originally discovered in nodules from M. lunatum in French Guiana, and most other strains in this lineage have been found associated with host legumes in the genus Mimosa, primarily in the Neotropics. Thus, the current results extend both the known host distribution and geographic range of this group of nodule symbionts. Whether B. phymatum is prevalent on rhizobial species within nodules of P. vulgaris in the geographic site where soil samples were taken cannot be elicited from the present results. Accordingly, it will be important in future work to survey additional sites both within the native geographic range and elsewhere to understand the consistency of the association between Phaseolus and Burkholderia.
Acknowledgments
This study was supported by FEDER-cofinanced grant CGL2006-06870 from Ministerio de Ciencia e Innovación; CVI-3177 and RNM-4746 from Consejería de Innovación, Ciencia y Empresa de la Junta de Andalucía; grant 107PICO312 from Programa Iberoamericano de Ciencia y Tecnología para el Desarrollo (CYTED); and 2005MX0032 from CONACYT/CSIC.
We are grateful to L. Martínez-Aguilar (CCG-UNAM) for technical assistance in PCR and SDS-PAGE assays and P. Estrada-de los Santos (CCG-UNAM) for performing phylogenetic tree determinations based on 16S rRNA, nodC, and nifH genes.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Amarger, N., V. Marcheret, and G. Laguerre. 1997. Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris. Int. J. Syst. Bacteriol. 47:996-1006. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, C. F., and M. A. Parker. 2005. Prevalence of Burkholderia sp. nodule symbionts on four mimosoid legumes from Barro Colorado Island, Panama. Syst. Appl. Microbiol. 28:57-65. [DOI] [PubMed] [Google Scholar]
- 3.Barrett, C. F., and M. A. Parker. 2006. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Appl. Environ. Microbiol. 72:1198-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, W. M., L. Moulin, and C. Bontemps. 2003. Legume symbiotic nitrogen fixation by β-proteobacteria is widespread in nature. J. Bacteriol. 185:7266-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, W. M., S. M. de Faria, R. Straliotto, R. M. Pitard, J. L. Simoes-Araujo, J.-H. Chou, Y.-J. Chou, E. Barrios, A. R. Prescott, G. N. Elliott, J. I. Sprent, J. P. W. Young, and E. K. James. 2005. Proof that Burkholderia strains form effective symbioses with legumes: a study of novel Mimosa-nodulating strains from South America. Appl. Environ. Microbiol. 71:7461-7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, W. M., E. K. James, T. Coenye, J. H. Chou, E. Barrios, S. M. de Faria, G. N. Elliott, S. Y. Sheu, J. I. Sprent, and P. Vandamme. 2006. Burkholderia mimosarum sp. nov. isolated from root nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 56:1847-1851. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W. M., S. M. de Faria, E. K. James, G. N. Elliott, K. Y. Lin, J. H. Chou, S. Y. Sheu, M. Cnockaert, J. I. Sprent, and P. Vandamme. 2007. Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int. J. Syst. Evol. Microbiol. 57:1055-1059. [DOI] [PubMed] [Google Scholar]
- 8.de Bruijn, F. J. 1992. Use of repetitive sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 58:2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delgado, M. J., J. Olivares, and E. J. Bedmar. 1989. Nitrate reductase activity of free living and symbiotic uptake hydrogenase-positive and uptake hydrogenase-negative strains of Bradyrhizobium japonicum. Arch. Microbiol. 151:166-170. [Google Scholar]
- 10.Eardly, B. D., F. S. Wang, T. S. Whittam, and R. K. Selander. 1995. Species limits in Rhizobium populations that nodulate the common bean (Phaseolus vulgaris). Appl. Environ. Microbiol. 61:507-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G. N., W.-M. Chen, J. H. Chou, H. C. Wang, S. Y. Sheu, L. Perin, V. M. Reis, L. Moulin, M. F. Simon, C. Bontemps, J. M. Sutherland, R. Bessi, S. M. de Faria, M. J. Trinick, A. R. Prescott, J. I. Sprent, and E. K. James. 2007. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 173:168-180. [DOI] [PubMed] [Google Scholar]
- 12.Estrada-de los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera-Cervera, J. A., J. Caballero-Mellado, G. Laguerre, H. V. Tichy, N. Requena, N. Amarger, E. Martínez-Romero, J. Olivares, and J. Sanjuan. 1999. At least five rhizobial species nodulate Phaseolus vulgaris in a Spanish soil. FEMS Microbiol. Ecol. 30:87-97. [Google Scholar]
- 14.Ishii, S., and M. J. Sadowsky. 2009. Application of the rep-PCR DNA fingerprinting technique to study microbial diversity ecology and evolution. Environ. Microbiol. 11:733-740. [DOI] [PubMed] [Google Scholar]
- 15.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 16.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Arizona State University, Tempe, AZ. [DOI] [PubMed]
- 17.Martínez-Aguilar, L., R. Díaz, J. J. Peña-Cabriales, P. Estrada-de los Santos, M. F. Dunn, and J. Caballero-Mellado. 2008. Multichromosomal genome structure and confirmation of diazotrophy in novel plant-associated Burkholderia species. Appl. Environ. Microbiol. 74:4574-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Romero, E., L. Segovia, and F. M. Mercante. 1991. Rhizobium tropici, a novel nodulating Phaseolus vulgaris L. bean and Leucaena sp. trees. Int. J. Syst. Bacteriol. 41:417-426. [DOI] [PubMed] [Google Scholar]
- 19.Mesa, S., J. D. Alché, E. J. Bedmar, and M. J. Delgado. 2004. Expression of nir, nor and nos denitrification genes from Bradyrhizobium japonicum in soybean root nodules. Physiol. Plant. 120:205-211. [DOI] [PubMed] [Google Scholar]
- 20.Mhamdi, R., G. Laguerre, M. E. Aouani, M. Mars, and N. Amarger. 2002. Different species and symbiotic genotypes of field rhizobia can nodulate Phaseolus vulgaris in Tunisian soils. FEMS Microbiol. Ecol. 41:77-84. [DOI] [PubMed] [Google Scholar]
- 21.Moulin, L., A. Munive, B. Dreyfus, and C. Boivin-Masson. 2001. Nodulation of legumes by members of the β subclass of proteobacteria. Nature 411:948-950. [DOI] [PubMed] [Google Scholar]
- 22.Perin, L., L. Martínez-Aguilar, G. Paredes-Valdez, J. I. Baldani, P. Estrada-de los Santos, V. M. Reis, and J. Caballero-Mellado. 2006. Burkholderia silvatlantica sp. nov., a diazotrophic bacterium associated with sugar cane and maize. Int. J. Syst. Evol. Microbiol. 56:1931-1937. [DOI] [PubMed] [Google Scholar]
- 23.Poly, F., L. J. Monrozier, and R. Bally. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152:95-103. [DOI] [PubMed] [Google Scholar]
- 24.Rasolomampianina, R., X. Bailly, R. Fetiarison, R. Rabevohitra, G. Béna, L. Ramaroson, M. Raherimandimby, L. Moulin, P. De Lajudie, B. Dreyfus, and J. C. Avarre. 2005. Nitrogen-fixing nodules from rose wood legume trees (Dalbergia spp.) endemic to Madagascar host seven different genera belonging to alpha- and beta-Proteobacteria. Mol. Ecol. 14:4135-4146. [DOI] [PubMed] [Google Scholar]
- 25.Regensburger, B., and H. Hennecke. 1983. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 135:103-109. [DOI] [PubMed] [Google Scholar]
- 26.Reis, V. M., P. Estrada-de los Santos, S. Tenorio-Salgado, J. Vogel, M. Stofels, S. Guyon, P. Mavingui, V. L. D. Baldani, M. Schmid, J. I. Baldani, J. Balandreau, A. Hartmann, and J. Caballero-Mellado. 2004. Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant associated bacterium. Int. J. Syst. Evol. Microbiol. 54:2155-2162. [DOI] [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 28.Sawada, H., L. D. Kuykendall, and J. M. Young. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J. Gen. Appl. Microbiol. 49:155-179. [DOI] [PubMed] [Google Scholar]
- 29.Segovia, L., J. P. W. Young, and E. Martínez-Romero. 1993. Reclassification of American Rhizobium leguminosarum biovar phaseoli type I strains as Rhizobium etli sp. nov. Int. J. Syst. Bacteriol. 43:374-377. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valdés, M., N. O. Pérez, P. Estrada-de los Santos, J. Caballero-Mellado, J. J. Peña-Cabriales, P. Normand, and A. M. Hirsh. 2005. Non-Frankia actinomycetes isolated from surface-sterilized roots of Casuarina equisetifolia fix nitrogen. Appl. Environ. Microbiol. 71:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandamme, P., B. Pot, M. Gillis, P. de Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandamme, P., J. Goris, W. M. Chen, P. de Vos, and A. Willems. 2002. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov. nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25:507-512. [DOI] [PubMed] [Google Scholar]
- 34.Versalovic, J. V., F. J. de Bruijn, and J. R. Lupski. 1998. Repetitive sequence-based PCR (rep-PCR) DNA fingerprinting of bacterial genomes, p. 437-454. In F. J. de Bruijn, J. R. Lupski, and G. M. Weinstock (ed.), Bacterial genomes: physical structure and analysis. Chapman and Hall, New York, NY.
- 35.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. IBP handbook 15. Blackwell Scientific Publications, Oxford, United Kingdom.
- 36.Willems, A. 2006. The taxonomy of rhizobia: an overview. Plant Soil 287:3-14. [Google Scholar]
- 37.Wong-Villarreal, A., and J. Caballero-Mellado. 2010. Rapid identification of nitrogen-fixing and legume-nodulating Burkholderia species based on PCR-16S rRNA species-specific oligonucleotides. Syst. Appl. Microbiol. 33:35-43. [DOI] [PubMed] [Google Scholar]
- 38.Zakhia, F., and P. de Lajudie. 2006. Modern bacterial taxonomy: techniques review application to bacteria that nodulate leguminous plants (BNL). Can. J. Microbiol. 52:169-181. [DOI] [PubMed] [Google Scholar]