Abstract
Influenza virus diagnosis has traditionally relied on virus isolation in chicken embryo or cell cultures. Many laboratories have adopted rapid molecular methods for detection of influenza viruses and discontinued routine utilization of the relatively slow viral culture methods. We describe an influenza A virus reporter cell line that contributes to more efficient viral detection in cell culture. Madin-Darby canine kidney (MDCK) cells were engineered to constitutively produce an influenza virus genome-like luciferase reporter RNA driven by the canine RNA polymerase I promoter. Induction of a high level of luciferase activity was detected in the Luc9.1 cells upon infection with various strains of influenza A virus, including 2009 H1N1 pandemic and highly pathogenic H5N1 virus. In contrast, infection with influenza B virus or human adenovirus type 5 did not induce significant levels of reporter expression. The reporter Luc9.1 cells were evaluated in neutralizing antibody assays with convalescent H3N2 ferret serum, yielding a neutralization titer comparable to that obtained by the conventional microneutralization assay, suggesting that the use of the reporter cell line might simplify neutralization assays by facilitating the establishment of infectious virus endpoints. Luc9.1 cells were also used to determine the susceptibility of influenza A viruses to a model antiviral drug. The equivalence to conventional antiviral assay results indicated that the Luc9.1 cells could provide an alternative cell-based platform for high-throughput drug discovery screens. In summary, the MDCK-derived Luc9.1 reporter cell line is highly permissive for influenza A virus replication and provides a very specific and sensitive approach for simultaneous detection and isolation of influenza A viruses as well as functional evaluation of antibodies and antiviral molecules.
Influenza viruses cause respiratory tract infections associated with substantial morbidity and mortality. Seasonal influenza epidemics affect between 5 and 15% of the world population, causing 3 to 5 million cases of severe disease and approximately 0.5 million deaths per year (54). Influenza pandemics have also caused sporadic large-scale morbidity and mortality in the past century (15). Type A influenza viruses are responsible for most of the influenza disease burden in human populations (46). A novel H1N1 virus that emerged in 2009 caused an ongoing pandemic with excess morbidity and mortality (52). The genome of influenza A viruses consists of eight negative-sense RNA molecules (35) with highly conserved termini comprising the core promoter for transcription and replication (9, 22, 30, 56). Each viral RNA segment associated with nucleoprotein (NP) and RNA polymerase subunits (PB2, PB1, and PA) forming ribonucleoprotein (RNP) complexes.
Clinical and public health reference laboratories generally rely on embryonated chicken eggs or cell cultures of mammalian origin for isolation and propagation of influenza viruses (41, 43, 47). However, culture of subtype H3N2 viruses from clinical specimens by inoculation into eggs is becoming increasingly problematic; currently very few specimens yield an isolate (34). Viral isolation in cell cultures is handicapped by the relatively longer times required to obtain test results (5 to 7 days) and substantial requirements for specialized materials, equipment, and labor (42), although culture systems such as R-Mix cells provide results faster (1 to 2 days) (2, 10). Cell lines expressing reporter genes inducible upon viral infection could mitigate this problem (23, 24, 32, 33). These reporter cells exploit the specificity of viral transcription factors for their target promoters in combination with the extreme sensitivity of reporter enzymes such as luciferase (31). This approach expedites detection of specific viruses and amplifies the virus present in the clinical specimen, providing a live virus stock to be stored for further analyses. However, reporter cell lines have not been widely used for influenza virus, perhaps because the available HEK-293T reporter cells are not a favored substrate for virus isolation due to their susceptibility to the toxic effects of trypsin, which is required for the production of infectious influenza viruses in cultured cell lines (16, 20). MDCK cells have become a most widely used substrate for isolation of influenza viruses since they are known to be highly permissive for propagation of influenza viruses (47) and resistant to the toxic effects of trypsin supplementation.
Some clinical virology laboratories continue to isolate influenza viruses in cultured cells, but the faster immunochromatographic or EIA (enzyme immunoassay)-based devices or PCR are most widely used for laboratory diagnosis of influenza (8, 17, 19, 28, 36, 44, 48, 55). This trend has become problematic for influenza surveillance programs because virus isolates are indispensable for monitoring antigenic drift, vaccine seed development, and drug sensitivity testing (12, 13, 34). In addition, phenotypic analyses of viral isolates are critical to fully interpret results from vaccine and antiviral effectiveness clinical trials (3). We report here on the development of a cell-based reporter system for influenza A virus using highly permissive MDCK cells expressing a luciferase-encoding amplicon controlled by canine RNA polymerase I (POL-I) promoter elements. This cell-based reporter system provides a sensitive method for the detection and isolation of influenza A viruses, and it is also useful for the screening of antiviral drugs or neutralizing antibody assays.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby canine kidney-London (MDCK-London) cells were obtained from the CDC Scientific Resources Program, and human lung adenocarcinoma (A549; CCL-185) cells were obtained from the American Type Culture Collection (Manassas, VA), cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum and 1% penicillin-streptomycin, and incubated at 37°C in a humidified CO2 incubator. Influenza type A viruses of subtypes H1N1 (A/WS/33, A/WSN/33, A/swine/Missouri/001187/06, A/Ohio/83, A/Solomon Islands/6/06, A/New Caledonia/20/99, A/Brisbane/59/07), 2009 pandemic H1N1 (A/New York/18/09, A/New York/4/09, A/Ohio/7/09, A/Michigan/10/09), H2N2 (cold-adapted A/Ann Arbor/6/60) (26), H3N2 (A/swine/Minnesota/001170/06, A/Memphis/102/72, A/Wisconsin/67/05), H3N8 (A/equine/Montana/07, A/canine/Florida/43/04), highly pathogenic H5N1 (A/Vietnam/1203/04, A/Vietnam/JP 12-2/05, A/Hong Kong/213/03), and H9N2 (A/turkey/Wisconsin/66) and type B viruses (B/Jiangsu/10/03, B/Brisbane/33/2008) were used in this study. Virus stocks were prepared in 10-day-old embryonated eggs or in MDCK cell culture as indicated. Influenza virus titers were determined by plaque assay or endpoint dilution on MDCK cells, whereas adenovirus titers were determined by plaque assay on A549 cells.
Plasmids and construction of reporter vectors.
pSV-Luc was obtained from Promega (Madison, WI). Plasmids expressing PB2, PB1, PA, and NP proteins of influenza virus A/WSN/33 were described previously (37). The canine RNA polymerase I (POL-I) promoter reporter plasmid was constructed by replacing the DNA region containing the chloramphenicol acetyltransferase (CAT) gene in the pCAT3-basic vector (Promega, Madison, WI) with the region containing the Renilla luciferase (Rluc) gene flanked by noncoding regions (NCR) from the NP segment of influenza virus A/WSN/33, with minor modification, as described previously (25, 30). To generate a viral RNA-like RLuc amplicon under the control of POL-I, the canine POL-I promoter and terminator sequences flanking RLuc were fused upstream to the 5′ NCR or downstream to the 3′ NCR, respectively (Fig. 1A). For selection of stable transfectant cells expressing the reporter amplicon, constitutively active transcriptional cassettes expressing the neomycin phosphotransferase gene and enhanced green fluorescent protein (EGFP) (derived from pEGFP-N1; Clontech, Mountain View, CA) were inserted into the pk9POLI-RLuc vector, resulting in pk9POLI-RLuc-NeoGFP.
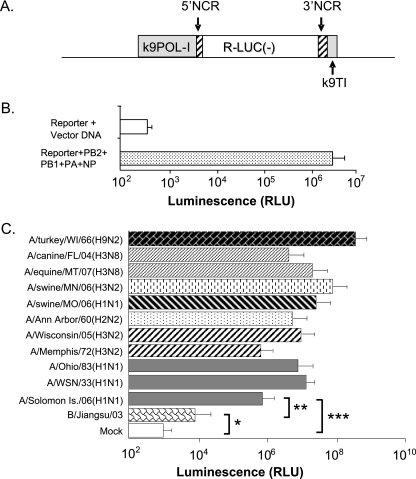
FIG. 1.
Functional analysis of the influenza virus reporter amplicon with a canine polymerase I promoter in MDCK cells. (A) The negative-sense Renilla luciferase [R-LUC(−)] coding region is flanked by modified noncoding regions (NCR; hatched boxes) from the nucleoprotein gene of A/WSN/33 virus (24, 30). The sequences of the canine POL-I promoter and the canine Pol-I terminator (k9POL-I and k9TI, respectively; gray box) were fused upstream to the 5′ NCR or downstream to the 3′ NCR, respectively. (B) MDCK cells were cotransfected with reporter plasmid pk9POLI-RLuc and influenza virus RNP-expressing plasmids or control plasmid. RLU, relative light units. (C) MDCK cells transfected with pk9POLI-RLuc plasmid for 24 h were infected with influenza viruses at an MOI of 0.001. Luciferase activities in whole-cell lysates collected at 24 h postinfection are shown as the average of luciferase activity of cells from three independent wells. The values shown are the Renilla firefly activities from 104 cells after normalization using firefly luciferase expression from a cotransfected plasmid to account for variation in transfection efficiency. Error bars depict standard errors, and brackets denote P values from Student's t test: *, P > 0.9; **, P < 0.05; and ***, P < 0.005.
Selection of stable transfectant MDCK cell lines.
For transient expression of the reporter amplicon and/or viral ribonucleoprotein (PB2, PB1, PA, and NP of WSN/33 virus), 106 MDCK cells seeded in each well of a 6-well cluster plate were transfected with 2.0 μg of each plasmid using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions (37). Reporter plasmid pSV-Luc (Promega, Madison, WI), which constitutively expresses firefly luciferase, was cotransfected (0.5 μg/well) as needed to normalize transfection efficiency. Twenty-four hours after the transfection, cells were rinsed with phosphate-buffered saline (PBS) and maintained in culture with fresh medium. Renilla luciferase activities were measured in lysates from cells harvested at 48 h after transfection.
Transiently transfected cells were inoculated with influenza viruses (multiplicity of infection [MOI] of 0.001) at 24 h after transfection. After virus adsorption for 1 h, cells were washed with PBS and then incubated for an additional 24 h in Opti-MEM medium (Invitrogen) containing 0.5 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Sigma, St. Louis, MO), followed by dual-luciferase reporter assays (Promega) or fluorescence microscopy analysis.
For development of stable MDCK transfectant cells that constitutively produce the viral RNA-like RLuc amplicon transcripts, 10 μg of linearized pk9POLI-RLuc-NeoGFP reporter plasmid DNA was transfected into MDCK cells as described above. The stably transfected cells were selected by adding 500 μg/ml of G418 (Invitrogen, Carlsbad, CA) to the culture medium with medium replacement every 3 days for a total of 2 weeks. G418-resistant cell colonies were detached from the dishes and cloned twice by use of cloning cylinders. Candidate cell clones were infected with A/WS/33 at an MOI of 0.01 to examine reporter gene inducibility by infection. Luciferase levels were measured at 24 to 72 h postinfection as appropriate.
Induction of luciferase expression by virus infection in the Luc9.1 cell line.
A stable MDCK transfectant cell clone expressing the k9POLI-RLuc amplicon, termed Luc9.1, was cultured in 100 μl DMEM (104 cells/well in a 96-well cluster plate) supplemented with 10% fetal bovine serum (FBS) and 500 μg/ml G418. Infections were performed 24 h later, by rinsing the monolayers with PBS followed by inoculation with 100 μl Opti-MEM containing 0.5 μg/ml TPCK trypsin and the appropriate amounts of virus (e.g., A/Ohio/83; within the desired multiplicity of infection range). Twenty-four hours after infection, the cell culture medium containing amplified virus was collected and stored for subsequent studies, whereas cell monolayers were rinsed once with PBS and harvested by lysis with Renilla luciferase detection reagent (Promega, Madison, WI) according to the manufacturer's instruction. For the time course study, Luc9.1 cells were infected with 10 PFU A/Ohio/83 (H1N1), A/Wisconsin/67/05(H3N2), or B/Jiangsu/10/03 virus (MOI of 0.001), and cell monolayers were harvested at 24, 48, and 72 h postinfection for Renilla luciferase activity measurements. Luminescence intensity was measured with a Centro LB960 luminometer (Berthold, Germany) or a Victor multilabel reader (Perkin-Elmer, MA). All experiments with live highly pathogenic H5N1 viruses were performed in biosafety level 3 containment, including enhancements required by the U.S. Department of Agriculture and the Select Agents program (http://www.cdc.gov/od/ohs/biosfty/bmbl5/bmbl5toc.htm).
Antibody neutralization assay.
Normal or convalescent (A/Sydney/5/97 (H3N2)) ferret sera were prepared as described previously and treated with receptor-destroying enzyme (RDE) (4, 53). Serial 2-fold dilutions of RDE-treated antiserum were carried out in Opti-MEM. A viral inoculum of 1,000 PFU diluted in Opti-MEM was added to each antibody dilution and incubated at 37°C for 1 h with brief gentle mixing at 15-min intervals. Luc9.1 cells in 96-well plates were rinsed with PBS, and virus-antibody mixtures supplemented with 0.5 μg/ml TPCK-treated trypsin were then added to the wells followed by incubation at 37°C. Intracellular Renilla luciferase activity was measured at 24 h after inoculation using a Renilla luciferase assay system according to the manufacturer's instructions. A conventional enzyme-linked immunosorbent assay (ELISA)-based microneutralization assay was performed as described by Rowe (40).
Antiviral activity assay.
Luc9.1 cells cultured in 96-well plates as described above were rinsed twice with PBS and cultured in 50 μl Opti-MEM containing the appropriate concentration of amantadine hydrochloride (Sigma, St. Louis, MO) for 30 min at 37°C. Next, a volume of 50 μl Opti-MEM containing 1,000 PFU of the appropriate virus and 0.5 μg/ml TPCK trypsin was added to the well, and the plate was incubated at 37°C. Luciferase activity was measured at 24 h after inoculation. Alternatively, parental MDCK cells were infected with viruses as described above. The cell culture supernatant was collected at 48 h after infection, and virus titer was determined by hemagglutination assay using 0.5% turkey red blood cells. Fifty percent inhibitory concentrations (IC50s), calculated from GraphPad Prism software, represent the concentration of drug that was required to inhibit virus-induced reporter activity to 50% of that of untreated cultures.
RESULTS
Functional characterization of an influenza A virus reporter amplicon in MDCK cells.
A human embryonic kidney cell line (HEK-293) was previously used as a platform to develop a reporter cell system based on a negative-sense RNA amplicon comprising a luciferase coding region flanked by the NCR (noncoding regions) of influenza virus genomic RNA segments (24). In this study, we aimed to develop a reporter cell system based on the Madin-Darby canine kidney (MDCK) cell line because it is a preferred substrate for influenza virus isolation in many clinical and research laboratories. Previous studies demonstrated that the widely used human RNA polymerase I (POL-I) promoter is virtually inactive in canine cells and most nonprimate cells (29, 49). To circumvent this restriction, we engineered an influenza virus NCR-luciferase reporter amplicon driven by the canine POL-I promoter (Fig. 1A). The reporter function of the resulting pk9POL-I plasmid was evaluated by transient transfection along with four other plasmids expressing the influenza A virus proteins required for viral transcription (PB2, PB1, PA, and NP) (25). Transfected MDCK cells yielded 8,000-fold-higher Renilla luciferase activity than control cells (without influenza virus RNP expression) (Fig. 1B). To determine whether luciferase reporter activity was also induced by viral infection, MDCK cells transfected 24 h earlier with pk9POLI-RLuc plasmid were infected with a panel of influenza A viruses (MOI of 0.001) of different subtypes. Viral infection induced increased luciferase expression between 100-fold and 20,000-fold in the transfected MDCK cells (P < 0.05) (Fig. 1C), indicating that the reporter amplicon is highly specific and responsive to influenza A virus infection (Fig. 1C) (data not shown).
Development of influenza virus reporter MDCK cell line Luc9.1.
In order to establish a reporter cell line for rapid isolation and detection of replicating influenza A virus, MDCK cells were transfected with an influenza virus luciferase reporter plasmid (pk9POLI-RLuc-NeoGFP) containing neomycin phosphotransferase and EGFP expression cassettes. Twenty- four cell colonies were individually recovered using cloning cylinders from cultures in G418 selective medium, which kills all cells that do not carry the transfected plasmid. The resulting cell clones were examined by fluorescence microscopy to determine expression of GFP indicative of plasmid retention in the cells and also tested for induction of luciferase activity by influenza A virus infection. Clone Luc9.1 was selected for further characterization because it showed the highest luciferase inducibility upon virus infection (data not shown).
Constitutive expression of an influenza virus amplicon RNA in the Luc9.1 cells might trigger host cell antiviral responses that could inhibit viral replication (45). In addition, the many population doublings (∼40) required for cloning from the original transfectant could reduce cellular permissiveness to influenza virus infection (14). To assess the susceptibility of the Luc9.1 reporter cell line to influenza viruses, we analyzed the plaquing efficiency and morphology of A/Ohio/83 (H1N1), A/Wisconsin/67/05 (H3N2), and B/Jiangsu/03 viruses on Luc9.1 cells. No significant differences in plaque number, size, and morphology were noted between Luc9.1 and parental MDCK cells, suggesting that amplicon expression and cloning procedures did not compromise their permissiveness for influenza virus replication (data not shown).
To determine the influenza virus-type specificity of luciferase induction in the Luc9.1 reporter cell line, we infected 104 cells with 1,000 PFU of A/Ohio/83 (H1N1), A/Wisconsin/67/05(H3N2), or B/Jiangsu/10/03 virus at an MOI of ∼0.1. Luciferase reporter induction by A/Wisconsin/67/05 infection (121-fold increase; P < 0.005) was similar to that of A/Ohio/83 virus (124-fold; P < 0.005) at 24 h after infection (Fig. 2A), while influenza B virus infection failed to induce significant reporter levels (2-fold). Luciferase induction by adenovirus, which is often detected in clinical specimens from patients with respiratory illness, was only 1- to 2-fold when infected with an infection dosage of 102 to 105 PFU (data not shown).
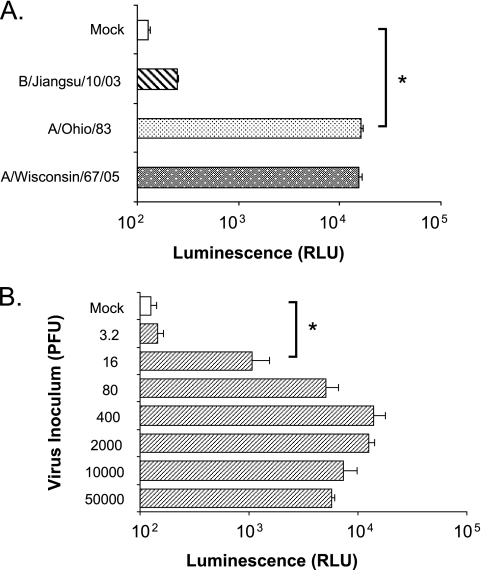
FIG. 2.
Induction of luciferase activity in the reporter Luc9.1 cells upon influenza virus infection. (A) Specificity of luciferase activation in Luc9.1 reporter cells. Luc9.1 cells were infected with A/Ohio/83 (H1N1), A/Wisconsin/67/05 (H3N2), or B/Jiangsu/10/03 virus at an MOI of 0.1. (B) Detection limits of Luc9.1 cells upon influenza virus infection. Luc9.1 cells were inoculated with the indicated amounts of infectious A/Ohio/83 virus. Luciferase activity in each culture was measured in lysates harvested at 24 h postinfection. Data represent average luciferase activity of 104 cells from three independent wells. Error bars depict standard error, and brackets denote P values from Student's t test. *, P < 0.005.
To determine the lower limit of virus detection by the Luc9.1 cells at 24 h after infection, we conducted an infection dose-response study with H1N1 influenza A virus. The maximum induction (110-fold) of luciferase activity was detected in cells infected with 400 PFU (MOI of ∼0.04) (Fig. 2B) of A/Ohio/83. Significant luciferase expression was still detectable with an infectious dose of 16 PFU (MOI of ∼0.0016) (P < 0.005). These results suggest that the stable reporter cell line Luc9.1 is highly sensitive for rapid detection (within 24 h) of low doses of infectious influenza A virus.
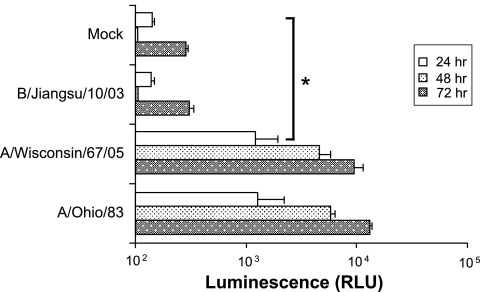
Luciferase levels from Luc9.1 cultures infected with ∼16 PFU of A/Ohio/83 were moderately increased (∼8-fold) (Fig. 2B), suggesting that longer incubation times may allow further viral replication and improve the reporter activity. Luc9.1 cells infected with 10 PFU of A/Ohio/83 (MOI of 0.001) revealed a 60-fold induction at 48 h postinfection (Fig. 3). Similarly, A/Wisconsin/67/05 (H3N2) infection yielded a 48-fold increase. A slight increase of luciferase activity was measured at 72 h postinfection. However, the increased background activity at 72 h results in similar or lower levels of induction: 46-fold for A/Ohio/83 and 33-fold for A/Wisconsin/67/05 (Fig. 3). These results indicated that with longer incubation, the Luc9.1 reporter cell line can detect as few as 10 PFU (MOI of 0.001) of influenza A virus from the two circulating influenza virus subtypes.
FIG. 3.
Time course of luciferase activity induction in the reporter Luc9.1 cells upon influenza virus infection. Luc9.1 cells were inoculated with A/Ohio/83(H1N1), A/Wisconsin/67/05 (H3N2), or B/Jiangsu/10/03 at an MOI of 0.001. Renilla luciferase activity was measured at different times after infection. Data represent normalized luciferase activity of 104 cells from three independent wells.
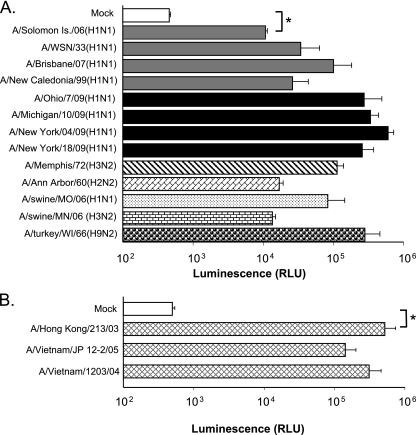
Next, we tested a set of 2009 pandemic H1N1 viruses and highly pathogenic avian H5N1 isolates in the Luc9.1 cells (Fig. 4). Cells were inoculated at an MOI of 0.01, and luciferase activity was measured after 24 h of incubation. We detected a 20- to 1,200-fold increase of luciferase activity depending on the virus isolates (Fig. 4A). Relatively high induction of luciferase activity is evident upon pandemic H1N1 virus infection. Infection of Luc9.1 cells with highly pathogenic H5N1 virus with an MOI of 0.01 induced 287- to 1,041-fold luciferase activity within 24 h of infection (Fig. 4B). These results corroborate the permissiveness of the Luc9.1 cell line to various subtypes of influenza A virus and its potential as an alternative method to detect influenza A virus infection.
FIG. 4.
Luciferase activity in Luc9.1 cells upon infection with seasonal, avian and 2009 pandemic H1N1 influenza A viruses. (A) Luc9.1 cells were infected with the seasonal H1N1, H3N2, pandemic H1N1, or virus isolated from animal species at an MOI of 0.01. Renilla luciferase activity was measured at 24 h postinfection. Error bars depict standard error, and brackets denote P values from Student's t test: *, P < 0.001. (B) Luc9.1 cells were inoculated with highly pathogenic H5N1 (A/Vietnam/1203/04, A/Vietnam/JP 12-2/05, and A/Hong Kong/213/03) at an MOI of 0.01. Renilla luciferase activity was measured at 24 h postinfection. Error bars depict standard error, and brackets denote significance per Student's t test. *, P < 0.05.
Antibody neutralization and drug sensitivity assays.
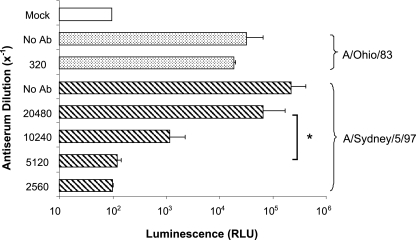
The most widely used methods to measure the specificity and potency of antiviral antibodies are neutralization assays that establish endpoints for reduction of plaque numbers or proportion of infected cultures in microtiter plates. However, plaque assays are extremely laborious and cannot be automated. Similarly, the 96-well microtiter plate assay is complicated by the need to perform a secondary assay such as ELISA to establish virus neutralization endpoints (40). Both methods pose a significant challenge, especially for studies involving large number of sera and multiple virus strains. We reasoned that the Luc9.1 MDCK cells could greatly simplify neutralizing antibody assays in clinical or research samples by facilitating the establishment of infectious virus endpoints. In this study, we performed a neutralization assay by the constant virus-variable serum approach. Convalescent H3N2 ferret serum dilutions were incubated with 1,000 PFU of A/Sydney/5/97 (H3N2) virus, and the presence of infectious virus was tested by inoculating these mixtures onto multiple cultures of Luc9.1 cells seeded in 96-well plates. Luc9.1 cell lysates revealed luciferase activity at high serum dilutions due to absence of virus neutralization by antibody. In contrast, no induction of reporter activity was recorded at low serum dilutions, indicative of the absence of virus infectivity due to neutralization (Fig. 5). Complete suppression of reporter activity was detected at dilutions of the H3N2 ferret serum of 1 in 5,120 or lower. This neutralization titer was comparable to that obtained by the conventional microneutralization assay (data not shown) (40). The H3N2 specificity of the ferret antisera was supported by the presence of high luciferase activity in Luc9.1 cells inoculated with A/Ohio/83 (H1N1) preincubated with ferret anti-H3N2 antiserum at dilutions of 320 or lower. These results indicated that the Luc9.1 MDCK cells provide a promising alternative to quantify neutralizing antibodies in biological samples.
FIG. 5.
Performance characteristics of the Luc9.1 cells in a virus neutralization assay. Two-fold dilutions of convalescent A/Sydney/5/97 ferret antiserum were incubated with 1,000 PFU of H1N1 (A/Ohio/83) or H3N2 (A/Sydney/5/97) virus. Intracellular Renilla luciferase was measured 24 h after infection. Data represent normalized luciferase activity of 104cells from three independent wells. Error bars depict standard error, and brackets denote P values from Student's t test. *, P < 0.05.
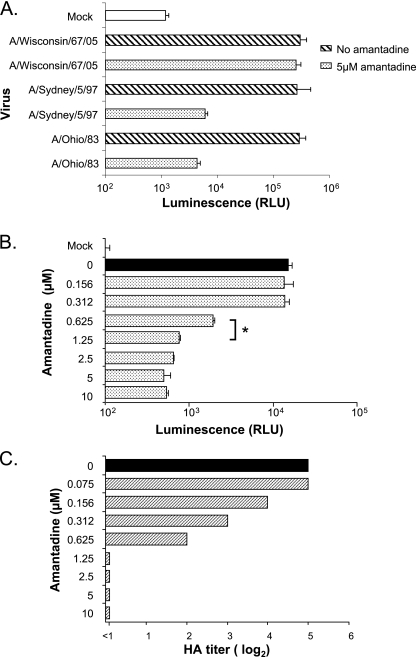
To investigate whether Luc9.1 MDCK cells are suitable to determine the susceptibility of influenza viruses to antiviral drugs, we used amantadine hydrochloride, a blocker of the influenza virus M2 protein ion channel as a model. Luc9.1 MDCK cells preincubated with 5 μM amantadine were infected with 1,000 PFU of A/Ohio/83, A/Sydney/5/97 (amantadine sensitive), or A/Wisconsin/67/05 virus, which carries an amantadine resistance mutation in M2. Luciferase activity was suppressed more than 40-fold for the amantadine-sensitive viruses, whereas A/Wisconsin/67/06 infection induced high levels of luciferase activity in treated Luc9.1 cells (Fig. 6A). To determine the median inhibitory concentration (IC50) of amantadine for A/Ohio/83 virus, different drug concentrations were preincubated with Luc9.1 MDCK cells. Cells treated with 0.625 μM amantadine showed an 80% reduction in luciferase activity at 24 h after A/Ohio/83 infection (Fig. 6B). However, further increases in amantadine concentration resulted in modest additional suppression of luciferase activity. The IC50 results from Luc9.1 cells (∼0.4 μM) were similar to those derived by the conventional hemagglutination titer reduction assay (Fig. 6C), indicating that the Luc9.1 cell line is a suitable reporter for antiviral sensitivity assays (6, 18).
FIG. 6.
Performance characteristics of the Luc9.1 cells in antiviral drug assays. (A) Amantadine-resistant virus (A/Wisconsin/67/05) and amantadine-sensitive viruses (A/Sydney/5/97 and A/Ohio/83) were incubated with 5 μM amantadine and added to Luc9.1 cells. Luciferase activity was measured at 24 h postinfection. (B and C) A/Ohio/83 virus was incubated with different concentrations of amantadine and added to Luc9.1 cells (B) and parental MDCK cells (C). Luciferase activity was measured 24 h postinfection from Luc9.1 cells. MDCK cell supernatants were collected 48 h later for HA titration. Data represent normalized luciferase activity of 104 cells from three independent wells. Error bars depict standard error, and brackets denote P values from Student's t test. *, P < 0.001.
DISCUSSION
Reporter cell lines expressing viral RNA-like amplicons provide an alternative approach to detect and isolate a variety of RNA viruses from diverse specimens (21). The sensitivity of reporter assays such as the luciferase assay reduces testing turnaround times. In addition, the simplicity of these assays relative to ELISA or real-time PCR reduces labor, supply, and space requirements. Despite these advantages, reporter cell lines have not been widely used for detection and isolation of influenza viruses (24). The high susceptibility of HEK-293T to trypsin toxicity may have discouraged the use of these reporter cells for influenza diagnosis. Multicycle replication of influenza A viruses requires trypsin to cleave the hemagglutinin (HA) of newly produced virions and enable a new round of infection (20). In the absence of proteases to cleave the HA, influenza virus reporter cell systems can only support a single round of infection, reducing their sensitivity. Thus, such systems are expected to require larger amounts of infectious virus to induce reporter gene expression.
In this study, we developed an alternative cell-based influenza virus reporter system based on MDCK cells, since the MDCK line is compatible with trypsin supplementation and reportedly a most permissive substrate for isolation and propagation of human influenza viruses (39). To this end, we first developed a plasmid that produces a transcript resembling an influenza virus genome segment directed by the strong constitutive canine RNA polymerase I promoter (25, 29, 38). This reporter plasmid was then modified to include a mammalian cell drug selection marker and used to identify a stable and sensitive influenza virus reporter cell line. Further characterization of the resulting Luc9.1 reporter cell line showed excellent sensitivity for isolation and detection of influenza A virus in samples with as few as 10 PFU with extended test time. These data represent a 1,000-fold gain in sensitivity relative to the previously reported 293T cell-based reporter system, providing improved signal-to-noise ratios for diagnosis (24). Diagnosis of influenza A virus using Luc9.1 cells can be achieved as early as 24 h postinfection with a virus inoculum of approximately 16 PFU.
Luc9.1 cells also provide an approach to simplify the detection of neutralizing antibodies in clinical specimens or research samples. The most widely used neutralization protocol with authentic influenza viruses relies on a laborious ELISA test (40). Replacement of conventional MDCK cells with the Luc9.1 cells eliminates the lengthy ELISA procedure for virus detection.
The MDCK reporter cell line generated in this study provides an alternative platform for high-throughput virus detection assays and a highly attractive antiviral assay for influenza virus drug discovery. The recent emergence of drug-resistant viruses, either spontaneously or after therapy, underscores for the need to identify novel antiviral drugs effective against influenza virus (7). In vivo cell-based screening approaches may yield broad antiviral lead compounds that target virus entry, endocytosis, and replication for further development.
In summary, the MDCK reporter cell line established in this study is highly permissive for influenza virus replication and provides a highly specific and sensitive approach for simultaneous detection and isolation of influenza viruses. Simplified neutralization assays and high-throughput antiviral drug screening can also be implemented using this reporter cell system.
Acknowledgments
These studies were supported in part by the National Vaccine Program Office, Department of Health and Human Services. We thank Alexander Klimov, Xiyan Xu, Marie Gramer, Thomas Chambers, Edward Dubovi, and Robert Webster for providing influenza virus isolates.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
Footnotes
Published ahead of print on 26 May 2010.
REFERENCES
- 1.Reference deleted.
- 2.Barenfanger, J., C. Drake, T. Mueller, T. Troutt, J. O'Brien, and K. Guttman. 2001. R-Mix cells are faster, at least as sensitive and marginally more costly than conventional cell lines for the detection of respiratory viruses. J. Clin. Virol. 22:101-110. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2008. Interim within-season estimate of the effectiveness of trivalent inactivated influenza vaccine—Marshfield, Wisconsin, 2007-08 influenza season. MMWR Morb. Mortal. Wkly. Rep. 57:393-398. [PubMed] [Google Scholar]
- 4.Chakraverty, P. 1971. Antigenic relationship between influenza B viruses. Bull. World Health Organ. 45:755-766. [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Deyde, V. M., M. Okomo-Adhiambo, T. G. Sheu, T. R. Wallis, A. Fry, N. Dharan, A. I. Klimov, and L. V. Gubareva. 2009. Pyrosequencing as a tool to detect molecular markers of resistance to neuraminidase inhibitors in seasonal influenza A viruses. Antiviral Res. 81:16-24. [DOI] [PubMed] [Google Scholar]
- 7.Dharan, N. J., L. V. Gubareva, J. J. Meyer, M. Okomo-Adhiambo, R. C. McClinton, S. A. Marshall, K. St. George, S. Epperson, L. Brammer, A. I. Klimov, J. S. Bresee, and A. M. Fry. 2009. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 301:1034-1041. [DOI] [PubMed] [Google Scholar]
- 8.Doller, G., W. Schuy, K. Y. Tjhen, B. Stekeler, and H. J. Gerth. 1992. Direct detection of influenza virus antigen in nasopharyngeal specimens by direct enzyme immunoassay in comparison with quantitating virus shedding. J. Clin. Microbiol. 30:866-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong, C. K., M. K. Lee, and B. P. Griffith. 2000. Evaluation of R-Mix FreshCells in shell vials for detection of respiratory viruses. J. Clin. Microbiol. 38:4660-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12.Gubareva, L. V., R. G. Webster, and F. G. Hayden. 2002. Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antiviral Res. 53:47-61. [DOI] [PubMed] [Google Scholar]
- 13.Hay, A. J., A. R. Douglas, D. B. Sparrow, K. R. Cameron, and J. J. Skehel. 1994. Antigenic and genetic characterization of current influenza strains. Eur. J. Epidemiol. 10:465-466. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, P., D. Marshall, Y. Reid, H. Parkes, and C. Gelber. 2007. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? Biotechniques 43:575, 577-578, 581-582. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, N. P., and J. Mueller. 2002. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76:105-115. [DOI] [PubMed] [Google Scholar]
- 16.Klenk, H. D., R. Rott, M. Orlich, and J. Blodorn. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68:426-439. [DOI] [PubMed] [Google Scholar]
- 17.Landry, M. L., and D. Ferguson. 2003. Suboptimal detection of influenza virus in adults by the Directigen Flu A+B enzyme immunoassay and correlation of results with the number of antigen-positive cells detected by cytospin immunofluorescence. J. Clin. Microbiol. 41:3407-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laplante, J. M., S. A. Marshall, M. Shudt, T. T. Van, E. S. Reisdorf, L. A. Mingle, P. A. Shult, and K. St. George. 2009. Influenza antiviral resistance testing in New York and Wisconsin, 2006-2008: methodology and surveillance data. J. Clin. Microbiol. 47:1372-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaSala, P. R., K. K. Bufton, N. Ismail, and M. B. Smith. 2007. Prospective comparison of R-mix shell vial system with direct antigen tests and conventional cell culture for respiratory virus detection. J. Clin. Virol. 38:210-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazarowitz, S. G., and P. W. Choppin. 1975. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68:440-454. [DOI] [PubMed] [Google Scholar]
- 21.Leland, D. S., and C. C. Ginocchio. 2007. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 20:49-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, X., and P. Palese. 1992. Mutational analysis of the promoter required for influenza virus virion RNA synthesis. J. Virol. 66:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo, M. K., M. Tilgner, and P. Y. Shi. 2003. Potential high-throughput assay for screening inhibitors of West Nile virus replication. J. Virol. 77:12901-12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutz, A., J. Dyall, P. D. Olivo, and A. Pekosz. 2005. Virus-inducible reporter genes as a tool for detecting and quantifying influenza A virus replication. J. Virol. Methods 126:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luytjes, W., M. Krystal, M. Enami, J. D. Pavin, and P. Palese. 1989. Amplification, expression, and packaging of foreign gene by influenza virus. Cell 59:1107-1113. [DOI] [PubMed] [Google Scholar]
- 26.Maassab, H. F., T. Francis, Jr., F. M. Davenport, A. V. Hennessy, E. Minuse, and G. Anderson. 1969. Laboratory and clinical characteristics of attenuated strains of influenza virus. Bull. World Health Organ. 41:589-594. [PMC free article] [PubMed] [Google Scholar]
- 27.Reference deleted.
- 28.Mills, R. D., K. J. Cain, and G. L. Woods. 1989. Detection of influenza virus by centrifugal inoculation of MDCK cells and staining with monoclonal antibodies. J. Clin. Microbiol. 27:2505-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami, S., T. Horimoto, S. Yamada, S. Kakugawa, H. Goto, and Y. Kawaoka. 2008. Establishment of canine RNA polymerase I-driven reverse genetics for influenza A virus: its application for H5N1 vaccine production. J. Virol. 82:1605-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann, G., and G. Hobom. 1995. Mutational analysis of influenza virus promoter elements in vivo. J. Gen. Virol. 76:1709-1717. [DOI] [PubMed] [Google Scholar]
- 31.Olivo, P. D. 1996. Transgenic cell lines for detection of animal viruses. Clin. Microbiol. Rev. 9:321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivo, P. D., P. L. Collins, M. E. Peeples, and S. Schlesinger. 1998. Detection and quantitation of human respiratory syncytial virus (RSV) using minigenome cDNA and a Sindbis virus replicon: a prototype assay for negative-strand RNA viruses. Virology 251:198-205. [DOI] [PubMed] [Google Scholar]
- 33.Olivo, P. D., I. Frolov, and S. Schlesinger. 1994. A cell line that expresses a reporter gene in response to infection by Sindbis virus: a prototype for detection of positive strand RNA viruses. Virology 198:381-384. [DOI] [PubMed] [Google Scholar]
- 34.Palese, P. 2006. Making better influenza virus vaccines? Emerg. Infect. Dis. 12:61-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palese, P., and J. L. Schulman. 1976. Mapping of the influenza virus genome: identification of the hemagglutinin and the neuraminidase genes. Proc. Natl. Acad. Sci. U. S. A. 73:2142-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasick, J. 2008. Advances in the molecular based techniques for the diagnosis and characterization of avian influenza virus infections. Transbound. Emerg. Dis. 55:329-338. [DOI] [PubMed] [Google Scholar]
- 37.Perez, D. R., and R. O. Donis. 2001. Functional analysis of PA binding by influenza a virus PB1: effects on polymerase activity and viral infectivity. J. Virol. 75:8127-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piccone, M. E., A. Fernandez-Sesma, and P. Palese. 1993. Mutational analysis of the influenza virus vRNA promoter. Virus Res. 28:99-112. [DOI] [PubMed] [Google Scholar]
- 39.Reina, J., V. Fernandez-Baca, I. Blanco, and M. Munar. 1997. Comparison of Madin-Darby canine kidney cells (MDCK) with a green monkey continuous cell line (Vero) and human lung embryonated cells (MRC-5) in the isolation of influenza A virus from nasopharyngeal aspirates by shell vial culture. J. Clin. Microbiol. 35:1900-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowe, T., R. A. Abernathy, J. Hu-Primmer, W. W. Thompson, X. Lu, W. Lim, K. Fukuda, N. J. Cox, and J. M. Katz. 1999. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 37:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, T. F., and L. Reichrath. 1974. Comparative recovery of 1972-1973 influenza virus isolates in embryonated eggs and primary rhesus monkey kidney cell cultures after one freeze-thaw cycle. Am. J. Clin. Pathol. 61:579-584. [DOI] [PubMed] [Google Scholar]
- 42.Suarez, D. L., A. Das, and E. Ellis. 2007. Review of rapid molecular diagnostic tools for avian influenza virus. Avian Dis. 51:201-208. [DOI] [PubMed] [Google Scholar]
- 43.Swenson, S. L., L. L. Vincent, B. M. Lute, B. H. Janke, K. E. Lechtenberg, J. G. Landgraf, B. J. Schmitt, D. R. Kinker, and J. K. McMillen. 2001. A comparison of diagnostic assays for the detection of type A swine influenza virus from nasal swabs and lungs. J. Vet. Diagn. Invest. 13:36-42. [DOI] [PubMed] [Google Scholar]
- 44.Taubenberger, J. K., and S. P. Layne. 2001. Diagnosis of influenza virus: coming to grips with the molecular era. Mol. Diagn. 6:291-305. [DOI] [PubMed] [Google Scholar]
- 45.Terenzi, F., M. J. deVeer, H. Ying, N. P. Restifo, B. R. Williams, and R. H. Silverman. 1999. The antiviral enzymes PKR and RNase L suppress gene expression from viral and non-viral based vectors. Nucleic Acids Res. 27:4369-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, W. W., L. Comanor, and D. K. Shay. 2006. Epidemiology of seasonal influenza: use of surveillance data and statistical models to estimate the burden of disease. J. Infect. Dis. 194(Suppl. 2):S82-S91. [DOI] [PubMed] [Google Scholar]
- 47.Tobita, K., A. Sugiura, C. Enomote, and M. Furuyama. 1975. Plaque assay and primary isolation of influenza A viruses in an established line of canine kidney cells (MDCK) in the presence of trypsin. Med. Microbiol. Immunol. (Berl.) 162:9-14. [DOI] [PubMed] [Google Scholar]
- 48.Tsukamoto, K., H. Ashizawa, K. Nakanishi, N. Kaji, K. Suzuki, M. Okamatsu, S. Yamaguchi, and M. Mase. 2008. Subtyping of avian influenza viruses H1 to H15 on the basis of hemagglutinin genes by PCR assay and molecular determination of pathogenic potential. J. Clin. Microbiol. 46:3048-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Z., and G. M. Duke. 2007. Cloning of the canine RNA polymerase I promoter and establishment of reverse genetics for influenza A and B in MDCK cells. Virol. J. 4:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reference deleted.
- 51.Reference deleted.
- 52.WHO. 2009. New influenza A (H1N1) virus: WHO guidance on public health measures, 11 June 2009. Wkly. Epidemiol. Rec. 84:261-264. [PubMed] [Google Scholar]
- 53.WHO. 2002. WHO manual on animal influenza diagnosis and surveillance. http://www.wpro.who.int/NR/rdonlyres/EFD2B9A7-2265-4AD0-BC98-97937B4FA83C/0/manualonanimalaidiagnosisandsurveillance.pdf.
- 54.WHO. March 2003, posting date. World Health Organization WHO fact sheet 211. Influenza, 2003. http://www.who.int/mediacentre/factsheets/fs211/en/.
- 55.Wright, K. E., G. A. Wilson, D. Novosad, C. Dimock, D. Tan, and J. M. Weber. 1995. Typing and subtyping of influenza viruses in clinical samples by PCR. J. Clin. Microbiol. 33:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamanaka, K., N. Ogasawara, H. Yoshikawa, A. Ishihama, and K. Nagata. 1991. In vivo analysis of the promoter structure of the influenza virus RNA genome using a transfection system with an engineered RNA. Proc. Natl. Acad. Sci. U. S. A. 88:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]