Abstract
Punctual mutations in the TEM-1 or TEM-2 gene may lead to inhibitor-resistant-TEM (IRT) β-lactamases with resistance to β-lactam-β-lactamase inhibitor combinations and susceptibility to cephalosporins. The aim of this work was to analyze the current epidemiology of IRT β-lactamases in contemporary clinical Escherichia coli. Isolates were prospectively collected in our hospital (2007 and 2008) from both outpatients (59.8%) and hospitalized patients (40.2%). The genetic relationships of the isolates were determined by XbaI pulsed-field gel electrophoresis, multilocus sequence typing, and phylogenetic group analysis. IRT genes were sequenced and located by hybridization, and the incompatibility group of the plasmids was determined. From a total of 3,556 E. coli isolates recovered during the study period, 152 (4.3%) showed reduced susceptibility to amoxicillin-clavulanate, with 18 of them producing IRT enzymes (0.5%). These were mostly recovered from urine (77.8%). A high degree of IRT diversity was detected (TEM-30, -32, -33, -34, -36, -37, -40, and -54), and the isolates were clonally unrelated but were mostly associated with phylogenetic group B2 (55.5%). In 12 out of 16 (75%) isolates, the blaIRT gene was plasmid located and transferred by conjugation in 9 of them, whereas chromosomal localization was demonstrated in 4 isolates (25%). The sizes of the plasmids ranged from 40 kb (IncN) to 100 kb (IncFII, IncFI/FIIA), and they showed different restriction patterns by restriction fragment length polymorphism analysis. Unlike extended-spectrum β-lactamase producers, the frequency of IRT producers remains low in both community and hospital settings, with most of them causing urinary tract infections. Although blaIRT genes are mainly associated with plasmids, they can be also located in the chromosome. Despite this situation, clonal expansion and/or gene dispersion was not observed, denoting the independent emergence of these enzymes.
Resistance to β-lactam-β-lactamase inhibitor combinations in Escherichia coli may be due to different mechanisms, including TEM-1 penicillinase hyperproduction, constitutive AmpC overproduction or plasmid AmpC production, OXA-type β-lactamase production, permeability deficiencies involving OmpF and/or OmpC porins, inhibitor-resistant TEM (IRT)- and complex mutant TEM (CMT)-like ß-lactamase production, and more recently, carbapenemase production (4).
IRT enzymes comprise a group of plasmid-encoding variants of TEM-1 and TEM-2 with decreased affinities for amino-, carboxy-, and ureidopenicillins and altered interaction with class A ß-lactamase inhibitors (6). IRT-producing isolates remain susceptible to cephalosporins, cephamycins, carbapenems, and in most cases, piperacillin-tazobactam. They are usually resistant to ampicillin-sulbactam and intermediate or resistant to amoxicillin-clavulanate combinations. IRT enzymes have previously been reported in different organisms, such as E. coli, Klebsiella spp., Enterobacter cloacae, Proteus mirabilis, Citrobacter freundii, and Shigella sonnei (4); but there are only a few recent epidemiological studies concerning these enzymes. Moreover, the population structure of IRT-producing E. coli isolates has not been addressed using a multilocus sequence typing (MLST) technique.
The aim of the present work was to analyze the current epidemiology of IRT β-lactamases in contemporary E. coli isolates with reduced susceptibility to amoxicillin-clavulanate recovered in our hospital in 2007 and 2008.
MATERIALS AND METHODS
Bacterial isolates.
E. coli clinical isolates with reduced susceptibility to amoxicillin-clavulanate (MICs ≥ 16/8 mg/liter), according to Clinical Laboratory Standards Institute (CLSI) recommendations (9), were prospectively collected between September 2007 and March 2008 in our institution from both outpatients and hospitalized patients. Bacterial identification and antibiotic susceptibility patterns were determined using the semiautomatic Wider system (Francisco Soria Melguizo, Madrid, Spain), which has been adapted to read MicroScan panels (Dade-MicroScan, West Sacramento, CA) (3). Only one isolate per patient was considered.
β-Lactamase detection, antimicrobial susceptibility testing, and phenotype classification.
In addition to susceptibility testing with the Wider system, agar dilution and disk diffusion methods with different β-lactam antibiotics and ß-lactamase inhibitors were performed and the results were interpreted according to CLSI guidelines (8); the resistance phenotypes were established following the recommendations of Livermore (21). Amoxicillin-clavulanate susceptibility tests were performed using both a fixed 2:1 ratio and a 2-mg/liter fixed clavulanate concentration by the agar dilution method. Disks (20 μg of amoxicillin and 10 μg of clavulanate) were purchased from Oxoid Ltd. (Basingstoke, England).
IEF.
Bacteria exponentially growing at 37°C in Luria-Bertani medium (Difco, St. Louis, MO) were harvested, and cell-free lysates were prepared by sonication. Isoelectric focusing (IEF) was performed by applying the crude sonic extract to Phast gels (pH 3 to 9) in a Phastsystem apparatus (Pharmacia AB, Uppsala, Sweden). β-Lactamases with known pIs (pIs 5.9, 5.4, 7.6, and 8.1) were used in parallel as controls. The gels were stained with nitrocefin (Oxoid) to identify the β-lactamase bands.
IRT identification.
Total bacterial DNA was obtained with a QIAamp DNA minikit (Qiagen, Hilden, Germany), and 2 μl of DNA was used as the template in each PCR. Primers for amplifying blaTEM genes (primer blaTEM-F, 5′-ATAAAATTCTTGAAGAC-3′; primer blaTEM-R, 5′-TTACCTGCTTAATC-3′) were used under the following conditions: 1 cycle of 5 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C; and a final extension of 10 min at 72°C (22). The amplicons were purified using the ExoSAP-IT reagent (Amersham, Bucks, United Kingdom), and they were further sequenced using an ABI Prism 377 automated sequencer (PE, Norwalk, CT). The nucleotide sequences obtained were compared with those available in the NCBI and Lahey databases.
Clonal relatedness.
E. coli major phylogenetic groups (groups A, B1, B2, and D) were determined by multiplex PCR, as described previously (7). Moreover, XbaI-digested genomic DNA was analyzed by pulsed-field gel electrophoresis (PFGE) using a CHEF-DRIII system (Bio-Rad, La Jolla, CA) under the following conditions: 14°C, 6 V/cm2, 10 to 40 s, and 24 h. The genetic diversity among the different band patterns was established using the software Phoretrix (version 5.0; Nonlinear Dynamics Ltd., United Kingdom) and dendrogram construction using the unweighted-pair group method using average linkages (UPGMA) method (11, 16). MLST was carried out using the primers and guidelines specified at the E. coli MLST website (http://mlst.ucc.ie/).
Location of IRT genes and plasmid characterization.
A filter mating method using LB agar plates supplemented with 50 mg/liter ampicillin was used to obtain transconjugants with IRT gene-containing plasmids. Plasmid size and plasmid typing were carried out with the transconjugants by comparing their RFLP patterns, as described previously (1, 31). Plasmids were classified according to their incompatibility group using the PCR replicon-typing scheme described by Carattoli et al. (5), followed by nucleotide sequencing of the amplicons obtained and hybridization with specific probes. The locations of the IRT genes were assessed by hybridization of I-CeuI-digested genomic DNA with blaTEM and 16S rRNA gene probes, as described previously (20).
RESULTS
Antibiotic susceptibility and IRT characterization.
A total of 3,556 E. coli isolates (46.3% from hospitalized patients and 53.7% from the community) were recovered from clinical samples in our department during the studied period (September 2007 to March 2008). Reduced susceptibility to amoxicillin-clavulanate was observed in 152 isolates (4.3%); 55 of them were fully resistant and 97 were intermediately resistant to this compound. Isolates were mainly recovered from urine (73.6%), followed by blood (9.8%), wounds (5.3%), respiratory secretions (4.6%), and other clinical samples (6.7%); and 59.8% belonged to outpatients and 40.2% to hospitalized patients.
Analysis of the phenotypic β-lactam resistance patterns of the 152 studied isolates revealed that reduced susceptibility to amoxicillin-clavulanate could be classified by phenotypic analysis as penicillinase TEM-1 hyperproduction (n = 92, 61%), extended-spectrum β-lactamase (ESBL) synthesis (n = 23, 15%), constitutive AmpC overproduction or plasmid AmpC presence (n = 19, 12%), and IRT synthesis (n = 18, 12%). Phenotypic β-lactam resistance was not associated with carbapenemase production in any of the isolates.
Presumptive IRT-producing E. coli isolates represented 0.5% (n = 18) of the total 3,556 E. coli isolates studied. Significant discrepancies in clinical categories were observed when different susceptibility testing methods were used. The IRT phenotype was detected better with a 2-mg/liter fixed clavulanate concentration (Table 1), as recommended by EUCAST (http://www.eucast.org/). Coresistance with non-β-lactam antibiotics is shown in Table 2 .
TABLE 1.
Results obtained with the different susceptibility methods used to test the 18 IRT-producing E. coli isolates
| Method | % (no.) of isolates |
||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Microdilution (2:1)a | 0 | 77.8 (14) | 22.2 (4) |
| Agar dilution (2:1)a | 11.1 (2) | 50 (9) | 38.9 (7) |
| Agar dilution (2 mg/liter)b | 0 | 27.8 (5) | 72.2 (13) |
| Disk diffusion (10 μg) | 5.5 (1) | 33.3 (6) | 61.2 (11) |
Fixed ratio of amoxicillin/clavulanate.
Fixed concentration of clavulanate.
TABLE 2.
Epidemiological characteristics of IRT-producing isolates
| IRT | pI | Isolate | Phylogroup | ST (CC) | Origin | Source | Antibiotic resistancea | blaIRT location | Plasmid size (kb)b |
Replicon type | Conjugation transfer | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | TC | |||||||||||
| TEM-30 (IRT-2) | 5.2 | 10924 | B2 | ST131 | Hospital | Respiratory tract | Nal, Cip | Plasmid | 60, 7, 5 | 50, 7, 5 | IncFII | + |
| 44965 | B2 | ST12 (CC12) | Community | Urine | Chromosome | 50 | − | |||||
| 29904 | D | ST38 (CC38) | Hospital | Respiratory tract | SxT | Plasmid | 100, 45 | IncFc | − | |||
| 46836 | D | ST393 (CC31) | Community | Urine | Plasmid | NOd | 40 | IncN | + | |||
| TEM-32 (IRT-3) | 5.4 | 34351 | B2 | ST1045 | Hospital | Blood | SxT | Plasmid | 145, 48 | 145, 48 | IncFII | + |
| 161490 | A | ST46 (CC46) | Community | Urine | Chromosome | 97 | − | |||||
| TEM-33 (IRT-5) | 5.4 | 45995 | B2 | ST95 (CC95) | Hospital | Blood | Nal, Cip | Plasmid | 97, 30 | 97 | IncFII | + |
| 20561 | B2 | ST131 | Hospital | Urine | Nal, Cip, SxT | Plasmid | 70 | 60 | IncFII | + | ||
| 127618 | D | ST12 (CC12) | Community | Urine | Nal, Cip | NO | 100 | − | ||||
| TEM-34 (IRT-6) | 5.4 | 132028 | D | ST1216 (CC38) | Hospital | Urine | Nal, Cip | Plasmid | 145, 30 | IncFIB/FIIA | − | |
| TEM-36 (IRT-7) | 5.2 | 41392 | A | ST617 (CC10) | Community | Urine | Nal, Cip | Plasmid | 97, 48 | 97,48 | IncFII | + |
| TEM-37 (IRT-8) | 5.2 | 43340 | B2 | ST131 | Community | Urine | Plasmid | 97, 40 | IncFII | − | ||
| 18204 | D | ST117 | Community | Urine | Nal | Chromosome | 97 | − | ||||
| TEM-40 (IRT-11) | 5.4 | 162266 | A | ST393 (CC31) | Hospital | Urine | Nal, Cip, SxT | NO | NO | IncFc | − | |
| 47499 | B2 | ST1217 | Community | Urine | SxT | Chromosome | 97, 40 | − | ||||
| TEM-54 | 5.4 | 138942 | B2 | ST1218 | Hospital | Urine | SxT | Plasmid | 50, 30, 20 | 50 | IncN | + |
| 115819 | B2 | ST95 (CC95) | Community | Urine | Nal, Cip, SxT | Plasmid | 145, 60 | 145, 50 | IncFII | + | ||
| 133972 | B2 | ST88 (CC23) | Community | Urine | Plasmid | NO | 50, 15 | + | ||||
Nal, nalidixic acid; Cip, ciprofloxacin; SxT; sulfamethoxazole-trimethoprim.
Plasmid size was determined by hybridization of S1-digested DNA. Underlining corresponds to plasmids with blaIRT-positive hybridization. WT, wild type; TC, transconjugant.
Double sequence.
NO, not obtained due to continuous DNA degradation.
The epidemiological characteristics of IRT-producing isolates are shown in Table 2. Characterization of these enzymes revealed a high degree of diversity, with TEM-30 (IRT-2) being the most prevalent (four patients, 22.2%), followed by TEM-33 (IRT-5) and TEM-54 (three patients each, 16.7%). Other IRT types identified were TEM-32 (IRT-3), TEM-37 (IRT-8), TEM-40 (IRT-11), TEM-34 (IRT-6), and TEM-36 (IRT-7). Resistance to nalidixic acid (55.5%) was more frequently observed in hospitalized patients (75%) than in community patients (40%).
Clonal relatedness.
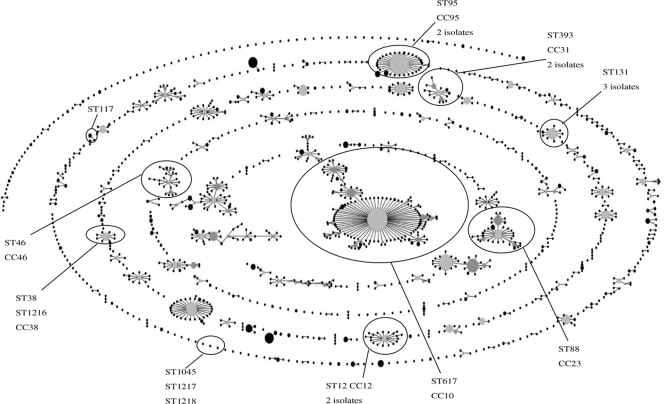
IRT-producing E. coli isolates corresponded to phylogroups B2 (10 isolates, 55.5%), D (5 isolates, 27.5%), and A (3 isolates, 16.7%). XbaI PFGE revealed unique band patterns for each of the 18 isolates. MLST results also confirmed that isolates were clonally unrelated (Table 2). Ten isolates were fit in well-defined clonal complexes (CCs) CC10, CC12, CC23, CC31, CC38, CC46, and CC95. The other eight strains were singletons; some of them were previously reported to be sequence type 131 (ST131), ST117, and ST1035; and three were newly described in this work as ST1216, grouping in CC38, ST1217, and ST1218 (Fig. 1).
FIG. 1.
Population structure determined by eBURST analysis of E. coli, based on MLST database. The STs identified in this study are indicated.
Plasmid characterization.
The blaIRT probe hybridized in a single plasmid in 12 isolates (66.6%), whereas hybridization experiments using I-CeuI-digested genomic DNA allowed us to locate the blaTEM-30 (IRT-2), blaTEM-32 (IRT-3), blaTEM-37 (IRT-8), and blaTEM-40 (IRT-11) genes in the bacterial chromosome. No clear hybridization was observed in two isolates. The transference of the blaIRT gene by conjugation was observed in only nine isolates (50%). Plasmid sizes ranged from 40 to 100 kb, with all of them showing different restriction patterns. The IRT plasmids displayed positive amplifications for the IncF (10 isolates) and IncN (2 isolates) replicons. Their nucleotide sequences revealed that both IncN plasmids were homologues to pR46 (GenBank accession number A4046276) and showed a 32045G → A mutation, while the sequences of the amplicons from the IncF plasmids were identical to the sequences of either IncFII-pRSB107 (n = 6, GenBank accession number AJ851089) or the recently described plasmids pECSF1, IncFI-FIIA-pEC14_114, and p1ESCUM (n = 1, GenBank accession numbers AP009379, GQ39086, and CU928148, respectively) (Table 2).
DISCUSSION
The IRT-producing E. coli isolates recovered in our hospital represent 0.5% of the total E. coli isolates identified over a 7-month period and 12% of all E. coli isolates with reduced susceptibility to amoxicillin-clavulanate. Nearly 80% of the isolates caused urinary tract infections, mainly in the community setting. Unlike ESBL-producing E. coli, the number of IRT producers has not substantially increased over time in our country (24). Different factors may have influenced this scenario, including the probable lower level of selective pressure exerted by β-lactam-β-lactamase inhibitor combinations compared with that exerted by extended-spectrum cephalosporins. This was particularly addressed in our study, where a polyclonal nature of the population structure was demonstrated.
The IRT phenotype confers resistance to β-lactam-β-lactamase inhibitor combinations, whereas the activity of cephalosporins normally remains unchanged. Detection of this phenotype is usually complicated due to the low level of resistance to amoxicillin-clavulanate combinations (26). The best approach to detection of the IRT phenotype is use of a 2-mg/liter fixed clavulanate concentration (Table 1), although other authors have used ampicillin-sulbactam combinations (4, 26). Nevertheless, our phenotypic approach may have some limitations to the detection of IRT producers due to the inconsistency of the IRT phenotype when other resistance mechanisms are present, such as TEM-1 or some other broad-spectrum β-lactamase, the concomitant presence of an ESBL, or even the hyperproduction of an IRT enzyme or the presence of a porin-deficient isolate.
Recently, the increment of ciprofloxacin-resistant ESBL-producing isolates, mostly of the CTX-M type, has been associated with specific epidemic multiresistant uropathogenic E. coli clones, which also include some of the clones identified in our study (28). Nevertheless, a great variety of STs were observed, including seven CCs and six singleton clones, with three of them being newly described in this work. A remarkable variety of MLST types among Spanish ESBL-producing E. coli isolates was also observed in a recent study (27), the most prevalent of which corresponded to CC10, CC23, ST131, and ST167 and some of which were associated with our IRT-producing isolates. ST131 was the most represented, with three isolates, followed by CC12 and CC95, with two isolates each. ST131-B2 is widely disseminated among both community and hospital patients and also in healthy persons and companion animals (15, 18, 29). This clone corresponds to a widespread uropathogenic E. coli strain causing urinary tract infections and bacteremia (17, 33, 34) and represents 3 out of 18 isolates (17%) of our collection. This result denotes that factors other than clonality might have an influence on the low level of representation of IRT producers, including plasmid epidemiology, the emergence of mutations leading to the IRT phenotype, or even the fitness cost associated with these mutations.
The most prevalent phylogroup among the community IRT isolates was group B2 (55%), followed by group D (28%) and group A (17%). Oteo et al. recently reported that ESBL-producing E. coli isolates in Spain usually correspond to phylogroup A (40%), followed by group B1 (28%), group D (18%), and finally, group B2 (14%) (27). B2 is mainly associated with extraintestinal pathogenic strains from the community and is frequently overrepresented in ESBL studies (2, 33). In our collection, B2 is represented in both community (all from urine samples) and hospital (two from urine, two from blood, and one from respiratory secretions) origins. Antibiotic resistance is also frequently linked to group B2, although in our case, only resistance to nalidixic acid, ciprofloxacin, or trimethoprim-sulfamethoxazole was observed, mostly in the nosocomial isolates. None of the 18 isolates showed resistance to aminoglycosides.
The large variety of IRT enzymes found in our study, eight different types in 18 E. coli unrelated clinical isolates, demonstrates the independent emergence of these enzymes, but they have less of a propensity for transmissibility or a selectable nature than ESBL-producing isolates. The chromosomal location of the blaIRT gene was demonstrated in four community isolates, a situation that has been scarcely described for TEM-type β-lactamases (19) and not previously described for IRT enzymes. Plasmids containing the blaIRT gene were identified as the narrow-host-range IncF (n = 7) or the broad-spectrum IncN (n = 2) plasmids. Recently, Marcadé et al. confirmed that IRT β-lactamases are mainly carried by IncF plasmids, which share the blaTEM-1 gene replicons and which, contrary to the blaIRT replicons, successfully disseminated in E. coli isolates around the world (23). Most of the IncF plasmids identified in our isolates (six out of seven) have identical nucleotide sequences in the amplified replicon and fit with the previously described IncFII pRSB107 (30). The first description of this plasmid was in an uncultured bacterium from a sewage treatment plant, and it had a 120- to 592-bp size. In our case, we observed different plasmid sizes, from 60 to 145 bp, but no additional antibiotic resistance determinants were observed.
One strain carried an IncFI/FIIA blaIRT plasmid that matched the pECSF1, pEC14-114, and p1ESCUM plasmids obtained from human commensal and uropathogenic E. coli strains (10, 12, 32). Finally, the blaIRT plasmids of two strains corresponded to previously described plasmid IncN-pR46, which is linked to antibiotic resistance, but in our case, a 32045G → A mutation was observed (13, 14, 25).
In summary, unlike ESBL producers, the frequency of IRT-producing isolates remains low in both community and hospital settings. The epidemiological characteristics of these isolates correspond to unrelated strains, with most of them causing urinary tract infections and being associated with the B2 phylogroup. Although blaIRT genes are mainly associated with either the IncF or the IncN plasmid, they can also be located in the chromosome. Despite this situation, clonal expansion and/or plasmid dispersion was not observed in our study. Moreover, the large variety of IRT enzymes within a complex population structure also denotes the independent emergence of these enzymes.
Acknowledgments
R.D.C. has a contract from the Spanish Ministry of Health (contract CB05/137). M.R.-D. has a post-MIR contract from the FIBIO-HRyC foundation. M.R.-B. has a contract from the Instituto de Salud Carlos III (FIS), project AI07/90034. A.V. has a contract with CIBERESP. This work was partially funded by a research grant from the CIBER en Epidemiología y Salud Pública (CIBERESP) and by the European Project TROCAR (HEALTH-F3-2008-223031).
Footnotes
Published ahead of print on 5 May 2010.
REFERENCES
- 1.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 2.Blanco, M., M. P. Alonso, M. H. Nicolas-Chanoine, G. Dahbi, A. Mora, J. E. Blanco, C. López, P. Cortés, M. Llagostera, V. Leflon-Guibout, B. Puentes, R. Mamani, A. Herrera, M. A. Coira, F. García-Garrote, J. M. Pita, and J. Blanco. 2009. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 63:1135-1141. [DOI] [PubMed] [Google Scholar]
- 3.Cantón, R., M. Pérez-Vázquez, A. Oliver, B. Sánchez Del Saz, M. O. Gutiérrez, M. Martínez-Ferrer, and F. Baquero. 2000. Evaluation of the Wider system, a new computer-assisted image-processing device for bacterial identification and susceptibility testing. J. Clin. Microbiol. 38:1339-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantón, R., M. I. Morosini, O. M. de la Maza, and E. G. de la Pedrosa. 2008. IRT and CMT β-lactamases and inhibitor resistance. Clin. Microbiol. Infect. 14(Suppl. 1):53-62. [DOI] [PubMed] [Google Scholar]
- 5.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 6.Chaïbi, E. B., D. Sirot, G. Pauland, and R. Labia. 1999. Inhibitor-resistant TEM β-lactamases: phenotypic, genetic and biochemical characteristics. J. Antimicrob. Chemother. 43:447-458. [DOI] [PubMed] [Google Scholar]
- 7.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Approved standard. Eighth edition. Document M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Clinical and Laboratory Standards Institute. 2009. Performance and standards for antimicrobial susceptibility testing. Nineteenth informational supplement. Document M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.DebRoy, C., M. S. Sidhu, U. Sarker, B. M. Jayarao, A. L. Stell, N. P. Bell, and T. J. Johnson. 2010. Complete sequence of pEC14_114, a highly conserved IncFIB/FIIA plasmid associated with uropathogenic Escherichia coli cystitis strains. Plasmid 63:53-60. [DOI] [PubMed] [Google Scholar]
- 11.Duck, W. M., C. D. Steward, S. N. Banerjee, J. E. McGowan, Jr., and F. C. Tenover. 2003. Optimization of computer software settings improves accuracy of pulsed-field gel electrophoresis macrorestriction fragment pattern analysis. J. Clin. Microbiol. 41:3035-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genoscope. 2008. Direct submission to the NCBI database of Escherichia coli UMN026 genome sequence project. Accession number CU928148. Centre National de Sequencage, Evry, France.
- 13.González-Zorn, B., A. Catalan, J. A. Escudero, L. Domínguez, T. Teshager, C. Porrero, and M. A. Moreno. 2005. armA and aminoglycoside resistance in Escherichia coli. Emerg. Infect. Dis. 11:954-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerineau, J. F., P. M. Mullineaux, and E. W. Parnell. Patent EP 0369637-A1. 23 May 1990. Sulfonamide resistance genes and their use. Advanced Technologies (Cambridge) Ltd.; Agricultural Genetics Company Limited; Biotal Limited; BP Nutrition Ltd.; Ciba-Geigy plc; Imperial Chemical Industries plc; Rhone-Poulenc Ltd.; Schering Agrochemicals Limited; Shell Research Limited; Twyford Seeds Ltd.; and Unilever United Kingdom Central Resources Ltd.
- 15.Johnson, J. R., S. Miller, B. Johnston, C. Clabots, and C. Debroy. 2009. Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and urovirulent E. coli strains among dogs and cats within a household. J. Clin. Microbiol. 47:3721-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufmann, M. E. 1998. Pulsed-field-electrophoresis, p. 33-50. In N. Woodford and A. P. Johnson (ed.), Molecular bacteriology. Protocols and clinical applications. Humana Press, Totowa, NJ.
- 17.Lau, S. H., S. Reddy, J. Cheesbrough, F. J. Bolton, G. Willshaw, T. Cheasty, A. J. Fox, and M. Upton. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J. Clin. Microbiol. 46:1076-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leflon-Guibout, V., J. Blanco, K. Amaqdouf, A. Mora, L. Guize, and M. H. Nicolas-Chanoine. 2008. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J. Clin. Microbiol. 46:3900-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lescat, M., A. Calteau, C. Hoede, V. Barbe, M. Touchon, E. Rocha, O. Tenaillon, C. Médigue, J. R. Johnson, and E. Denamur. 2009. A module located at a chromosomal integration hot spot is responsible for the multidrug resistance of a reference strain from Escherichia coli clonal group A. Antimicrob. Agents Chemother. 53:2283-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mabilat, C., and S. Goussard. 1995. PCR detection and identification of genes for extended-spectrum ß-lactamases, p. 553-557. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology. Principles and applications. American Society for Microbiology, Washington, DC.
- 23.Marcadé, G., C. Deschamps, A. Boyd, V. Gautier, B. Picard, C. Branger, E. Denamur, and G. Arlet. 2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum β-lactamases. J. Antimicrob. Chemother. 63:67-71. [DOI] [PubMed] [Google Scholar]
- 24.Miró, E., F. Navarro, B. Mirelis, M. Sabaté, A. Rivera, P. Coll, and G. Prats. 2002. Prevalence of clinical isolates of Escherichia coli producing inhibitor-resistant β-lactamases at a University Hospital in Barcelona, Spain, over a 3-year period. Antimicrob. Agents Chemother. 46:3991-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novais, A., R. Cantón, R. Moreira, L. Peixe, F. Baquero, and T. M. Coque. 2007. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob. Agents Chemother. 51:796-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver, A., M. Pérez-Vázquez, M. Martínez-Ferrer, F. Baquero, L. de Rafael, and R. Cantón. 1999. Ampicillin-sulbactam and amoxicillin-clavulanate susceptibility testing of Escherichia coli isolates with different β-lactam resistance phenotypes. Antimicrob. Agents Chemother. 43:862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oteo, J., K. Diestra, C. Juan, V. Bautista, A. Novais, M. Pérez-Vázquez, B. Moyá, E. Miró, T. M. Coque, A. Oliver, R. Cantón, F. Navarro, J. Campos, and Spanish Network in Infectious Pathology Project (REIPI). 2009. Extended-spectrum β-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34:173-176. [DOI] [PubMed] [Google Scholar]
- 28.Pitout, J. D., D. B. Gregson, L. Campbell, and K. B. Laupland. 2009. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli causing bacteraemia in the Calgary Health Region 2000-2007: the emergence of clone ST131 as a cause of community-acquired infections. Antimicrob. Agents Chemother. 53:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pomba, C., J. D. da Fonseca, B. C. Baptista, J. D. Correia, and L. Martínez-Martínez. 2009. Detection of the pandemic O25-ST131 human virulent Escherichia coli CTX-M-15-producing clone harboring the qnrB2 and aac(6′)-Ib-cr genes in a dog. Antimicrob. Agents Chemother. 53:327-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szczepanowski, R., S. Braun, V. Riedel, S. Schneiker, I. Krahn, A. Pühler, and A. Schlüter. 2005. The 120.592 bp IncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron-acquisition systems and other putative virulence-associated functions. Microbiology 151:1095-1111. [DOI] [PubMed] [Google Scholar]
- 31.Threlfall, E. J., and N. Woodford. 1995. Plasmid profile typing and plasmid fingerprinting. Methods Mol. Biol. 46:225-236. [DOI] [PubMed] [Google Scholar]
- 32.Toh, H., K. Oshima, A. Toyoda, Y. Ogura, T. Ooka, H. Sasamoto, S. H. Park, S. Iyoda, K. Kurokawa, H. Morita, K. Itoh, T. D. Taylor, T. Hayashi, and M. Hattori. 2010. Complete genome sequence of the wild-type commensal Escherichia coli strain SE15, belonging to phylogenetic group B2. J. Bacteriol. 192:1165-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodford, N., A. Carattoli, E. Karisik, A. Underwood, M. J. Ellington, and D. M. Livermore. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 53:4472-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yumuk, Z., G. Afacan, M. H. Nicolas-Chanoine, A. Sotto, and J. P. Lavigne. 2008. Turkey: a further country concerned by community-acquired Escherichia coli clone O25-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 62:284-288. [DOI] [PubMed] [Google Scholar]