Abstract
After interruption of highly active antiretroviral therapy, 15 out of 53 patients with the X4 HIV strain had a significantly larger decrease in CD4+ T cell count (P = 0.001) and shorter length of treatment interruption (P = 0.02) than patients with the R5 strain. At treatment resumption, HIV inferred tropism switched from the X4 strain to the R5 variant in 9 patients (60%). These patients had a prolonged length of treatment interruption compared to that of those who still carried the X4 strain.
The SMART study (17) demonstrated that the continuous use of antiretroviral therapy is superior to its episodic use guided by CD4+ T cell counts. However, cumulative data from other studies (11, 13, 15) suggest that CD4+ T cell-guided treatment interruptions (TIs) may be prolonged without major clinical complications in some well-controlled and otherwise healthy HIV-1-infected patients. To date, no study has evaluated the influence of HIV tropism on the efficacy of TIs.
This study was designed to assess the relevance of HIV tropism in 53 patients with persistently high CD4+ T cell counts and undetectable viremia who spontaneously decided to suspend treatment.
The patients had absolute CD4+ T cell counts of >500/μl as measured within the previous 6 months and CD4+ T cell nadir counts of >100/μl and HIV RNA levels of <50 copies/ml at study entry. Patients were restarted on therapy when their CD4+ T cell counts fell below 350/μl or upon the development of any AIDS-defining illness or any acute severe clinical event during the study.
To determine HIV-1 coreceptor use, blood samples were collected on the day of TI and when drug therapy resumed. V3 sequences were amplified using nested PCR with 1F1 and 1R1 as outer primers and 3F3 and 2R2 as inner primers (2). The V3 sequences generated were then interpreted using three different genotypic predictors, available at two websites as follows: PSSMX4R5 and PSSMSINSI at http://indra.mullins.microbiol.washington.edu/webpssm/ and geno2pheno coreceptor http://coreceptor.bioinf.mpi-inf.mpg.de/ (5, 7, 8). PSSM interprets a propensity to use CXCR4, giving low, intermediate, and high scores for R5, R5/X4, and X4 viruses, respectively (7). PSSMX4R5 and PSSMSINSI automatically report different cutoffs and assign predictions (0 = R5 or 1 = X4). According to the recommendations on the geno2pheno website, all predictions were made using the maximum sensitivity value for recognizing X4; when choosing the significance levels, we selected a 10% false-positive rate (10% probability of classifying an R5 virus falsely as X4). In our series, there was total agreement between the results for PSSMX4R5 and PSSMSINSI, whereas on two occasions (at resumption of therapy), geno2pheno predicted an X4 coreceptor in contrast to an R5 result from PSSM. Both viral strains were classified as using the R5 coreceptor at the time of TI. In these cases, we maintained the conservative R5 prediction made by PSSM, given the lower specificity of geno2pheno compared to PSSM for the detection of X4 viruses (14). Geno2pheno was useful for obtaining predictions in three cases (all R5) in which a PSSM prediction was not possible due to the presence of too many nucleotide mixtures. In summary, we determined a clear viral switch only with concordant predictions. Finally, X4 and R5/X4 dually tropic viruses have different scores according to PSSM but were all considered X4 viruses.
The chi square and Kruskal-Wallis tests were used to assess differences between patient groups with reference to categorical and continuous nonparametric variables. A multiple logistic regression model was used to obtain an adjusted analysis, accounting for all possible risk factors. All the analyses were performed on an intention-to-treat basis using SPSS 13.0 for Windows (SPSS, Inc., Chicago, IL).
The HIV inferred tropism study using peripheral blood mononuclear cells (PBMCs) demonstrated that 38 patients harbored R5 viruses, while 15 patients had X4 variants. There were no significant differences in gender, age, risk factor for HIV infection, nadir CD4+ T cell count, or treatment history between the two groups of patients (Table 1). The 38 patients carrying the R5 strain had a significantly lower median CD4+ T cell count at the time of TI than the 15 patients with the X4 variant (711 and 921 cells/μl, respectively; P = 0.008).
TABLE 1.
Characteristics of HIV-infected patients with respect to coreceptor phenotype at the time of treatment interruption
| Characteristica | Overall (n = 53) | Patients with X4 strain (n = 15) | Patients with R5 strain (n = 38) | P value |
|---|---|---|---|---|
| No. (%) of males | 37 (69.8) | 10 (66.7) | 27 (69.8) | 0.7 |
| No. (%) with indicated HIV risk factor | 0.8 | |||
| IVDU | 8 (15.4) | 3 (20.0) | 5 (13.5) | |
| Unprotected sex (heterosexual) | 24 (46.2) | 7 (46.7) | 17 (45.9) | |
| Unprotected sex (homosexual) | 20 (38.5) | 5 (33.3) | 15 (40.5) | |
| Median (IQR) age (yr) | 42 (37-47.5) | 41 (39-44) | 42 (37-48.2) | 0.7 |
| No. (%) at CDC stage A3 or B3 or C | 16 (30.2) | 4 (26.7) | 12 (31.6) | 0.7 |
| Median (IQR) CD4+ T cell count (cells/μl) at nadir | 290 (161-393) | 308 (136-396) | 279 (170-393) | 0.6 |
| Median (IQR) CD4+ T cell count (cells/μl) at baseline | 801 (625-987) | 921 (801-1120) | 711 (588-940) | 0.008 |
| No. (%) with more or less than median CD4+ T cell count (>807 CD4+ T cells/μl) at baseline | 25 (47.2) | 11 (73.3) | 14 (36.8) | 0.03 |
| No. (%) currently receiving indicated therapy | ||||
| NNRTI | 26 (49.1) | 7 (46.7) | 19 (50.0) | 0.8 |
| PI | 19 (35.8) | 7 (46.7) | 12 (31.6) | 0.3 |
| NRTI only | 8 (15.1) | 1 (6.7) | 7 (18.4) | 0.3 |
| Median (IQR) length (mo) of current therapy | 26.5 (11.7-50.2) | 31 (9-53) | 25 (15-47) | 0.9 |
| Median (IQR) total drug exposure (mo) | 84 (62.2-103.2) | 83 (65-104) | 85 (53-102) | 0.9 |
| Median (IQR) amt of HIV DNA at baseline (copies/106 PBMCs) | 330 (146.5-658.5) | 330 (239-551) | 343 (126-668) | 0.8 |
IVDU, intravenous drug user; IQR, interquartile range; NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor; NRTI, nucleoside reverse transcriptase inhibitor.
Patients with the R5 strain had a longer duration of TI than patients with the X4 variant (average, 115 days; range, 72.5 to 288 days, and average, 99 days; range, 69 to 206 days, respectively; P = 0.3). To match patients with similar immunological parameters, only patients with a baseline CD4 cell count of ≥636 (the lowest baseline CD4 cell count level in patients with X4 virus) were selected. In this analysis, the length of TI was significantly shorter for the 15 patients with the X4 strain than for 24 out of 38 (63%) patients with the R5 viral strain (160 ± 127 [mean ± standard deviation] and 533 ± 418 days, respectively; P = 0.02). The 4 patients that continued TI for longer than 942 days had the R5 strain.
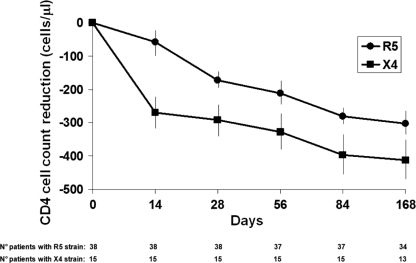
After TI, there was a significant loss in CD4 cells for patients with the R5 or X4 strains on days 14 (−58 and −270 CD4/μl, respectively; P = 0.001) and 28 (−172 and −291 CD4/μl, respectively; P = 0.04) (Fig. 1). HIV coreceptor tropism switched to the R5 variant in 9 of 15 patients (60%) with the X4 strain and to the X4 variant in 2 of 38 patients (5%) with the R5 strain.
FIG. 1.
Decreases in CD4+ T cell counts during 168 days of treatment interruption in 38 patients harboring the R5 strain and in 15 patients with the X4 strain. The number of patients evaluated at each time point is indicated at the bottom of the figure. Error bars show standard deviations.
There was no association between a switch in coreceptor usage and gender, age, transmission route, nadir or baseline CD4+ T cell count, or the number of HIV DNA copies (data not shown). The duration of TI was longer for the 9 patients showing an X4-to-R5 tropism switch than for the patients who continued to carry the X4 strain (164 ± 97 and 128 ± 127 days, respectively; P = 0.5).
On days 14 and 28, patients who switched from X4 to R5 had greater decreases in CD4+ T cell counts than patients who maintained the X4 HIV inferred tropism. On day 14 of TI, X4-to-R5 patients showed greater increases in HIV RNA plasma viremia than patients who maintained the X4 inferred tropism (26,024 and 401 HIV RNA copies/ml, respectively; P = 0.3). There were no further differences in plasma viral load between the two groups after 2 months of TI (data not shown).
In this study, the majority of patients (71.7%) who had successfully undergone prolonged highly active antiretroviral therapy (HAART) harbored an R5 HIV variant. This result could be explained by the high nadir CD4+ T cell counts in the study population.
In the first month of TI, patients harboring the X4 strain had significantly greater decreases in their CD4+ T cell counts than patients with the R5 strain. Multiple properties of X4 variants may contribute to the decline of the CD4+ T cell population, including increased replication rate, pathogenicity, and syncytium-inducing capacity in immortalized CD4+ T cell lines (1, 6, 10, 16). However, it is unknown whether X4 variants are inherently more pathogenic and directly responsible for a more rapid disease progression or whether they emerge as a consequence of progressive immune dysfunction. Nevertheless, the association of the X4 virus with a poorer prognosis remains an important issue in clinical practice.
In our study, as demonstrated by other authors (18), patients with the R5 strain remained drug free for longer periods of time than patients with the X4 variant, and only patients with the R5 strain were able to maintain TI for more than 2 years. These observations confirm specific data from other studies on the natural course of HIV infection, in which smaller decreases in CD4+ T cell counts and fewer AIDS-related clinical events were detected in patients harboring the R5-tropic virus (9).
In the absence of treatment, a switch from X4 to R5 inferred tropism was observed in 60% of patients, whereas the inverse switch (R5 to X4) was detected in only 5% of patients. Changes in coreceptor use by HIV have previously been demonstrated during HAART in the presence or absence of a CCR5 coreceptor antagonist (4, 12). In these cases, the switch is the result of an X4 variant present before treatment that reemerges from the reservoirs (4, 12, 19). It is possible that the absence of antiviral selective pressure during TI induced the emergence of minority quasispecies that were already present in the proviral DNA.
Finally, the lack of phenotypic evaluation of viral tropism could represent a bias; nevertheless, the inability to study the virus in plasma of patients with undetectable viremia justified the use of genotypic tests on plasma and PBMCs, allowing for comparison among data obtained with the same method.
The potential clinical impacts of changes in viral tropism during periods of treatment interruption are difficult to predict at this time, and the pathogenic consequences are not clear. Further studies using randomized controlled designs are urgently needed to address the potential impacts of HIV tropism switches during periods of TI on prognostic outcomes and the use of coreceptor antagonists.
Acknowledgments
This work was supported by European AIDS Treatment Network (NEAT) contract number LSHT/CT/2006/037570.
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Blaak, H., A. B. van't Wout, M. Brouwer, B. Hooibrink, E. Hovenkamp, and H. Schuitemaker. 2000. In vivo HIV-1 infection of CD45RA(R) CD4(R) T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4(R) T cell decline. Proc. Natl. Acad. Sci. U. S. A. 97:1269-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brumme, Z. L., W. W. Dong, B. Yip, B. Wynhoven, N. G. Hoffman, R. Swanstrom, et al. 2004. Clinical and immunological impact of HIV envelope V3 sequence variation after starting initial triple antiretroviral therapy. AIDS 18:F1-F9. [DOI] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Ercoli, L., L. Sarmati, E. Nicastri, G. Giannini, C. Galluzzo, S. Vella, and M. Andreoni. 1997. HIV phenotype switching during antiretroviral therapy: emergence of saquinavir-resistant strains with less cytopathogenicity. AIDS 11:1211-1217. [DOI] [PubMed] [Google Scholar]
- 5.Garrido, C., V. Roulet, N. Chueca, E. Poveda, A. Aguilera, K. Skrabal, et al. 2008. Evaluation of eight different bioinformatics tools to predict viral tropism in different human immunodeficiency virus type 1 subtypes. J. Clin. Microbiol. 46:887-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jekle, A., O. T. Keppler, E. De Clercq, D. Schols, M. Weinstein, and M. A. Goldsmith. 2003. In vivo evolution of human immunodeficiency virus type 1 toward increased pathogenicity through CXCR4-mediated killing of uninfected CD4 T cells. J. Virol. 77:5846-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen, M., F. Li, A. van't Wout, et al. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen, M., M. Coetzer, A. van't Wout, D. C. Nickle, D. Shriner, H. X. He, et al. 2006. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. J. Virol. 80:4698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, et al. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 10.Kreisberg, J. F., D. Kwa, B. Schramm, V. Trautner, R. Connor, H. Schuitemaker, et al. 2001. Cytopathicity of human immunodeficiency virus type 1 primary isolates depends on coreceptor usage and not patient disease status. J. Virol. 75:8842-8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maggiolo, F., D. Ripamonti, G. Gregis, G. Quinzan Callegaro, and F. Suter. 2004. Effect of prolonged discontinuation of successful antiretroviral therapy on CD4s: a controlled, prospective trial. AIDS 18:439-446. [DOI] [PubMed] [Google Scholar]
- 12.Moncunill, G., M. Armand-Ugon, E. Pauls, B. Clotet, and J. A. Este. 2008. In vitro HIV-1 escape to CCR5 coreceptor antagonism through selection of CXCR4-using variants. AIDS 22:23-31. [DOI] [PubMed] [Google Scholar]
- 13.Mussini, C., A. Bedini, V. Borghi, G. Guaraldi, R. Esposito, E. Barchi, R. Enilia, A. Cozzi-Lepri, A. N. Philips, P. Ortolani, G. Bratt, L. E. Eriksson, L. Sighinolfi, A. Cossarizza, A. d'Arminio Monforte, A. De Luca, S. Di Giambenedetto, and A. Antinori for the International Study Group on CD4-monitored Treatment Interruptions. 2005. CD4 cell-monitored treatment interruption in patients with a CD4 cell count > 500 × 106 cells/l. AIDS 19:287-294. [PubMed] [Google Scholar]
- 14.Raymond, S., P. Delobel, M. Mavigner, M. Cazabat, C. Souyris, K. Sandres-Sauné, L. Cuzin, B. Marchou, P. Massip, and J. Izopet. 2008. Correlation between genotypic predictions based on V3 sequences and phenotypic determination of HIV-1 tropism. AIDS 22:F11-F16. [DOI] [PubMed] [Google Scholar]
- 15.Sarmati, L., C. Andreoni, E. Nicastri, C. Tommasi, A. Buonomini, G. D'Ettorre, et al. 2009. Prognostic factors of long-term CD4+ count-guided interruption of antiretroviral treatment. J. Med. Virol. 81:481-487. [DOI] [PubMed] [Google Scholar]
- 16.Schuitemaker, H., N. A. Kootstra, R. E. de Goede, F. de Wolf, F. Miedema, and M. Tersmette. 1991. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J. Virol. 65:356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarwater, P., M. Parish, and J. Gallant. 2003. Prolonged treatment interruption after immunologic response to highly active therapy. Clin. Infect. Dis. 37:1541-1548. [DOI] [PubMed] [Google Scholar]
- 18.Toulson, A. R., R. Harrigan, K. Heath, B. Yip, Z. L. Brumme, M. Harris, R. S. Hogg, and J. S. G. Montaner. 2005. Treatment interruption of highly active antiretroviral therapy in patients with nadir CD4 cell counts >200 cells/mm3. J. Infect. Dis. 192:1787-1793. [DOI] [PubMed] [Google Scholar]
- 19.Westby, M., M. Lewis, J. Whitcomb, M. Youle, L. Anton Pozniak, Ian T. James, et al. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1(HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]