Abstract
Cupriavidus pauculus is a water microorganism rarely isolated from clinical specimens. We describe a pseudo-outbreak in which multiple strains that were associated with moistening of culturette swabs with tap water were isolated from a single clinic before collecting the patient specimen.
CASE REPORT
In a period of 6 weeks, 27 skin and superficial site swab specimens were submitted from an outpatient clinic to the clinical microbiology laboratory for bacterial culture. The specimens were plated onto blood agar (BA; Becton Dickinson, MD), MacConkey (Becton Dickinson, MD), and chocolate agar (CA; Becton Dickinson, MD) per routine procedure. Eleven of the clinical specimens grew an unusual bacterium. After 24 h, the colonies were smooth, round, and colorless on BA and CA and nonfermentative on MacConkey agar. The cultures were negative for common skin flora, such as coagulase negative Staphylococcus or Corynebacterium spp., and other commonly isolated wound pathogens, such as Staphylococcus aureus or Pseudomonas aeruginosa (6). The organisms were found to be Gram negative and catalase and oxidase positive and were reported as “Pseudomonas-like” nonfermenting bacilli by the MicroScan WalkAway system (Siemens Healthcare Diagnostics, IL).
Because of the uncommon nature and the presence of this bacterium in multiple specimens in a short span of time, further investigation was undertaken. All the specimens originated from a single hospital-affiliated outpatient clinic and were submitted by the same physician's office. The swab specimens had been collected with double culturette swabs (BBL CultureSwab; BD Diagnostics, Sparks, MD) from various skin and wound sites from patients with various comorbidities and of different age groups. The clinic was contacted, and unopened culturette swabs of the same lot number were requested to rule out contamination. The uninoculated culturettes plated onto BA, CA, MacConkey, and colistin nalidixic acid (CNA; Remel, KS) plates did not yield any growth, thereby ruling out contamination. The clinic was contacted again for a detailed account of specimen collection methods. Upon further inquiries, it was discovered that the culturette swabs were moistened with tap water before collecting the patient specimen. A tap water sample from the clinic sink was requested and plated onto BA, CA, MacConkey, and CNA plates by using a swab, and it grew a “Pseudomonas-like” nonfermenting bacilli morphologically similar to the patient isolates.
A total of six isolates and the water isolate were separately referred to the Ohio Department of Health Laboratory and to the Centers for Disease Control and Prevention (CDC) reference laboratories for further identification. The isolates were identified as Cupriavidus (Ralstonia, Wautersia) pauculus based on morphology and biochemical tests. The following tests were reported positive: oxidase, catalase, citrate, and urea. The following were negative: indole, nitrate, methyl red-Voges-Proskauer, gelatin, and esculin. There was no acid production from glucose, xylose, mannitol, lactose, sucrose, or maltose. There was no growth in 6% NaCl, in the presence of cetrimide, or on Pseudo F and P agar. Triple sugar iron and litmus milk were alkaline, and motility was observed.
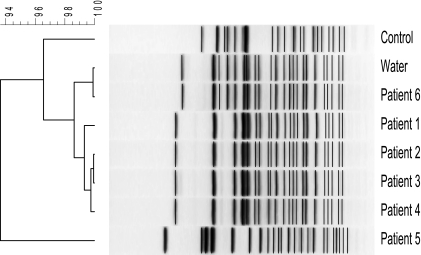
To determine if these isolates were related, pulsed-field gel electrophoresis (PFGE) analysis was performed on six available patient isolates, the tap water isolate, and an unrelated Cupriavidus pauculus control isolate (Fig. 1). Salmonella enterica serotype Braenderup was restricted for 4 h at 37°C by using XbaI (Roche Molecular Biochemical, Indianapolis, IN) and used as the normalization standard for gel analysis. Cupriavidus isolates were restricted for 4 h at 37°C using SpeI (New England BioLabs, Ipswich, MA). The restriction fragments were separated in a contour-clamped homogeneous electric field (CHEF) Mapper unit (Bio-Rad Laboratories, Hercules, CA) for 24 h, using the following running parameters: 6 V/cm; initial switch, 2 s; and final switch, 60 s. The restricted DNA was embedded in a 1.6% SeaKem Gold agarose gel and run on a Bio-Rad CHEF Mapper unit in 0.5× Tris-borate-EDTA. Interpretation of PFGE profiles followed the description by Tenover et al. (14). Four distinct strain types were identified among the patient and tap water samples based on their electrophoresis banding pattern. The results of the analysis indicate that the PFGE patterns of isolates from patients 1, 2, and 3 were indistinguishable, as the isolates shared an identical banding pattern. The isolate from patient 4 was probably related to the isolates from patients 1, 2, and 3, as its banding pattern differed by one band. The PFGE pattern of the isolate from patient 5 was unique, as it was different from the PFGE patterns of the isolates from patients 1 through 4 and patient 6. The PFGE patterns of the isolates from patient 6 and the tap water were indistinguishable, and these isolates' patterns showed similarity to those from patients 1 though 4. The banding patterns of patient isolates 1 through 4 were the most closely related, and the clinical specimens were also temporally related. Patient isolate number 6 and the tap water isolate were both sent on the same day, roughly 6 weeks after the patient number 5 isolate, and their banding patterns were identical. We infer that the slight changes in banding pattern seen on PFGE represent different genotypes of C. pauculus present in the clinic's tap water source. Additionally, only one distinct strain was identified in the tap water sample. Given the epidemiologic context, we conclude that the patient isolates depicted the C. pauculus strain present in the tap water at various times of specimen collection. The control isolate was retrieved from a previously archived unrelated laboratory strain of C. pauculus.
FIG. 1.
PFGE pattern and dendrogram of C. pauculus isolates. Six patient and water C. pauculus isolates were restricted, and fragments were separated by electrophoresis. The gel was stained with ethidium bromide, visualized using a Bio-Rad GelDoc 2000 to create a TIF image, and analyzed using Applied Maths BioNumerics version 5.1 software to produce the dendrogram. The tree was calculated using Pearson's pairwise correlation with the average linkage method. Numbers at the upper left of the figure represent percent similarity.
Retrospectively, we found that 46 specimens had been submitted by the physician's office in a period of 7 months. During this time period, 24 wound specimens from 11 different patients grew an isolate identified as Gram-negative bacilli, nonfermentative bacilli, or Pseudomonas-like by the MicroScan WalkAway system. Six isolates that were sent to the reference laboratory were subsequently identified as C. pauculus and were from this location, and C. pauculus was the only microorganism recovered. The remaining five isolates had been reported as Pseudomonas-like, and/or several of these cultures grew additional isolates, including Escherichia coli, Staphylococcus aureus, and coagulase-negative Staphylococcus. The clinic was contacted to report our findings and to advise proper specimen collection procedures. Cultures were repeated on three of the original patients, and none of the patients from whom C. pauculus was isolated grew this organism in any subsequent clinical specimens. Six months after the “pseudo-outbreak,” no additional C. pauculus isolates have been recovered from any further specimen from this clinic.
Cupriavidus spp. are ubiquitous environmental organisms that are mostly found in soil, in water, and on plants. In 1995, the genus Ralstonia was established to include species formerly known as Alcaligenes eutrophus, Burkholderia solanacearum, and Burkholderia pickettii (20). In 1999, Vandamme et al. described it as Ralstonia paucula (17); the description is identical to the one given by Vaneechoutte et al. in 2004 as Wautersia paucula (18), and they were finally renamed C. pauculus (16). Cupriavidus species are Gram-negative, aerobic, non-spore-forming, motile bacilli. They are catalase and oxidase positive, are nonfermentative on MacConkey agar, oxidize glucose, and degrade nitrate. Cupriavidus species are nutritionally versatile and mesophilic, though slight differences in optimal growth temperatures can help to distinguish between species (17). Colonies are round, smooth, convex, and nonpigmented (17).
Species known to cause human disease include Ralstonia pickettii, Ralstonia gilardii, Ralstonia mannitolilytica, and C. pauculus (formerly CDC group IV c-2). Though Cupriavidus species are not commonly isolated from clinical specimens, C. pauculus is the species most likely to be isolated (17). C. pauculus can be distinguished from R. pickettii, R. gilardii, and R. mannitolilytica by biochemical tests (nitrate reduction, carbohydrate acidification, urease, and fatty acid profile) and conclusively by DNA sequencing (17).
Historically, C. pauculus has rarely been identified as a pathogen in patients. It can, however, cause significant disease with significant comorbidities, especially in immunocompromised individuals, such as patients with hematologic malignancies and AIDS. There have been case reports of bacteremia, septicemia, peritonitis, abscess, and tenosynovitis (1-10, 13, 15, 19-21). In many cases, the source of the microorganism remained undetermined, but for cases in which the source was found, a contaminated water source was identified (9, 10). In the literature, hydrotherapy pools (5), nebulization solution, and even bottled mineral water (8) have been recognized as potential sources of contamination (12). Though generally considered a nosocomial infection, there is a case report of community-acquired nonfatal septicemia in a 37-year-old man with plasma cell leukemia (5). Additionally, Noyola and Edwards reported a case of bacteremia in an immunocompetent 6-month-old infant (11).
Though we describe several cases of C. pauculus or isolates reported as Pseudomonas-like and likely to be C. pauculus, it is unlikely that these isolates were actual pathogens. None of the patients in question were immunocompromised, and none, upon reculture, grew C. pauculus a second time. In a different setting, i.e., an inpatient ward, a contaminated water source such as this may have serious implications for certain patient populations. Ultimately, this “pseudo-outbreak” of Cupriavidus emphasizes the importance of proper specimen collection, physician education, and trend recognition in the clinical microbiology laboratory.
Footnotes
Published ahead of print on 5 May 2010.
REFERENCES
- 1.Anderson, R. R., P. Warnick, and P. C. Schreckenberger. 1996. Recurrent CDC group IV c-2 bacteremia in a human with AIDS. J. Clin. Microbiol. 35:780-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arduino, S., H. Villar, M. T. Veron, B. Koziner, and M. Dictar. 1993. CDC group IV c-2 as a cause of catheter-related sepsis in an immunocompromised patient. Clin. Infect. Dis. 17:512-513. [DOI] [PubMed] [Google Scholar]
- 3.Aspinall, S. T., and R. Graham. 1989. Two sources of contamination of a hydrotherapy pool by environmental organisms. J. Hosp. Infect. 14:285-292. [DOI] [PubMed] [Google Scholar]
- 4.Crowe, H. M., and S. M. Brecher. 1987. Nosocomial septicemia with CDC group IV c-2, an unusual gram-negative bacillus. J. Clin. Microbiol. 25:2225-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dan, M., S. A. Berger, D. Aderka, and Y. Levo. 1986. Septicemia caused by the gram-negative bacterium CDC IV c-2 in an immunocompromised human. J. Clin. Microbiol. 23:803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giacometti, A., O. Cirioni, A. M. Schimizzi, M. S. Del Prete, F. Barchiesi, M. M. D'Errico, E. Petrelli, and G. Scalise. 2000. Epidemiology and microbiology of surgical wound infections. J. Clin. Microbiol. 38:918-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen, W., and P. D. Y. Glupczynski. 1985. Group IV c-2 associated peritonitis. Clin. Microbiol. Newsl. 7:43-44. [Google Scholar]
- 8.Manaia, C. M., O. C. Nunes, P. V. Morias, and M. S. da Costa. 1990. Heterotrophic plate counts and the isolation of bacteria from mineral waters on selective and enrichment media. J. Appl. Bacteriol. 69:871-876. [DOI] [PubMed] [Google Scholar]
- 9.Moissenet, D., M. Tabone, J. Girardet, G. Leverger, A. Garbarg-Chenon, and H. Vu-Thien. 1996. Nosocomial CDC group IV c-2 bacteremia: epidemiological investigation by randomly amplified polylmorphic DNA analysis. J. Clin. Microbiol. 34:1264-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musso, D., M. Drancourt, J. Bardot, and R. Legre. 1994. Human infection due to the CDC group IV c-2 bacterium: case report and review. Clin. Infect. Dis. 18:482-484. [DOI] [PubMed] [Google Scholar]
- 11.Noyola, D. E., and M. S. Edwards. 1999. Bacteremia with CDC group IV c-2 in an immunocompetent infant. Clin. Infect. Dis. 29:1572. [DOI] [PubMed] [Google Scholar]
- 12.Oie, S., D. Makieda, S. Ishida, Y. Okano, and A. Kamiya. 2006. Microbial contamination of nebulization solution and its measures. Biol. Pharm. Bull. 29:503-507. [DOI] [PubMed] [Google Scholar]
- 13.Ramos, J. M., F. Soriano, M. Bernacer, J. Esteban, and J. Zapardiel. 1993. Infection caused by the nonfermentative gram-negative bacillus CDC group IV c-2: case report and literature review. Eur. J. Clin. Microbiol. Infect. Dis. 12:456-458. [DOI] [PubMed] [Google Scholar]
- 14.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thayu, M., R. S. Baltimore, B. J. Slight, M. Reyes-Mugica, and P. Hotez. 1999. CDC group IV c-2 bacteremia in a child with recurrent acute monoblastic leukemia. Pediatr. Infect. Dis. J. 18:397-398. [DOI] [PubMed] [Google Scholar]
- 16.Vandamme, P., and T. Coenye. 2004. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. 54:2285-2289. [DOI] [PubMed] [Google Scholar]
- 17.Vandamme, P., J. Goris, T. Coenye, B. Hoste, D. Janssens, K. Kersters, P. De Vos, and E. Falsen. 1999. Assignment of Centers for Disease Control group IVc-2 to the genus Ralstonia as Ralstonia paucula sp. nov. Int. J. Syst. Bacteriol. 49:663-669. [DOI] [PubMed] [Google Scholar]
- 18.Vaneechoutte, M., P. Kämpfer, T. De Baere, E. Falsen, and G. Verschraegen. 2004. Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al. 1990) comb. nov. Int. J. Syst. Evol. Microbiol. 54:317-327. [DOI] [PubMed] [Google Scholar]
- 19.Vay, C., S. García, G. Alperovich, M. Almuzara, M. Lasala, and A. Famiglietti. 2007. Bacteremia due to Cupriavidus pauculus (formerly CDC group IVc-2) in a hemodialysis patient. Clin. Microbiol. Newsl. 29:30-32. [Google Scholar]
- 20.Yabuuchi, E., Y. Kosako, I. Yano, H. Hotta, and Y. Nishiuchi. 1995. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol. Immunol. 39:897-904. [DOI] [PubMed] [Google Scholar]
- 21.Zapardiel, J., G. Blum, C. Caramelo, R. Fernandez-Roblas, J. L. Rodriguez-Tudela, and F. Soriano. 1991. Peritonitis with CDC group IV c-2 bacteria in a patient on continuous ambulatory peritoneal dialysis. Eur. J. Clin. Microbiol. Infect. Dis. 10:509-511. [DOI] [PubMed] [Google Scholar]