Abstract
Several methods have been used for typing of Streptococcus agalactiae (group B streptococci [GBS]). Methods currently in use may provide inadequate resolution (e.g., typing of capsular polysaccharides and surface protein) or are labor-intensive and expensive (e.g., multilocus sequence typing [MLST] or pulsed-field gel electrophoresis). This work describes the construction and use of a multiple-locus variant-repeat assay (MLVA) on 126 well-characterized human GBS strains, consisting mostly of invasive Norwegian strains and international reference strains. Based on in silico whole-genomic analysis of the genomes of strains A909, NEM316, and 2603V/R, 18 candidate loci were selected and investigated by PCR. Eleven loci showed diversity, and the five most diverse loci were used for the construction of an MLVA, consisting of a multiplex PCR followed by fragment analysis with capillary electrophoresis. The assay generated clusters which corresponded well with those observed by other methods. However, it provided a considerably higher degree of diversity, with 70 different MLVA types compared to 36 types generated by MLST. Simpson's index of diversity for the 5-locus MLVA was 0.963, compared to 0.899 for the MLST in this strain collection. MLVA results will generally be available within 2 days, which is usually faster than MLST. In our hands, MLVA of GBS represents a rapid, easy, and comparably inexpensive method for high-resolution genotyping of GBS.
Streptococcus agalactiae is the only species within the group B streptococci (GBS) of Lancefield's classification of hemolytic streptococci. It is well recognized as a human pathogen, especially for causing septicemia, meningitis, and other serious invasive disease in neonates; it is also associated with stillbirth. In addition, it is known to cause serious maternal infections and serious infections in elderly and immunocompromised patients.
Different approaches for typing of GBS have been developed through the years. Most commonly, GBS strains are assigned to one of the 10 capsular polysaccharide (CPS) types. Typing of surface proteins and analysis of the antibiotic resistance pattern can add further resolution to typing of phenotypic features. Examination of CPS or surface proteins can be done by immunological methods or, more recently, by detecting the corresponding genes by PCR (6, 8, 10). Multilocus sequence typing (MLST) was developed to study the long-term evolutionary development of a species. The total number of GBS sequence types (STs) is currently around 490 (http://pubmlst.org/sagalactiae/). Pulsed-field gel electrophoresis (PFGE) based on macrorestriction fragment analysis of genomic DNA is considered highly discriminatory. For GBS, the restriction enzyme SmaI is often used, generating only 8 to 12 restriction fragments, which may be suboptimal with respect to discriminating capacity (2, 19, 21). Since PFGE is an image-based method, it may be difficult to compare results between different laboratories. Both MLST and PFGE are labor-intensive techniques requiring experienced personnel and usually several days of work before results are available.
DNA loci consisting of repeated sequences are widespread throughout the genome of bacteria and have a tendency to vary in repeat number from strain to strain, so-called “variable number of tandem repeats” (VNTR). The combination of several VNTR loci in an assay (multiple-locus variant-repeat assay [MLVA]) has been shown in various bacterial species to generate strain-specific profiles that can be compared, exchanged, and reproduced in different laboratories. The technique has been employed successfully for the typing of an increasing number of bacterial species (23). The aim of the present study was to explore the GBS genome for variably repeated genetic regions and to investigate the feasibility of an MLVA method for typing the bacterium.
MATERIALS AND METHODS
Selection, culture, and DNA extraction of GBS strains.
A panel of 126 GBS strains was selected to represent a broad range of CPS and surface proteins and sequence types. Most of these (n = 113) were invasive strains submitted to the Norwegian reference laboratory for GBS. They were also used in a previous study of invasive GBS in infants (3). In addition, nine international reference strains were included, among them all eight fully sequenced strains. Two Zimbabwean colonizing strains and two Danish CPS type IX strains were also included. The distribution of CPS types was as follows: Ia, 12 strains; Ib, 10 strains; II, 6 strains; III, 55 strains; IV, 9 strains; V, 30 strains; VI, 1 strain; and IX, 3 strains. Strains stored at −80°C were grown overnight on blood agar plates. For nucleic acid extraction, 4 to 5 colonies were added to 200 μl lysis solution containing 12 μl (20 mg/ml) lysozyme (Sigma-Aldrich Corp., St. Louis, MO), 4.8 μl (20 mg/ml) proteinase K (Sigma), 4.8 μl (10,000 U/ml) mutanolysin (Sigma), and 178.4 μl Tris-EDTA (TE) buffer and incubated at 37°C and 65°C for 15 min each. DNA was purified on a Qiagen BioRobot M48 instrument using the MagAttractDNA Mini 48 kit (Qiagen, Hilden, Germany) and eluted in a volume of 50 μl. The eluate was diluted 1:10 for PCR.
Detection of VNTRs, PCR protocols, and analysis of fragments.
Genomes were analyzed using the Tandem Repeats Finder program, version 4.00 (1). The Variable Region Finder available at the Health Protection Agency website (http://www.hpa-bioinformatics.org.uk/variable_region_finder/index.html) was also used. Primers were designed using Oligo 6.71 software (Molecular Biology Insights, Inc., Cascade, CO). The loci were consecutively termed TR1 through TR18, while the five loci proposed for the subsequent MLVA were renamed SATR1 to SATR5 in the later phase of the study. Analysis of the repeats was generally done with single PCRs in 25 μl of reaction mixture containing 50 μM (each) dATP, dCTP, dGTP, and dTTP; forward and reverse primers (0.5 μM); 2.5 μl 10× PCR buffer with 15 mM MgCl and 0.75 U AmpliTaq Gold DNA polymerase (both of the latter by Applied Biosystems, Foster City, CA); a 1-μl aliquot of the purified, diluted DNA template; and RNase-free water. The PCR conditions were 5 min at 95°C for activation of the polymerase and then 35 cycles of 30 s at 95°C, 30 s at annealing temperature, and 60 s at 72°C. The annealing temperature used initially was that proposed by the primer design software, but the temperature was later changed to 55°C. The PCR was performed on an MJ Research PTC-200 instrument (MJ Research, Inc., Watertown, MA). Amplicon size was determined with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) using the DNA 1000 chip. Later, amplicon size was determined by either 2% agarose gel electrophoresis for the VNTRs that showed only two or three alleles or by fragment analysis with capillary electrophoresis. The Qiagen multiplex PCR kit was used for the subsequent multiplex PCR. The manufacturer's recommendations were followed, except for the annealing temperature, which was set to 55°C. In addition, the concentrations of the individual primers in the primer mix were adjusted to 1.5 μM for SATR1 and -5, 1 μM for SATR2 and -3, and 0.5 μM for SATR4, and the total PCR volume was reduced to 25 μl.
Fragment analysis was done on an ABI 3130xl genetic analyzer. Hi-Di formamide, GeneScan 1200 LIZ size standard, and the PCR product were mixed (9 plus 0.5 plus 0.5 μl). The standard protocol for fragment analysis with 36-cm capillaries and POP7 polymer was used. The product sizes were analyzed using the GeneMapper 4.0 software. (All products used for fragment analysis were from Applied Biosystems.)
For analysis of the stability of the alleles, three strains (06/14, 06/123, and 07/15) were passaged on blood agar plates 40 times over a period of 4 months.
Serotyping and MLST.
Typing of CPS and surface proteins was done by PCR (6, 11, 24), except that immunofluorescence microscopy was used for the nine Norwegian strains collected in 2005 (4). MLST was performed as described elsewhere (9). Results were available for all 126 of the strains. STs of most of the Norwegian strains were available from a previous study (3), whereas the ST of the international reference strains was available from a recent publication (22).
Analysis of results.
For cluster analysis, BioNumerics 6.0 software (Applied Maths, Sint-Martens-Latem, Belgium) was used. Simpson's index of diversity was calculated either via the VNTR DIversity and Confidence Extractor (V-DICE) software at the Health Protection Agency website (Health Protection Agency, Colindale, London, United Kingdom; http://www.hpa-bioinfotools.org.uk/cgi-bin/DICI/DICI.pl) or manually (18). The number of repeats was determined by subtracting the offset (the number of nucleotides between the primers and the start/end of the repeat) and dividing the remaining number of nucleotides by the repeat length (Table 1).
TABLE 1.
Properties of 18 assumed variable genetic loci in Streptococcus agalactiae
| Namea | Strainb | Position in strain genome | Repeat size and count in original strain (offset size) | Annotation in genome |
|---|---|---|---|---|
| TR1 (SATR1) | 2603 V/R | 143000 | 60 bp × 3 (139 bp) | Hypothetical protein SAG0134, CRISPR |
| TR2* | A909 | 115500 | 9 bp × 4 | |
| TR3 (SATR2) | A909 | 746100 | 18 bp × 6 (85 bp) | Hypothetical protein SAK_0805 |
| TR4* | A909 | 1159300 | 18 bp × 2 | |
| TR5* | A909 | 1256100 | ||
| TR6 (SATR3) | A909 | 1993600 | 12 bp × 23 (92 bp) | Surface adhesion protein gbs2018 in NEM316 |
| TR7* | A909 | 2071160 | ||
| TR8 (SATR4) | NEM316 | 29700 | 18 bp × 3 (99 bp) | pcsB protein |
| TR9 | NEM316 | 107750 | 24 bp × 2 | Chaperone protein, dnaJ |
| TR10* | NEM316 | 476780 | ||
| TR11* | NEM316 | 1547230 | ||
| TR12 (SATR5) | A909 | 1118500 | 48 bp × 14 (161 bp) | Putative fibrinogen-binding protein, SAK_1142 |
| TR13 | 2603 V/R | 451800 | 45-bp insert upstream of alp proteins | |
| TR14 | NEM316 | 857700 | 53-bp insert, gene gbs0831 | |
| TR15* | NEM316 | 917600 | ||
| TR16 | NEM316 | 1010700 | 66-bp insert, hypothetical protein gbs0966 | |
| TR17 | NEM316 | 1269700 | 45-bp deletion in A909, intergenetic between gbs1230 and ung | |
| TR18 | NEM316 | 1624650 | 44-bp deletion, gbs1557 |
*, not surveyed further after initial screening.
The strain column refers to the strain in which the locus was found originally.
Sequence verification.
For DNA sequencing, loci were amplified in PCRs with unlabeled primers as described above. The PCR products were purified by using ExoSAP-IT (USB Corporation, Cleveland, OH) following the instructions of the manufacturer. BigDye 3.1 and BigDye terminator 3.1 (Applied Biosystems) reagents were used according to the manufacturer's instructions for sequencing PCR and subsequent purification. Capillary electrophoresis of sequencing PCR products was performed on an ABI 3130xl genetic analyzer (Applied Biosystems). The resulting sequences were analyzed using Sequencher 4.2 software (Gene Codes Corporation, Ann Arbor, MI).
RESULTS
Identification of tandem repeats in GBS.
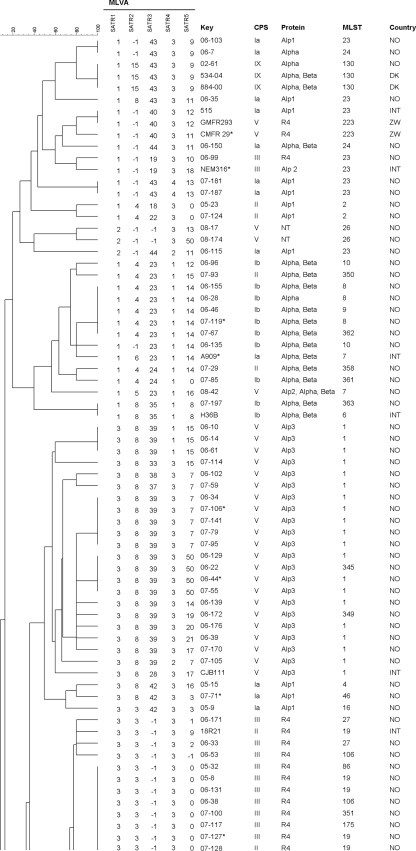
The sequences of the three complete genomes of strains A909, NEM316, and 2603 V/R were analyzed in silico using the Tandem Repeats Finder program or the Variable Region Finder website. Candidate loci were compared via the BLAST website with the eight published whole-genome sequences, namely, the three above mentioned and five not fully assembled and annotated shotgun sequences (strains 515, H36B, 18RS21, COH1, and CJB111) (22). Most loci were excluded from further investigation for reasons such as large size, poorly conserved repeats, or poorly conserved flanking sequences. For the 18 remaining loci (Table 1), primers were designed. Their presence and their suitability for an MLVA were tested in 11 GBS strains. Seven loci (marked with asterisks in Fig. 1 ) were found unsuitable because they did not show variability or were not amplified even after several primer pairs were designed and tested.
FIG. 1.
Cluster analysis of 126 GBS strains using categorical values and the UPGMA (unweighted-pair group method using average linkages) algorithm, generated with Bionumerics 6.0 software. Numbers in the SATR1 to -5 columns indicate repeat count (−1 repeat, no PCR product). CPS, capsular polysaccharide. The country column gives the origin of the strain: NO, Norway; INT, international reference strain; DK, Denmark; and ZW, Zimbabwe. Strains marked with asterisks were used in the initial analysis of 18 variable loci. There is an overlap of three strains between the two panels.
The remaining 11 loci were investigated in all 126 strains. Four of the loci had four or more alleles, and three had three alleles, while the remaining four loci had two alleles in this strain collection. Six of the 11 diverse loci did not generate a PCR product in some strains (indicated as −1 repeats in Fig. 1). Amplification of SATR4 and SATR5 generated a PCR product devoid of the repeat in 1 strain and 21 strains, respectively (indicated as 0 repeats in Fig. 1). The number of alleles for the 11 loci and their calculated diversity index are shown in Table 2. All 11 loci were stable in three strains that were passaged 40 times.
TABLE 2.
Primers, fluorescent dyes, and Simpson's index of diversity for 11 VTNR loci in GBS
| Locus | Sequence (5′→3′)a |
Diversity indexb | 95% CI | Kc | max(pi)d | |
|---|---|---|---|---|---|---|
| Forward primer | Reverse primer | |||||
| SATR5 (TR12) | 6-FAM-AGTCACCTTGACTAGAGTG | AATAAAATAGGTTTTAGAACTTGG | 0.913 | 0.903-0.923 | 24 | 0.173 |
| SATR3 (TR6) | VIC-CCGCCAAGTTTCCGCTAG | AGCTAACAAGCTACGTGATGC | 0.869 | 0.856-0.881 | 19 | 0.220 |
| SATR2 (TR3) | PET-GTTGATAAAGTTGATGTTCCG | AGCCTTCTTCAACTATAGGTG | 0.703 | 0.675-0.732 | 9 | 0.472 |
| SATR1 (TR1) | NED-ACTTGTGAAAGTGTATCGGT | AAATCAGTGTTTTAACAGCAGC | 0.643 | 0.626-0.661 | 3 | 0.465 |
| SATR4 (TR8) | VIC-AAAGCATCTTTAATTCAGGCA | CTGTGGCAGACTCAACTTGTG | 0.633 | 0.608-0.658 | 5 | 0.504 |
| TR9 | TGTCTAAGAAATACCATCCAG | TTCAAAACTTAAATTAACACG | 0.470 | 0.444-0.496 | 3 | 0.643 |
| TR13 | TGTTTCTAAGAAAAAAGAGG | ACCTTACATTTGATTACACC | 0.470 | 0.444-0.496 | 3 | 0.643 |
| TR16 | ATCAAAAAACCAGATACTGC | CAATGCTTTAACCGAACCAG | 0.455 | 0.431-0.478 | 2 | 0.651 |
| TR14 | ATTATAGAGGACGTTACTCG | CAACCATCCGACCTGTAATA | 0.172 | 0.133-0.212 | 2 | 0.905 |
| TR18 | ATGACAGTTGAACATGTGGA | TACCATGTACCAACAGACTG | 0.031 | 0.011-0.051 | 2 | 0.984 |
| TR17 | AATTTGGTTCCGTTTGGCT | ATGCTCGAAAGAAAAAGTCG | 0.016 | 0.001-0.030 | 2 | 0.992 |
Certain primers were labeled with the fluorescent dyes 6-FAM (6-carboxyfluorescein), VIC, PET, and NED, as indicated.
The diversity index (for VNTR data) is a measure of the variation of the number of repeats at each locus, ranging from 0.0 (no diversity) to 1.0 (complete diversity). The most diverse loci are shown at the top.
K, number of different repeats present at this locus in a collection of 126 GBS strains.
max(pi), fraction of samples that have the most frequent repeat number in this locus (range, 0.0 to 1.0).
Construction of an MLVA.
The five most diverse loci were chosen for the construction of a GBS MLVA with fluorescently labeled primers and size estimation by capillary electrophoresis. The five proposed MLVA loci (SATR1 to -5) were amplified in one multiplex PCR. Due to uneven amplification efficiency, the primer concentration had to be adjusted as described above. Fragment sizes were estimated on an ABI 3130xl Genetic Analyzer using the 1200 LIZ size standard. This standard was chosen because SATR3 and SATR5 had large PCR products in some strains: up to around 750 bp in SATR3 and above 1,200 bp in the latter.
The five loci are described in more detail as follows.
(i) SATR1 (TR1).
This SATR1 repeat locus consisted of well-conserved repeats of 60 bp and was present in all strains investigated with one to three repeats. This locus was previously described by Dmitriev et al. (7, 17) as consisting of 16-bp direct repeats and 44-bp spacer regions.
(ii) SATR2 (TR3).
The SATR2 repeats were well conserved, consisting of 18 bp, and the strains contained 3 to 15 repeats. All sequenced strains had an insert of 9 bp found between repeats 2 and 3. The only strains having 15 repeats were the three strains in this study of the recently described serotype IX (20). In 14 strains (11%), no PCR product was detected.
(iii) SATR3 (TR6).
The SATR3 repeat locus gave comparably large products from 297 to 750 bp. The repeat unit of 12 bp was quite well conserved. The repeat count was computed to 18 to 54 repeats; however, sequencing demonstrated that the strains with the largest amplicons had 18 repeats and an inserted sequence of two 216-bp repeats. This combination was found in 33 strains (26.4%), all but one of which belonged to MLST clonal complex 17. A similar observation was made by Lamy et al. (12), as discussed below. SATR3 was not amplified in 25 strains (20%), most of which were of serotype III/R4 and MLST clonal complex 19.
(iv) SATR4 (TR8).
The SATR4 repeat gave the smallest PCR products, ranging between 99 and 168 bp. The underlying repeat of 18 bp was quite degenerated; in some strains, some or all of the repeats were only 15 bp. For convenience of typing, all alleles are assigned as multiples of 18 bp in the calculation of repeat numbers. A PCR product was observed in all 126 strains, and five different alleles were found. The repeat is located in the pcsB gene, encoding a protein which is important for cell wall separation (15).
(v) SATR5 (TR12).
The SATR5 locus is the most diverse repeat locus in this study, with 24 different alleles in the 126 strains and a Simpson's index of diversity of 0.913 (confidence interval [CI], 0.900 to 0.922). The repeat of 48 bp was quite well conserved, and an amplification product was detected in all but one strain. Of 21 strains with a PCR product size equivalent to the offset (0 repeats), 13 were of ST19. A result of six repeats was found in 22 strains, 18 of which belonged to ST17. Some strains had an insert after the repeat locus, causing a variable offset. However, for typing purposes, the offset was set to 161 bp with the primers chosen. The larger products of this locus, especially those above 900 bp, did give weak signals in spite of attempts to optimize both multiplex PCR and fragment analysis. If SATR5 did not yield a product in the multiplex PCR, it was repeated in a single PCR. In five strains, the SATR5 locus was calculated to be more than 1,400 bp by the GeneMapper software, although this was above the upper limit of the 1200 LIZ size standard and therefore unreliable. When the five strains containing SATR5 amplicons above 1,200 bp were analyzed by gel electrophoresis, they showed diversity. In the analysis, however, these five strains were assigned a repeat count of 50. SATR5 is located in the gene encoding the fibrinogen binding protein FbsA (16).
Comparison between MLVA, serotyping, and MLST.
MLVA analysis with the five proposed loci resolved the 126 strains into 70 different types with a Simpson's index of diversity of 0.963, while MLST separated them into 36 STs with an index of 0.899. The two largest groups of identical MLVA profiles consisted of 19 strains and 13 strains, most of which belonged to ST17 and ST19, respectively. These two MLST types are common subgroups of CPS type III (Fig. 1). All 27 of the ST17 strains in this work clustered together but were resolved into 12 different MLVA types within this cluster; 16 of them had identical profiles (Fig. 2). Similarly, all 11 CPS III/ST19 strains belonged to the same MLVA cluster and were resolved into four MLVA types; 7 of them had identical profiles. CPS type V has previously been described as relatively homogenous, possibly due to a recent introduction of this group as a human pathogen (5). Most of the type V strains belonged to ST1 (21 out of 30 type V strains); 20 of these 21 were in a cluster consisting of 12 MLVA types, mostly because of heterogeneity in SATR5.
FIG. 2.
MLVA cluster analysis of 27 ST17 strains included in the study using categorical values and the UPGMA algorithm, generated with Bionumerics 6.0 software. Numbers in the SATR1 to -5 columns indicate the repeat count.
A simpler MLVA analysis using SATR1 to -4 resulted in 43 different types for the 126 strains (Simpson's index of diversity, 0.926), also corresponding very well with the clustering by the other typing methods.
DISCUSSION
This study identified VNTR loci in the genome of GBS that are suitable for MLVA typing. MLVA is a relatively new typing method with several advantages compared to other methods. Mainly, MLVA is easy to perform at moderate costs, and results are in most cases available within 2 days, which will usually be more rapid than MLST or PFGE. Since MLVA generates numeric results, comparison of strains between different laboratories may easily be done. In the present study, 18 candidate loci were identified in the GBS genome by computational studies. Initial laboratory analysis found seven of these not suitable. The remaining 11 loci were tested in 126 strains, and the five most diverse loci were selected for an MLVA based on multiplex PCR and capillary electrophoresis for determination of the number of repeats.
The strain collection analyzed consisted of Norwegian invasive GBS strains and international reference strains which together represented a wide selection of CPS types and STs. The Norwegian strains, mostly from neonates, had been submitted to the national reference laboratory from hospitals throughout the country in the years 2006 and 2007. It is therefore reasonable to assume that there was no epidemiological association between most patients. Based on the high discriminatory power of the MLVA shown in this study, the method may well be suited for outbreak investigation. This should be addressed in a study with epidemiologically related strains.
Typing with MLVA in this study did generally correspond with the clustering of strains observed in CPS/surface protein typing or MLST. The 5-locus MLVA provided a very high degree of resolution, discriminating the 126 GBS strains into 70 different MLVA types (Fig. 1). The discriminatory power of this MLVA is superior to those of both MLST and combined CPS/protein typing, which resolved the strains into 36 and 19 types, respectively. The strains could have been resolved further if SATR5 alleles above 1,200 bp had been differentiated, which could be necessary under certain circumstances. If required, this will imply the use of other methods for exact size enumeration of SATR5 and consequently a delay of results. For SATR4, the diversity due to the degenerate character of the repeat should be kept in mind. In addition to the repeat count, the fragment size may be considered, especially if the MLVA is used in epidemiological investigations. As an alternative option for providing rapid results, a 4-locus MLVA, consisting of the loci SATR1 to -4 was also analyzed. Even this 4-locus MLVA was capable of providing higher resolution than MLST or CPS/protein typing. Conversely, some or all the six loci mentioned in Table 2 but not included in the 5-locus MLVA analysis (e.g., TR9, TR13, and TR16) could be added to the MLVA if an even higher degree of resolution is desirable.
Some of the loci selected for the 5-locus MLVA have been investigated previously for other purposes. SATR1 has been addressed by a Russian and Slovakian group (7, 17) who found eight alleles in 112 bovine strains. Strictly speaking, this locus is a clustered regularly interspaced short palindromic repeat (CRISPR) but worked very well as component of this MLVA. SATR3 is located in the gene of surface adhesion protein gbs2018 previously analyzed by Lamy et al. (12). They described three alleles, one of which included the insert of 216 bp with two repeats mentioned above and found it as well strongly correlated with ST17. GBS belonging to ST17 account for a high proportion of newborn infections (3, 9, 14); however, whether these GBS represent a hypervirulent cluster is under discussion (13). SATR5 is the most diverse repeat locus and is responsible for a substantial part of the overall resolution obtained with this assay. Analyzed alone, it did cluster some of the MLST groups such as ST17 and ST19 well, while others (e.g., CPS type V/ST1) were more dispersed. On the other hand, SATR5 resolved CPS-type V/ST1 into many different types in the MLVA, while other typing techniques tend to give a much more homogeneous picture of this subgroup. In SATR2, the finding of 15 repeats was indicative for CPS type IX. This was also observed in three additional type IX strains not included in this study. Results that were indicative of certain sequence types were, e.g., the finding of 54 or 55 repeats in SATR3 for ST17 (Fig. 2). Similarly, an ST19 could be expected when SATR3 had no PCR product (−1 repeats) and SATR5 had 0 repeats.
To summarize, the present study identified VNTR loci in the genome of Streptococcus agalactiae suitable for MLVA analysis. Five loci were selected for a multiplex PCR protocol followed by capillary electrophoresis, enabling a very discriminatory GBS typing assay. The 5-locus MLVA resolved a strain collection of 126 GBS strains considerably better than MLST and with less workload. This method is well suited for typing of GBS, e.g., by national reference laboratories and for research purposes. Further work with this method would include refinement of the method, comparison with PFGE with epidemiologically related strains, as well as analysis of other strain collections, such as non-Norwegian strains and noninvasive and animal strains, and establishment of a website for international collaboration and strain comparison.
Acknowledgments
This work was supported by grants from the Clinic of Laboratory Medicine, St. Olav's University Hospital, Trondheim, and the Central Norway Regional Health Authority. A.R. was supported by a Ph.D. grant from the Norwegian University of Science and Technology, Department of Laboratory Medicine, Children's and Women's Health.
Kirsti Løseth is gratefully acknowledged for expert laboratory work.
Footnotes
Published ahead of print on 26 May 2010.
REFERENCES
- 1.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, J. A., and P. Ferrieri. 2001. Rapid pulsed-field gel electrophoresis method for group B streptococcus isolates. J. Clin. Microbiol. 39:3006-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergseng, H., J. E. Afset, A. Radtke, K. Loeseth, R. V. Lyng, M. Rygg, and K. Bergh. 2009. Molecular and phenotypic characterization of invasive group B streptococcus strains from infants in Norway 2006-2007. Clin. Microbiol. Infect. 15:1182-1185. [DOI] [PubMed] [Google Scholar]
- 4.Bevanger, L., and J. A. Maeland. 1977. Type classification of group B streptococci by the fluorescent antibody test. Acta Pathol. Microbiol. Scand. B 85B:357-362. [DOI] [PubMed] [Google Scholar]
- 5.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, A. Schuchat, W. Baughman, and M. M. Farley. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173:365-373. [DOI] [PubMed] [Google Scholar]
- 6.Creti, R., F. Fabretti, G. Orefici, and C. von Hunelstein. 2004. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42:1326-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dmitriev, A. V., A. D. Shen, and A. A. Totolian. 2007. Streptococcus agalactiae sak0192 gene contains direct repeats and spacers that are genetic markers for characterizing the strains. Mol. Gen. Mikrobiol. Virusol. 4:37-41. (In Russian.) [PubMed] [Google Scholar]
- 8.Imperi, M., M. Pataracchia, G. Alfarone, L. Baldassarri, G. Orefici, and R. Creti. 2010. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J. Microbiol. Methods 80:212-214. [DOI] [PubMed] [Google Scholar]
- 9.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong, F., and G. L. Gilbert. 2006. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB)—a practical epidemiological and diagnostic tool. Nat. Protoc. 1:2668-2680. [DOI] [PubMed] [Google Scholar]
- 11.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamy, M. C., S. Dramsi, A. Billoet, H. Reglier-Poupet, A. Tazi, J. Raymond, F. Guerin, E. Couve, F. Kunst, P. Glaser, P. Trieu-Cuot, and C. Poyart. 2006. Rapid detection of the “highly virulent” group B Streptococcus ST-17 clone. Microbes Infect. 8:1714-1722. [DOI] [PubMed] [Google Scholar]
- 13.Lin, F., V. Sintchenko, F. Kong, G. L. Gilbert, and E. Coiera. 2009. Commonly used molecular epidemiology markers of Streptococcus agalactiae do not appear to predict virulence. Pathology 41:576-581. [DOI] [PubMed] [Google Scholar]
- 14.Luan, S. L., M. Granlund, M. Sellin, T. Lagergard, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinscheid, D. J., B. Gottschalk, A. Schubert, B. J. Eikmanns, and G. S. Chhatwal. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenau, A., K. Martins, S. Amor, F. Gannier, P. Lanotte, M. van der Mee-Marquet, L. Mereghetti, and R. Quentin. 2007. Evaluation of the ability of Streptococcus agalactiae strains isolated from genital and neonatal specimens to bind to human fibrinogen and correlation with characteristics of the fbsA and fbsB genes. Infect. Immun. 75:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozhdestvenskaya, A., M. Bhide, J. Mikula, Jr., A. Dmitriev, and I. Mikula. 2008. Structural and sequence polymorphism of SAK0192 gene of bovine Streptococcus agalactiae strains as molecular epidemiological marker, p. p321. In XVII Lancefield International Symposium on Streptococci & Streptococcal Diseases, Porto Heli, Greece.
- 18.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 19.Skjaervold, N. K., K. Bergh, and L. Bevanger. 2004. Distribution of PFGE types of invasive Norwegian group B streptococci in relation to serotypes. Indian J. Med. Res. 119(Suppl.):201-204. [PubMed] [Google Scholar]
- 20.Slotved, H. C., F. Kong, L. Lambertsen, S. Sauer, and G. L. Gilbert. 2007. Serotype IX, a proposed new Streptococcus agalactiae serotype. J. Clin. Microbiol. 45:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yazdankhah, S. P., and B. A. Lindstedt. 2007. Variable number tandem repeat typing of bacteria. Methods Mol. Biol. 396:395-405. [DOI] [PubMed] [Google Scholar]
- 24.Zeng, X., F. Kong, J. Morgan, and G. L. Gilbert. 2006. Evaluation of a multiplex PCR-based reverse line blot-hybridization assay for identification of serotype and surface protein antigens of Streptococcus agalactiae. J. Clin. Microbiol. 44:3822-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]