Abstract
It has been demonstrated that smoking cessation alters the subgingival microbial profile; however, the response of individual bacteria within this ecosystem has not been well studied. The aim of this investigation, therefore, was to longitudinally examine the effect of smoking cessation on the prevalence and levels of selected subgingival bacteria using molecular approaches for bacterial identification and enumeration. Subgingival plaque was collected from 22 smokers at the baseline and 12 months following periodontal nonsurgical management and smoking cessation counseling. The prevalence and abundance of selected organisms were examined using nested PCR and multiplexed bead-based flow cytometry. Eleven subjects successfully quit smoking over 12 months (quitters), while 11 continued to smoke throughout (smokers). Smoking cessation led to a decrease in the prevalence of Porphyromonas endodontalis and Dialister pneumosintes at 12 months and in the levels of Parvimonas micra, Filifactor alocis, and Treponema denticola. Smoking cessation also led to an increase in the levels of Veillonella parvula. Following nonsurgical periodontal therapy and smoking cessation, the subgingival microbiome is recolonized by a greater number of health-associated species and there are a significantly lower prevalence and abundance of putative periodontal pathogens. The results indicate a critical role for smoking cessation counseling in periodontal therapy for smokers in order to effectively alter the subgingival microbiome.
It is established that bacterial consortia within the subgingival microbiome play a critical role in the etiology of chronic periodontitis. Although tobacco smoking has been shown to preferentially enrich this microbiome for pathogenic species (26, 29), it is not known if smoking cessation is capable of reversing this pathogenic colonization, since current evidence is based only on cross-sectional comparisons of former and current smokers (10, 25). A longitudinal examination of subgingival bacteria following smoking cessation would allow us to determine how the individual bacteria within this ecosystem respond to smoking cessation.
We have previously shown, using a cultivation-independent, open-ended approach, that smoking cessation results in a shift in the subgingival microbial profile during recolonization following periodontal therapy (7). In order to identify the individual organisms that contributed to this compositional shift, it is necessary to use a targeted, sensitive, and quantitative approach to measure the levels of these species.
Bead-based flow cytometry has recently been used to measure the abundance of target organisms in complex bacterial communities (24, 28). This cultivation-independent approach is capable of simultaneously measuring the levels of several organisms, both cultivated and uncultivated. It also has a wide dynamic range; that is, it is capable of quantifying both the less numerous and the numerically dominant members of a microbial community.
The goals of the present investigation were to identify the effects of smoking cessation on the prevalence and abundance of selected bacteria using a novel method for the simultaneous quantification of cultivated and uncultivated organisms.
MATERIALS AND METHODS
Subject selection and study design.
Ethical approval for this study was obtained from the Newcastle Research Ethics Committee and the Office of Responsible Research Practices at The Ohio State University. The study population, study design, and inclusion and exclusion criteria have been described previously (20). Briefly, smokers with a pretreatment diagnosis of moderate to severe chronic periodontitis who expressed a desire to quit smoking were selected. All subjects were treated nonsurgically (oral hygiene instruction, root surface instrumentation, periodontal maintenance care) and given smoking cessation counseling. Clinical data and subgingival plaque samples were collected at the baseline and 12 months later. Probing depths and clinical attachment levels were recorded with a disk probe (Florida Probe Corp., Gainesville, FL). Compliance with smoking cessation was assessed by measuring exhaled carbon monoxide and salivary cotinine levels. For the purposes of this study, 22 subjects were selected on the basis of a review of their smoking status over the 12-month study period.
Sample collection and DNA isolation.
Six nonadjacent sites, one at each of six teeth with probing depths of 4 mm or greater, were identified and isolated for sampling. Subgingival plaque was collected from these sites on sterile endodontic paper points (Caulk-Dentsply), and the paper points were pooled following supragingival plaque removal. The samples were placed in 1.5-ml microcentrifuge tubes and frozen at −80°C until further analysis. Bacteria were removed from the paper points by adding 200 μl of phosphate-buffered saline to the tubes and vortexing. The points were then removed, and the DNA was isolated with a Qiagen DNA MiniAmp kit (Qiagen, Valencia, CA) using the tissue protocol, according to the manufacturer's instructions.
Prevalence of candidate pathogens and beneficial bacteria.
The prevalence of 13 organisms, including some as-yet-uncultivated phylotypes, were assessed at the baseline (when all the patients were still smokers) and at 12 months (when 11 patients were quitters and 11 were smokers) using a nested PCR approach described previously (12). Briefly, the 16S rRNA gene and the intergenic spacer region (ISR) were amplified by PCR with broad-range primers targeted to the 16S and 23S regions of the ribosomal operon (positions 422 and 785). A second, species-specific PCR was performed with 2 μl of the amplicons using a species-specific forward primer and a universal 23S reverse primer (primer L189). The primer sequences are listed in Table 1. The resultant amplicons were separated by 1% agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV light transillumination. The presence of a clear band of the expected molecular size was scored as positive for the species. All reactions were replicated.
TABLE 1.
Primer and probe sequences
| Primer/probe or species name | Sequence | Reference or source |
|---|---|---|
| Broad-range primers | ||
| 785 | GGA TTA GAT ACC CTG GTA GTC | This study |
| 422 | GGA GTA TTT AGC CTT | This study |
| 785R | GAC TAC CAG GGT ATC TAA TCC | 12 |
| A18 | TT TGA TCC TGG CTC AG | 12 |
| Species-specific primers | ||
| Dialister oral clone GBA27 | CAG AAA TGC GGA GTT CTT CTT CG | This study |
| Veillonella parvula | AGA CGG AAG CGA GAT CGC GAG ATG | 12 |
| Campylobacter gracilis | GAA TGC GAA ATT CGC TAC C | This study |
| Parvimonas micra | AAC GAG AAG CGA GAT AGA GAT GTT A | This study |
| Desulfobulbus oral clone R004 | CCC ATG AAA GTG GGT GGT GCC TTC | This study |
| Filifactor alocis | ACA TAC CAA TGA CAG CCT TTT AA | This study |
| Megasphaera oral clone MBB166 | CGG GTA GAG ATA CCT GGT TCT TCT TCG | This study |
| Porphyromonas gingivalis | CAT CGG TAG TTG CTA ACA GTT TTC | This study |
| Porphyromonas endodontalis | TTT AGA TGA TGG CAG ATG AGA G | This study |
| Treponema denticola | CAA GAG CAA TGA CAT AGA GAT ATG G | This study |
| Dialister pneumosintes | CCT TGA CAT TGA TCG CAA TCC ATA GAA ATA | This study |
| Tannerella forsythia | TGC GAT ATA GTG TAA GCT CTA CAG | This study |
| Fusobacterium nucleatum | TTC GGG GAA ACC TAA AGA CAG GTG G | This study |
| Species-specific probes | ||
| Parvimonas micra | GTA AGA AGG GCT CGC GTC TG | 12 |
| Treponema denticola | GCA GAT GAA GAA TAA GAA G | 12 |
| Campylobacter gracilis | TTT ACC ATA AGA TAA A | 12 |
| Filifactor alocis | CGA CTA TTA ACA GAA CCT TTC GGG GCG AAG | 12 |
| Desulfobulbus oral clone R004 | CAT TTC ACA GGT AAG G | 12 |
| Tannerella forsythia | GAT GGT AGC AAT ACC TGT C | 12 |
| Fusobacterium nucleatum | AGG CAT CTT AGA ATT ATG AAA GCT ATA AGC A | 12 |
| Veillonella parvula | AGC TAT CAC TGA AGG AGG GGA | 12 |
| Porphyromonas gingivalis | CAC TTG TAT TAT TGC ATG ATA TTA | 12 |
Bacterial prevalence analysis.
The prevalences of the 13 candidate organisms at the baseline and 12 months later were compared for each subject to identify the frequency of the loss or acquisition of species. The frequency of loss was computed as the percentage of subjects in whom the target species was detected at the baseline but not at 12 months. The frequency of gain was similarly computed. These frequencies were compared between smokers and quitters using chi-square analysis.
Quantitative analysis of candidate pathogens and beneficial bacteria.
The levels of nine organisms that were detected in both smokers and quitters at 12 months were compared using mutiplexed bead-based flow cytometry.
Amplification of DNA.
The 16S rRNA genes were amplified from the community DNA and from bacterial standards using 22 cycles of PCR with universal primers 785R and A18. The reverse primer was modified to contain a 5′ biotin, 12-carbon linker and five phosphorothioate bonds (Integrated DNA Technologies, Skokie, IL). The cycling conditions and protocols have been described previously (13). Our previous experiments using 16S rRNA cloning and sequencing have provided us with a clone library of 16S rRNA genes (1,500 bp) from cultivated and as-yet-uncultivated organisms. Serial dilutions of these rRNA genes were used as internal bacterial standards. The amplicons were purified using a QIAquick kit (Qiagen, Valencia, CA), and the amount of DNA was measured with a spectrophotometer (NanoDrop 1000; Thermo Scientific, Wilmington, DE).
Preparation of capture bead-probes.
Species-specific probes were designed by comparing closely related sequences downloaded from GenBank to identify unique signature sequences in the target species. The specificity of the probes was tested by PCR amplification and sequencing of the resultant amplicons. The probes were synthesized with a 5′ amino unilinker modification (Integrated DNA Technologies). The probe sequences are listed in Table 1. Nine types of carboxylated microspheres with different intensities of red and infrared internal fluorescence were selected (Invitrogen Molecular Probes, Carlsbad, CA). Each species-specific probe was bound to a single bead type, creating nine unique bead-probe complexes.
DNA amplification and hybridization.
The amplicons from the community DNA and the bacterial standards were digested with T7 exonuclease to remove the unlabeled (nonbiotinylated) strand. Differences in probe hybridization efficiencies and matrix effects within individual samples were accounted for by using internal standards, as described previously (15). Briefly, each sample was used in four hybridization reactions with 200 fmol of each sample and either 0, 5, 15, or 30 fmol of each standard. Samples were hybridized to the bead-probes; a no-DNA control was included, and all reactions were replicated. The DNA-bead-probe hybrids were further labeled with a secondary reporter molecule, phycoerythrin-streptavidin (SAPE) complex. This binding was facilitated by the biotin on the DNA molecule binding to the streptavidin in the SAPE reporter.
Flow cytometric detection.
The beads were analyzed using a Becton-Dickinson FACSCalibur flow cytometer. The fluorescence from phycoerythrin (reporter signal), as well as the internal fluorescence from the microspheres (classification signal), was measured on different channels. The fluorescence from the beads was quantified on the basis of the fluorescence of the reference bead standards (Bangs Laboratories, Fishers, IN).
Data analysis.
Each bead population was visualized by plotting the side scatter (SSC) versus the classification (FL3 or FL4) signals for the monomer population. The intensity of the reporter signal from each of these bead types was measured on the FL2 channel and was converted to standard fluorescence units on the basis of the reference bead standards. The background signal was then subtracted using the no-DNA control and a standard curve computed with the internal standards. The concentration of each bead (and therefore the level of each species in a sample) was calculated from the standard curves using linear regression (28). The FlowJo program (Tree Star, Ashland, OR) was used for data analysis.
Statistical analysis.
Statistical analyses were carried out with the JMP program (SAS Institute Inc., Cary, NC). Eight data points were available for each sample (four dilutions and replicates), and the level of each species in a sample was computed by averaging these data points. The mean levels of nine candidate species were compared between smokers and quitters using Kruskal-Wallis analysis.
RESULTS
This study was designed to examine the effects of smoking cessation on the prevalence and abundance of selected bacterial species over 12 months. Subjects were divided into smokers and quitters on the basis of their compliance with smoking cessation. Eleven subjects successfully quit smoking for the 12-month period, while 11 smoked throughout this period. As previously reported, there were no statistically significant differences in the age, gender, duration of smoking, and baseline levels of the amount of smoking between the smokers and quitters (20).
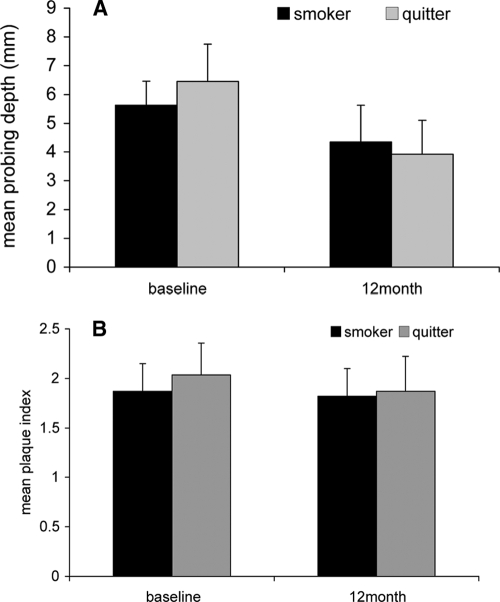
Figure 1 shows the mean probing depths (Fig. 1A) and the plaque index (Fig. 1B) of the six sites selected for sampling at the baseline and at 12 months. The mean ± standard deviation probing depths at the baseline for quitters and smokers were 6.5 ± 1.3 mm and 5.6 ± 0.8 mm, respectively. The mean probing depths for quitters and smokers at 12 months were 4.4 ± 1.3 mm and 3.9 ± 1.2 mm, respectively. The probing depths decreased significantly from those at the baseline (P ≤ 0.01, repeated-measures analysis of variance [ANOVA]) for both quitters and smokers; however, there were no differences in mean probing depths at the sampling sites between smokers and quitters at 12 months (P > 0.05). The baseline and 12-month plaque scores at the sampled sites were also not different between smokers and quitters (P > 0.05).
FIG. 1.
Clinical status of smokers and quitters at the baseline and 12 months. The mean probing depths (A) and mean plaque index (B) at the six sites selected for sampling are shown. There were no differences in the clinical parameters recorded at the plaque sampling sites between smokers and quitters at any of the time points (P > 0.05, ANOVA).
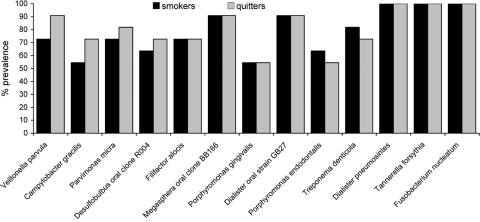
The baseline prevalence of 13 species and phylotypes is shown in Fig. 2. Fusobacterium nucleatum, Tannerella forsythia, and Dialister pneumosintes were detected in all subjects at the baseline, while the prevalence of the other organisms ranged from 40 to 90%.
FIG. 2.
Percent prevalence of 13 organisms in smokers and quitters at the baseline. Nested PCR was used to examine samples for the presence of selected species. No significant differences were detected between the two groups (P > 0.05, Fisher's exact test).
Several changes in the detection frequencies of these species were observed in both groups. The mean numbers of species/phylotypes lost were 1.9 ± 1.1 for smokers and 3.5 ± 0.9 for quitters, while the mean numbers of species acquired were 1.0 ± 0.7 for smokers and 1.4 ± 0.6 for quitters. These differences were not statistically significant by ANOVA. However, 41% of smokers lost or gained species, while 59% of quitters demonstrated a similar change in prevalence. This difference was statistically significant (P = 0.04).
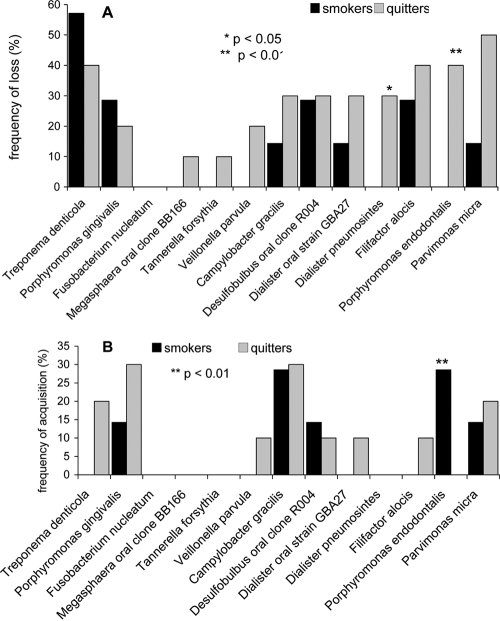
Figure 3 shows the loss or acquisition of organisms in smokers and quitters after 12 months of recolonization. Percent loss is shown in Fig. 3A and percent acquisition in Fig. 3B. Fusobacterium nucleatum was the only species that was present in all subjects at the baseline and 12 months. Smokers showed no change in prevalence of Veillonella parvula, Tannerella forsythia, Dialister pneumosintes, and Megasphaera oral clone BB166. Porphyromonas endodontalis was lost only in quitters and was acquired only by those who were continuous smokers, a statistically significant difference (P < 0.001, Fisher's exact test). Dialister pneumosintes was lost only by quitters (P < 0.05).
FIG. 3.
Loss or acquisition of species following 12 months of recolonization in smokers and quitters. The percentage of subjects who lost (A) or acquired (B) species along with significant differences between the two groups are shown.
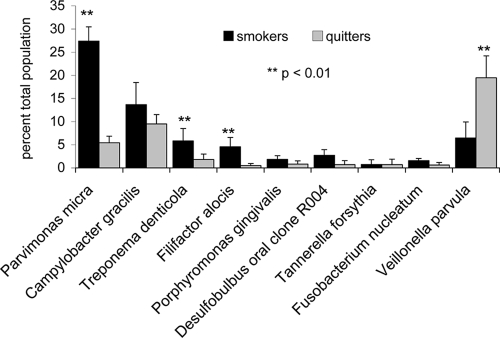
Figure 4 shows a comparison between the levels of nine bacterial species and phylotypes that were prevalent in both smokers and quitters at month 12. The levels of Parvimonas micra (formerly Peptostreptococcus micros), Filifactor alocis, and Treponema denticola were significantly elevated in smokers compared to those in quitters, while Veillonella parvula was found in significantly higher levels in quitters (P < 0.01, Kruskal-Wallis analysis).
FIG. 4.
Abundance of nine species and phylotypes following 12 months of recolonization in smokers and quitters. The levels of each species are shown as a percentage of the total population with standard deviation bars and significance levels. Species are arranged in a gradient from those present at higher levels in smokers on the left and those predominant in quitters on the right.
DISCUSSION
The subgingival microbiome is complex, with several uncultivated and as-yet-unrecognized members (13, 17), and hence, cultivation-based approaches have not been able to comprehensively examine the effects of smoking cessation on this ecosystem. We have previously reported that smoking cessation alters subgingival microbial profiles, determined using a comprehensive, open-ended molecular approach (7). However, it was not within the scope of that study to examine the changes in individual species that contributed to this shift. The objective of the present study, therefore, was to longitudinally examine the effect of smoking cessation on the prevalence and abundance of candidate pathogens and beneficial species.
Interbacterial interactions play an important role in determining the composition of a community, and hence, the presence of a preexisting established subgingival flora poses a challenge when the changes occurring in the subgingival microbiome are examined longitudinally. Periodontal nonsurgical therapy significantly reduces this baseline flora, thereby allowing an examination of the microbiome as it recolonizes (9, 16, 18). Monitoring the microbial changes during recolonization from this clean-slate baseline also allows comparisons between individuals to be made over time. Hence, we used this recolonization model to examine the effects of smoking cessation on target species in the subgingival biofilm.
Recent studies on several host-associated ecosystems have revealed that smoking contributes to preferential colonization by certain bacteria in these niches (2-4, 6, 27). In the subgingival environment, several cultivated and uncultivated species belonging to Parvimonas, Fusobacterium, Bacteroides, Porphyromonas, and Campylobacter have been detected in greater abundance in smokers with periodontitis than in never smokers (26, 29). Our own investigations using 16S rRNA cloning and sequencing have revealed that smokers with periodontitis demonstrated greater abundances of Parvimonas, Fusobacterium, Campylobacter, Bacteroides, and Treponema and lower levels of Veillonella, Neisseria, and Streptococcus (22a). The detection frequencies of these genera, however, were not different between the two groups. In order to investigate if smoking cessation is capable of altering this baseline community, candidate species for this study were selected from among those organisms exhibiting the greatest difference between current and never smokers.
Flow cytometry has traditionally been used in immunological studies to identify immune cells on the basis of their fluorescent characteristics (5, 19). Recent investigations have modified this methodology to quantify fluorescent polycarboxylate microspheres that are bound to DNA (bead-based flow cytometry). This method offers several advantages over currently available assays; it provides quantitative data on multiple cultivated and uncultivated bacteria simultaneously, permits analysis of large numbers of samples, and can detect both numerically dominant and minor species in a community.
Smoking cessation does not appear to affect the prevalence of Fusobacterium nucleatum (Fig. 2 and 3). Our results corroborate the findings of recent studies suggesting that it is a ubiquitous species that forms a bridge between early and late colonizers during plaque development (11, 12). Although higher levels of F. nucleatum have previously been associated with periodontitis in smokers (26), there was no difference in the levels between smokers and quitters in the present study (Fig. 4). A possible explanation could be that this Gram-negative anaerobe is metabolically versatile (22) and is capable of using several different nutrient sources for growth during de novo biofilm formation.
Treponema denticola, Filifactor alocis, and Parvimonas micra showed similar prevalences among smokers and quitters at 12 months; however, the levels of these species were significantly lower in quitters than in smokers (Fig. 4). There were no differences in probing depths or plaque levels at the sampled sites between the two groups (Fig. 1), suggesting that smoking cessation per se may have contributed to a decrease in their levels during recolonization.
Porphyromonas endodontalis was the only species to be lost by quitters and gained by smokers during recolonization (Fig. 3A and B). However, Porphyromonas gingivalis, a closely related species, showed no change in prevalence or abundance following smoking cessation. The implication of this finding is not clear; while it appears to be logical that smoking cessation influences the subgingival microbiome through several different mechanisms, it is possible that P. gingivalis is metabolically more versatile than P. endodontalis. P. endodontalis is a Gram-negative, obligate anaerobe that has previously been identified in periodontal pockets as well as endodontic infections (12, 21, 23). This strong temporal association between smoking and the prevalence of P. endodontalis suggests that determining the role of this species in the plaque biofilm could have important implications for monitoring periodontitis in smokers.
Another species lost only by quitters, Dialister pneumosintes, has previously been reported to be significantly associated with periodontitis in smokers (8). A closely related phylotype, Dialister oral strain GBA27, did not show a similar association with smoking cessation (Fig. 3), suggesting that phylogenetic relatedness does not necessarily reflect environmental clustering.
Veillonella parvula increased significantly in abundance following smoking cessation, accounting for over 20% of the microbial community in quitters. There is growing evidence from molecular investigations, including the present study, that members of the genus Veillonella form a major component of the subgingival microbiome in periodontal health (1, 13, 14). This ability of Veillonella to saturate the subgingival niche could play an important role in preventing colonization by pathogens and deserves further investigation as an indicator of clinical health.
In summary, the results presented here provide evidence that following nonsurgical therapy and smoking cessation, the subgingival microbiome undergoes a compositional shift resulting in colonization by health-associated species and a significantly lower prevalence and abundance of pathogens than those observed in smokers who receive nonsurgical therapy but continue to smoke. This shift toward a health-compatible profile may contribute to the clinical improvements in periodontal status associated with smoking cessation. The results also indicate that there is very little alteration in the microbiome following periodontal therapy in continuous smokers, suggesting an urgent need for smoking cessation counseling in conjunction with active periodontal therapy.
Acknowledgments
This study was funded by a start-up grant awarded to Purnima S. Kumar by the College of Dentistry, The Ohio State University. This study was also supported by a summer research award to Suzanne L. Delima from the College of Dentistry, The Ohio State University, and an NIH road map award to Robert K. McBride.
Footnotes
Published ahead of print on 21 April 2010.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson, M. D., S. T. Weiss, R. L. Ben, and A. L. Komaroff. 1982. Association between cigarette smoking and acute respiratory tract illness in young adults. JAMA 248:181-183. [PubMed] [Google Scholar]
- 3.Brook, I., and A. E. Gober. 2005. Recovery of potential pathogens and interfering bacteria in the nasopharynx of smokers and nonsmokers. Chest 127:2072-2075. [DOI] [PubMed] [Google Scholar]
- 4.El Ahmer, O. R., S. D. Essery, A. T. Saadi, M. W. Raza, M. M. Ogilvie, D. M. Weir, and C. C. Blackwell. 1999. The effect of cigarette smoke on adherence of respiratory pathogens to buccal epithelial cells. FEMS Immunol. Med. Microbiol. 23:27-36. [DOI] [PubMed] [Google Scholar]
- 5.Erciyas, K., R. Orbak, F. Kavrut, T. Demir, and H. Kaya. 2006. The changes in T lymphocyte subsets following periodontal treatment in patients with chronic periodontitis. J. Periodontal Res. 41:165-170. [DOI] [PubMed] [Google Scholar]
- 6.Ertel, A., R. Eng., and S. M. Smith. 1991. The differential effect of cigarette smoke on the growth of bacteria found in humans. Chest 100:628-630. [DOI] [PubMed] [Google Scholar]
- 7.Fullmer, S. C., P. M. Preshaw, P. A. Heasman, and P. S. Kumar. 2009. Smoking cessation alters subgingival microbial recolonization. J. Dent. Res. 88:524-528. [DOI] [PubMed] [Google Scholar]
- 8.Gomes, S. C., F. B. Piccinin, R. V. Oppermann, C. Susin, C. I. Nonnenmacher, R. Mutters, and R. A. Marcantonio. 2006. Periodontal status in smokers and never-smokers: clinical findings and real-time polymerase chain reaction quantification of putative periodontal pathogens. J. Periodontol. 77:1483-1490. [DOI] [PubMed] [Google Scholar]
- 9.Haffajee, A. D., M. Patel, and S. S. Socransky. 2008. Microbiological changes associated with four different periodontal therapies for the treatment of chronic periodontitis. Oral Microbiol. Immunol. 23:148-157. [DOI] [PubMed] [Google Scholar]
- 10.Haffajee, A. D., and S. S. Socransky. 2001. Relationship of cigarette smoking to the subgingival microbiota. J. Clin. Periodontol. 28:377-388. [DOI] [PubMed] [Google Scholar]
- 11.Kolenbrander, P. E., R. N. Andersen, and L. V. Moore. 1989. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338-344. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, P. S., A. L. Griffen, M. L. Moeschberger, and E. J. Leys. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar, P. S., E. J. Leys, J. M. Bryk, F. J. Martinez, M. L. Moeschberger, and A. L. Griffen. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 44:3665-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe, M., A. Spiro, Y. Z. Zhang, and R. Getts. 2004. Multiplexed, particle-based detection of DNA using flow cytometry with 3DNA dendrimers for signal amplification. Cytometry A 60:135-144. [DOI] [PubMed] [Google Scholar]
- 16.Magnusson, I., J. Lindhe, T. Yoneyama, and B. Liljenberg. 1984. Recolonization of a subgingival microbiota following scaling in deep pockets. J. Clin. Periodontol. 11:193-207. [DOI] [PubMed] [Google Scholar]
- 17.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersilka, G. J., B. Ehmke, and T. F. Flemmig. 2002. Antimicrobial effects of mechanical debridement. Periodontol. 2000 28:56-71. [DOI] [PubMed] [Google Scholar]
- 19.Petrunov, B., S. Marinova, R. Markova, P. Nenkov, S. Nikolaeva, M. Nikolova, H. Taskov, and J. Cvetanov. 2006. Cellular and humoral systemic and mucosal immune responses stimulated in volunteers by an oral polybacterial immunomodulator “Dentavax.” Int. Immunopharmacol. 6:1181-1193. [DOI] [PubMed] [Google Scholar]
- 20.Preshaw, P. M., L. Heasman, F. Stacey, N. Steen, G. I. McCracken, and P. A. Heasman. 2005. The effect of quitting smoking on chronic periodontitis. J. Clin. Periodontol. 32:869-879. [DOI] [PubMed] [Google Scholar]
- 21.Rocas, I. N., and J. F. Siqueira, Jr. 2008. Root canal microbiota of teeth with chronic apical periodontitis. J. Clin. Microbiol. 46:3599-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers, A. H., J. Chen, P. S. Zilm, and N. J. Gully. 1998. The behaviour of Fusobacterium nucleatum chemostat-grown in glucose- and amino acid-based chemically defined media. Anaerobe 4:111-116. [DOI] [PubMed] [Google Scholar]
- 22a.Shchipkova, A. S., H. N. Nagaraja, and P. S. Kumar. Subgingival microbial profiles of smokers with periodontitis. J. Dent. Res., in press. [DOI] [PMC free article] [PubMed]
- 23.Siqueira, J. F., Jr., I. N. Rocas, J. C. Oliveira, and K. R. Santos. 2001. Detection of putative oral pathogens in acute periradicular abscesses by 16S rDNA-directed polymerase chain reaction. J. Endod. 27:164-167. [DOI] [PubMed] [Google Scholar]
- 24.Spiro, A., and M. Lowe. 2002. Quantitation of DNA sequences in environmental PCR products by a multiplexed, bead-based method. Appl. Environ. Microbiol. 68:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoltenberg, J. L., J. B. Osborn, B. L. Pihlstrom, M. C. Herzberg, D. M. Aeppli, L. F. Wolff, and G. E. Fischer. 1993. Association between cigarette smoking, bacterial pathogens, and periodontal status. J. Periodontol. 64:1225-1230. [DOI] [PubMed] [Google Scholar]
- 26.van Winkelhoff, A. J., C. J. Bosch-Tijhof, E. G. Winkel, and W. A. van der Reijden. 2001. Smoking affects the subgingival microflora in periodontitis. J. Periodontol. 72:666-671. [DOI] [PubMed] [Google Scholar]
- 27.Weinberg, E. D. 2000. Microbial pathogens with impaired ability to acquire host iron. Biometals 13:85-89. [DOI] [PubMed] [Google Scholar]
- 28.Wireman, J., M. Lowe, A. Spiro, Y. Z. Zhang, A. Sornborger, and A. O. Summers. 2006. Quantitative, longitudinal profiling of the primate fecal microbiota reveals idiosyncratic, dynamic communities. Environ. Microbiol. 8:490-503. [DOI] [PubMed] [Google Scholar]
- 29.Zambon, J. J., S. G. Grossi, E. E. Machtei, A. W. Ho, R. Dunford, and R. J. Genco. 1996. Cigarette smoking increases the risk for subgingival infection with periodontal pathogens. J. Periodontol. 67:1050-1054. [DOI] [PubMed] [Google Scholar]