Abstract
Assessing the relatedness of strains isolated from patients and their environment is instrumental in documenting the source of preventable health care-associated life-threatening Aspergillus flavus human infection clusters. The present study aimed at identifying and selecting suitable microsatellite markers for A. flavus typing. This typing scheme was then applied to investigate the A. flavus epidemiology within a hematology unit in Sfax, Tunisia. Use of a combination of five markers made it possible to discern clusters of isolates and to substantiate the genetic diversity of A. flavus within clusters. Isolates from Tunisia and Marseille, France, displayed distinct haplotypes, indicating a highly significant geographical structuring of A. flavus. The typing of clinical and environmental A. flavus isolates in a hematology unit provided insights into its hospital epidemiology. From a heterogeneous genetic background, a cluster indicative of a clonal propagation episode within the unit could be identified. In two patients with invasive aspergillosis, the same genotype was found in clinical and environmental isolates, indicating hospital-acquired colonization and infection. In further studies, this novel microsatellite typing scheme might be instrumental in illuminating important epidemiological issues about A. flavus population genetics or epidemiology, including tracing the sources and routes of transmission.
Invasive aspergillosis is a life-threatening disease in hematological units. Typing studies aiming at assessing the relatedness of strains isolated from patients and their environment are instrumental in understanding the epidemiology of this mold and documenting the source of preventable health care-associated life-threatening human infections. Most of the studies have focused on Aspergillus fumigatus, the medically most important species. Briefly, typing studies showed that environmental outdoor A. fumigatus populations are genetically very heterogeneous (3, 4). However, aspergillosis outbreaks due to clonal, i.e., genotypically identical, A. fumigatus strains have been documented in hospitals (2). Very few data exist on non-A. fumigatus Aspergillus species that are also involved in human infections. In particular, Aspergillus flavus has been described to be an emerging species second only to the A. fumigatus pathogenic Aspergillus species (17). Furthermore, A. flavus has been described as the main etiological agent of human aspergillosis in Saudi Arabia and in Tunisia, Sudan, and other African countries (10, 12). Three molecular methods have been proposed for A. flavus typing: restriction fragment length polymorphism (15, 19), randomly amplified polymorphic DNA (20), and random amplified microsatellite polymorphism analysis (11). When it was applied to A. fumigatus, microsatellite typing (22) showed a higher discriminatory power than other typing methods (13, 15, 19), including the other popular exact typing method used to date: multilocus sequence typing (MLST) (1). Recently, Tran-Dinh and Carter have described seven polymorphic microsatellite markers that may be used to type aflatoxin-producing A. flavus and A. parasiticus (21), but to our knowledge, this typing scheme has not been used either in population genetic or in epidemiological studies.
The study described here aimed at identifying and selecting microsatellite markers and at applying this typing scheme in order to investigate the epidemiology of A. flavus in one hematology unit.
MATERIALS AND METHODS
Genomic sequences of Aspergillus flavus were formed from published sequences (http://www.Aspergillusflavus.org/). The sequences were analyzed for the presence of short tandem repeats by using the Tandem Repeats Finder software (http://tandem.bu.edu/trf/trf.html). Only di-, tri-, and tetramicrosatellite loci that had perfect repeat sequences (having 100% identity between repeats units) and that were highly repeated (with a copy number of >12) were selected; imperfect repeats containing point mutations and/or insertions or deletions or having mismatches in their repeated units were excluded. This software made it possible to give a flanking sequence for these selected microsatellites. Then, primers were designed using Primer (version 3) software (http://frodo.wi.mit.edu) and verified for their Aspergillus flavus specificity using a BLASTn search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). We additionally used five markers described by Tran-Dinh and Carter (21).
The 63 Aspergillus flavus isolates used in this study originated from either Tunisia or France. Fifteen isolates were collected from three patients and their environment during the investigation of a single epidemic episode at Timone Hospital in Marseille, France. The characteristics of the 48 isolates collected from 14 patients with proven and probable invasive aspergillosis hospitalized in the hematology ward of the Hedi Cheker Hospital in Sfax, Tunisia, and their environment over a period of 3 years (2004 to 2007) are summarized in Table 1. This study design has been described elsewhere (12). Briefly, A. flavus infections were diagnosed in high-risk patients hospitalized in one hematology ward where neither high-efficiency particulate air filtration nor an aspergillosis prophylaxis policy was implemented. The 35 clinical and 13 environmental isolates sampled in the patients' rooms are detailed in Table 1. These A. flavus strains were identified on the basis of macroscopic and microscopic morphological characteristics. The morphological identification of the Aspergillus flavus isolates was verified by internal transcribed spacer 1 (ITS1), 5.8S, and ITS2 region rRNA sequence analysis, as described previously (5).
TABLE 1.
Origins of A. flavus isolates collected from patients with aspergillosis and their hospital environment in the hematology ward of the University Hospital of Sfax, Tunisia
| Patient no. | Isolate no. | Date (day-mo-yr) | Sample source |
|
|---|---|---|---|---|
| Clinical | Environmental | |||
| Patient 1 | 0588 | 02-05-05 | Sputum | |
| 0590 | 06-05-05 | Sputum | ||
| 0658 | 06-05-05 | Air conditioner | ||
| 0704 | 15-05-05 | Nasal | ||
| 0870 | 15-05-05 | Table | ||
| Patient 2 | 0601 | 15-05-05 | Table | |
| 0605 | 18-05-05 | Nasal | ||
| 0603 | 15-05-05 | Sputum | ||
| 0604 | 15-05-05 | Nasal | ||
| 0626 | 18-05-05 | Sputum | ||
| 0662 | 21-05-05 | Air conditioner | ||
| 0948 | 25-05-05 | Sputum | ||
| LBA2 | 21-05-05 | BAL fluid | ||
| Patient 4 | 1058 | 05-06-05 | Sputum | |
| 1069 | 08-06-05 | Nasal | ||
| 1268 | 22-06-05 | Sputum | ||
| 1080 | 15-06-05 | Bed | ||
| LBA4 | 11-06-05 | BAL fluid | ||
| 1075 | 11-06-05 | Air conditioner | ||
| Patient 5 | 1169 | 24-11-05 | Nasal | |
| 1240 | 30-11-05 | Nasal | ||
| 1289 | 03-12-05 | Sputum | ||
| 1211 | 27-11-05 | Bed | ||
| Patient 6 | 0019 | 05-05-06 | Sputum | |
| 0016 | 09-05-06 | Air conditioner | ||
| Patient 7 | 0922 | 22-03-06 | Sputum | |
| 0944 | 25-03-06 | Nasal | ||
| 0938 | 29-03-06 | Sputum | ||
| Patient 8 | 1056 | 27-02-06 | Bed | |
| 1119 | 01-03-06 | Nasal | ||
| 1128 | 01-03-06 | Sputum | ||
| 1090 | 9-03-06 | Sputum | ||
| Patient 9 | 1403 | 23-04-07 | Sputum | |
| 0811 | 27-04-07 | Bed | ||
| Patient 10 | 1073 | 12-08-05 | Table | |
| 1076 | 16-08-05 | Nasal | ||
| 1368 | 16-08-05 | Sputum | ||
| Patient 11 | 1369 | 16-11-07 | Sputum | |
| 0486 | 26-11-07 | Lung biopsy | ||
| LBA33 | 19-11-07 | BAL fluid | ||
| Patient 12 | LBA16 | 23-06-06 | BAL fluid | |
| Patient 13 | 1331 | 27-03-07 | Sputum | |
| 0887 | 24-03-07 | Nasal | ||
| 0890 | 24-03-07 | Bed | ||
| Patient 14 | 0587 | 13-03-07 | Sputum | |
| 0889 | 16-03-07 | Table | ||
| 0855 | 21-03-07 | Nasal | ||
| Patient 15 | LBA31 | 27-12-05 | BAL fluid | |
DNA was extracted by using a Nucleospin kit (Machery-Nagel, France), as indicated by the manufacturer's instructions, and eluted with 70 μl of sterile water. In each multiplex PCR, different fluorescent labels (6-carboxyfluorescein, PET [Applied Biosystems], NED [Applied Biosystems], VIC) were used for the different markers in order to distinguish the amplification products from each other. PCRs were performed in a final volume of 25 μl containing 1 ng of genomic DNA, 1 μM all amplification primers, 0.2 mM each deoxynucleoside triphosphate, 3 mM MgCl2, and 1 U of Ampli Taq DNA polymerase (Applied Biosystems, Courtaboeuf, France) in 1× reaction buffer (Applied Biosystems). Thermocycling was performed in a T1 thermocycler (Biometra, Göttingen, Germany) using the following protocol: 5 min of initial denaturation at 94°C, followed by 30 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 54°C, and 30 s of extension at 72°C, finally followed by 30 min at 72°C. The PCR products were diluted 10-fold with formamide. One microliter of the diluted PCR products was combined with 15 μl of formamide and 0.5 μl of LIZ 500 marker (Applied Biosystems Inc., CA). Following denaturation, the PCR products were resolved by capillary electrophoresis with polymer POP-7 in an ABI Prism 3130 genetic analyzer (Applied Biosystems Inc.). The injection and running parameters were those according to the recommendations of the manufacturer (Applied Biosystems Inc.). Analyses were performed with Gene Mapper software (Applied Biosystems Inc.). The repeatability of microsatellite typing was evaluated by using five different DNA preparations of the same isolate and by 10 repeated analyses of the same DNA preparation.
The Simpson index of diversity, D (14), was computed for each marker and each possible marker combination, with the aim of determining the most parsimonious combination yielding a D value of > 0.95, a sufficiently high discriminatory power recommended for typing experiments. The degree of similarity was calculated by applying the Dice coefficient test. This was performed using an NTSYS-PC numerical taxonomy and multivariate analysis system (version 2.1; Exeter Software, Setauket, NY). A dendrogram was generated using neighbor-joining methods. The fixation index (FST) on all loci was estimated from 1,000 bootstrap repetitions using the ARLEQUIN software package (9). Isolates possessing alleles with the same number of repeat units in all loci were defined as a clonal cluster.
RESULTS
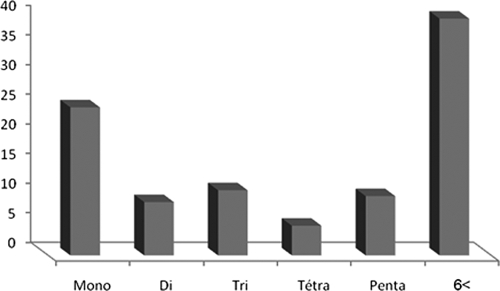
Using the Tandem Repeats Finder software package, we screened the 6,972,320 bp of A. flavus genomic sequence available (www.Aspergillusflavus.org, accessed in June 2007). We found 338 microsatellites; in 60% repeat units, they ranged from 1 to 6 nucleotides, and in 40%, they were greater than 6 nucleotides (Fig. 1). We finally retained seven markers: two dinucleotides, four trinucleotides, and one tetranucleotide matched our selection criteria (>12 repeats and 100% match). Each marker was tested in five repeated assays with different DNA preparations of the same isolate as well as with the same DNA preparation. Each marker's reproducibility, i.e., the ability to assign an identical type to the same isolate, was 100%. These markers were specific to A. flavus. The primers used to amplify the microsatellite flanking regions (Table 2) were selected on the basis of in silico specificity to A. flavus. In vitro PCR amplification was not observed either with A. fumigatus and A. niger or with Penicillium sp.
FIG. 1.
Distribution frequency of microsatellites in A. flavus genome.
TABLE 2.
Features of the 12 polymorphic microsatellite sequences of A. flavus upon analysis of 63 isolates
| Primer name | Primer sequence (5′ to 3′) | Repeat unit | Fragment size (bp) | No. of alleles | D |
|---|---|---|---|---|---|
| AFLA1 | CGTTGGCATGTTATCGTCAC | AC | 249-291 | 7 | 0.795 |
| CTACTGAATGGCGGGACCTA | |||||
| AFLA2 | GAGCACGTGCGATTTAGTCA | CTT | 282-291 | 3 | 0.414 |
| TATCTACTCCGGCCAACTCG | |||||
| AFLA3 | CTGAAAGGGTAAGGGGAAGG | TAGG | 174-229 | 5 | 0.703 |
| CACGCGAACTTATGGGACTT | |||||
| AFLA4 | TGCTTAAGTGACCCCAATCC | ||||
| CAGTTGATTTAAGGGGCAACA | CT | 212-242 | 4 | 0.554 | |
| AFLA5 | GTGAGAGCAATTGGGAAACC | GAG | 197-209 | 3 | 0.531 |
| TGGCATTTGATCTCTTGCAG | |||||
| AFLA6 | CATAAGCGCTCCGAAGTCAT | CTT | 238-250 | 3 | 0.533 |
| CCACCGATGATGGAAAAGAT | |||||
| AFLA7 | GCGGACACTGGATGAATAGC | TAG | 261-354 | 7 | 0.738 |
| AACAAATCGGTGGTTGCTTC | |||||
| AFPM2 | CTGGACGGAGATCACGAC | (AC)5T(CTC)4 | 206-266 | 5 | 0.455 |
| CCACGCTCCTCAAATACG | |||||
| AFPM3 | CCTTTCGCACTCCGAGAC | (AT)6AAGGGCG(GA) | 199-217 | 5 | 0.740 |
| CACCACCAGTGATGAGGG | |||||
| AFPM4 | AGCGATACAGTTTTAACACC | CA | 179-206 | 4 | 0.634 |
| TCTTGCTATACATATCTTCACC | |||||
| AFPM5 | TCCTACCCGAGAGAGTCTG | (AG)5AC(AG)2 | 210-338 | 2 | 0.493 |
| CCATTATGACATGTGGTTAAGAG | |||||
| AFPM7 | TTGAGGCTGCTGTGGAACGC | AC | 215-276 | 7 | 0.633 |
| CAAATACCAATTACGTCCAACAAGGG |
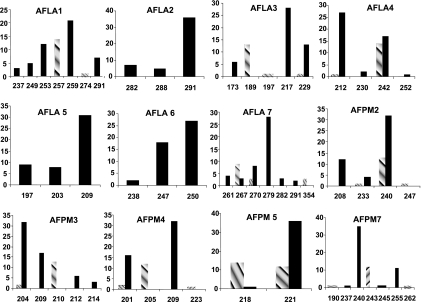
Upon the analysis of 63 isolates (48 from Sfax and 15 from Marseille), 2 to 7 distinct alleles were detected for each microsatellite marker (Fig. 2). The highest discriminatory power for a single locus was obtained with the AFLA1 marker, which had seven distinct alleles and a D value of 0.795 (Table 2). The combination of all 12 markers yielded 35 different haplotypes with a D value of 0.97. A five-marker combination (AFLA1, AFLA7, AFLA3, AFPM3, and AFPM7) yielded 27 different alleles with a D value of 0.952 (Table 3). This five-marker combination was selected, as it was the most parsimonious panel achieving a D value of >0.95.
FIG. 2.
Allele size distribution of A. flavus at 12 microsatellites upon analysis of 15 isolates from Marseille (hatched bars) and 48 isolates from Tunisia (black bars). x axis, number of isolates; y axis, allele size.
TABLE 3.
Discriminatory indices of different microsatellite marker combinations
| Locus combination | No. of profiles | D |
|---|---|---|
| AFLA3-AFLA7 | 12 | 0.840 |
| AFLA1-AFLA3 | 14 | 0.845 |
| AFLA3-AFPM3 | 10 | 0.853 |
| AFLA7-AFPM3 | 10 | 0.858 |
| AFLA1-AFLA7 | 15 | 0.882 |
| AFLA1-AFPM3 | 15 | 0.892 |
| AFLA1-AFLA3-AFPM3 | 18 | 0.911 |
| AFLA1-AFLA7-AFPM3 | 18 | 0.920 |
| AFLA1-AFLA7-AFLA3-AFPM3 | 22 | 0.927 |
| AFLA1-AFLA7-AFLA3-AFPM3-AFPM7 | 27 | 0.952 |
| 12 microsatellites | 35 | 0.970 |
Use of this microsatellite panel analysis revealed a marked genetic heterogeneity of A. flavus within each geographic area, with some alleles being present in the isolates from only one country (Fig. 2). A 0.424 FST index indicated a highly significant (P < 10−5) geographical genetic structure in the A. flavus population studied.
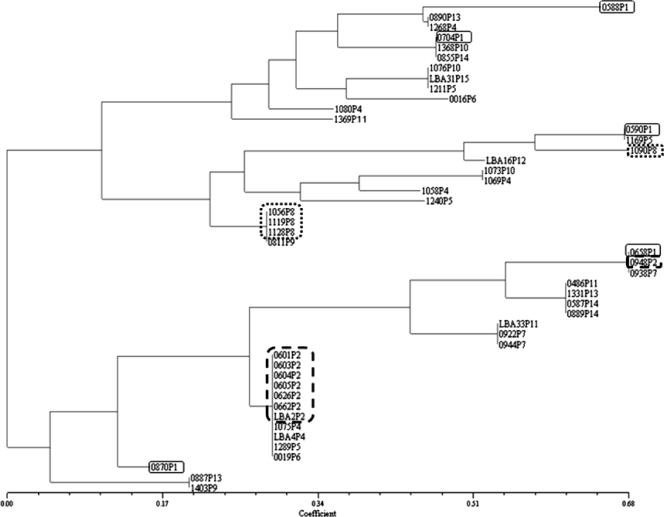
From the 48 clinical and environmental A. flavus isolates sampled in the hematology ward in Sfax, 35 distinct genotypes could be identified. The dendrogram (Fig. 3) illustrates this genetic heterogeneity, where each isolate from one patient and that patient's room had distinct genotypes. For instance, all five isolates sampled from patient 1 (P1) and the environment of P1 were genetically unrelated. However, in contrast to this very heterogeneous genetic background, we were able to identify a clonal cluster that included two out of three environmental isolates and all five clinical isolates of P2, who was hospitalized concomitantly with P1 (Table 1). The four remaining isolates of this cluster were concomitantly isolated from P4 or from two patients (P5 and P6) hospitalized up to 1 year after P2 was hospitalized (Table 1). Three out of four isolates from P8 formed another clonal cluster that included two clinical isolates (nasal and sputum) and one environmental isolate (from the patient's bed). This cluster also included one unrelated environmental isolate. In P11, two closely related isolates (recovered from a lung biopsy specimen and bronchoalveolar lavage [BAL] fluid) differed in only a single (AFPM3) marker genotype.
FIG. 3.
Neighbor-joining dendrogram based on the Dice similarity coefficient upon analysis of five microsatellite makers in 48 A. flavus isolates from a hematology ward in Tunisia. The isolates are identified by the isolate number followed by the patient number (P1 to P15). The isolates are described in Table 1. The highlighted isolates from three patients and their environment illustrate two epidemiological scenarios: for P1 (square with solid-line border), each isolate (clinical and environmental) has a distinct genotype, indicating exposure to a genetically highly heterogeneous A. flavus population; for P2 (square with dashed-line border) and P8 (square with dotted-line border), clinical and environmental isolates share the same genotype, indicating exposure to a clonal A. flavus population.
DISCUSSION
We present here a novel typing scheme that uses microsatellite markers highly polymorphic for A. flavus. Use of this five-microsatellite-marker combination yielded a high discriminatory power and could easily distinguish epidemiologically related A. flavus isolates within two distinct geographic areas (Tunisia and France). In addition to a high degree of discrimination, reproducibility, and typeability, microsatellite analysis has several other advantages over other DNA-based typing assays. Because the assay is PCR based, microsatellite analysis requires relatively small amounts (∼1 ng per reaction) of template DNA, and it can be simplified by using multiplex assays with primers labeled with different fluorescent dyes, resulting in higher throughput (6, 23). Finally, microsatellite analysis allows the detection of mixed cultures, which would appear as a PCR profile harboring two or more microsatellite alleles from a single DNA sample of the haploid genome of A. flavus. The strengths and pitfalls of microsatellite-based typing have been comprehensively described for A. fumigatus (8). With respect to A. fumigatus, microsatellites have proven to offer the best available typing option by outperforming MLST in terms of speed, throughput, costs, and discriminatory power (16). The major pitfall is the interlaboratory reproducibility of PCR fragment sizing. However, this was overcome for A. fumigatus typing by using allelic ladders (7), which then make it possible to deposit the microsatellite allelic profiles in a global typing database similar to those developed for several pathogens using MLST (18).
This 3-year longitudinal genotyping survey provided insights into the A. flavus epidemiology in one hematology unit where patients who are at high risk of invasive aspergillosis are hospitalized. Microsatellite polymorphism analysis revealed a genetically very heterogeneous A. flavus population in this hospital ward. This finding is in line with the A. fumigatus data (3, 4). Indoor and outdoor aerocontamination are probably strongly correlated in the absence of air filtration systems, as was the case in this hospital. Therefore, the genetic heterogeneity of these isolates probably reflects the diversity of airborne A. flavus spores in the hospital area. In this setting it seems particularly challenging to trace patients' exposure to a particular clone disseminated in the hospital environment. However, outbreaks due to clonal A. fumigatus strains have been documented (2). In our survey, there was no cluster of aspergillosis and we did not identify an invasive aspergillosis outbreak. However, we could detect a temporally related clonal cluster of A. flavus. This situation, where over a short time period one genotype becomes predominant, indicates that clonal reproduction, which we would like to refer to as a clonal burst, of the fungus occurred within the ward. Here this clonal burst was detected and probably occurred in P2's room. The genotype involved in this clonal burst was then occasionally identified in isolates sampled in the ward up to 1 year after this episode. Regrettably, aerobiological monitoring was lacking. Therefore, neither a concomitant increase in the number of airborne A. flavus spores in the ward nor the cause of this clonal burst could be documented to support this hypothesis.
Clinical and environmental isolates were independent in the majority of the patients. However, in P2 and P8 clinical and environmental isolates had identical genotypes, suggesting that A. flavus colonization and infection were hospital acquired in these two patients. Moreover, in P2 invasive aspergillosis can be traced to the above-mentioned clonal burst genotype. This finding supports the hypothesis that aerobiological monitoring might help promote actions to restrict, if not prevent, similar clonal burst episodes inside the wards. From this perspective, highly discriminating typing methods such as the microsatellite typing scheme are advantageous in limiting false results and helping to locate the origin of outbreaks.
In conclusion, the novel microsatellite typing scheme described here demonstrated a significant geographical genetic structure in A. flavus and provided further insight into the hospital epidemiology of this emerging human pathogen. In the hematology unit described here, invasive aspergillosis could be traced to two main epidemiological scenarios. The first and most common scenario was colonization and infection by multiple genotypes, reflecting the high genetic diversity of A. flavus outdoors. The second was colonization and infection by a single genotype, most probably indicating a clonal burst episode. This clonal burst can be triggered by favorable local conditions, such as a water pipe leak in the patient's room. The first scenario points to the necessity of air filtration systems in rooms where high-risk patients are hospitalized. The hypotheses underlying the second scenario point to the utility of implementing aerobiological monitoring in those rooms, but they need to be confirmed in further epidemiological studies.
Acknowledgments
Inès Hadrich received travel grants from the Université de la Méditerranée.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print on 21 April 2010.
REFERENCES
- 1.Bain, J. M., A. Tavanti, A. D. Davidson, M. D. Jacobsen, D. Shaw, N. A. Gow, and F. C. Odds. 2007. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J. Clin. Microbiol. 45:1469-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balajee, S. A., H. A. de Valk, B. A. Lasker, J. F. Meis, and C. H. Klaassen. 2008. Utility of a microsatellite assay for identifying clonally related outbreak isolates of Aspergillus fumigatus. J. Microbiol. Methods 73:252-256. [DOI] [PubMed] [Google Scholar]
- 3.Chazalet, V., J. P. Debeaupuis, J. Sarfati, J. Lortholary, P. Ribaud, P. Shah, M. Cornet, H. Vu Thien, E. Gluckman, G. Brucker, and J. P. Latge. 1998. Molecular typing of environmental and patient isolates of Aspergillus fumigatus from various hospital settings. J. Clin. Microbiol. 36:1494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debeaupuis, J. P., J. Sarfati, V. Chazalet, and J. P. Latge. 1997. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect. Immun. 65:3080-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Hoog, G. S., A. S. Nishikaku, G. Fernandez-Zeppenfeldt, C. Padin-Gonzalez, E. Burger, H. Badali, N. Richard-Yegres, and A. H. van den Ende. 2007. Molecular analysis and pathogenicity of the Cladophialophora carrionii complex, with the description of a novel species. Stud. Mycol. 58:219-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Ruiter, M. T., H. A. de Valk, J. F. Meis, and C. H. Klaassen. 2007. Retrotransposon insertion-site context (RISC) typing: a novel typing method for Aspergillus fumigatus and a convenient PCR alternative to restriction fragment length polymorphism analysis. J. Microbiol. Methods 70:528-534. [DOI] [PubMed] [Google Scholar]
- 7.de Valk, H. A., J. F. Meis, S. Bretagne, J. M. Costa, B. A. Lasker, S. A. Balajee, A. C. Pasqualotto, M. J. Anderson, L. Alcazar-Fuoli, E. Mellado, and C. H. Klaassen. 2009. Interlaboratory reproducibility of a microsatellite-based typing assay for Aspergillus fumigatus through the use of allelic ladders: proof of concept. Clin. Microbiol. Infect. 15:180-187. [DOI] [PubMed] [Google Scholar]
- 8.de Valk, H. A., J. F. Meis, and C. H. Klaassen. 2007. Microsatellite based typing of Aspergillus fumigatus: strengths, pitfalls and solutions. J. Microbiol. Methods 69:268-272. [DOI] [PubMed] [Google Scholar]
- 9.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 10.Falvey, D. G., and A. J. Streifel. 2007. Ten-year air sample analysis of Aspergillus prevalence in a university hospital. J. Hosp. Infect. 67:35-41. [DOI] [PubMed] [Google Scholar]
- 11.Guarro, J., M. Sole, R. Castany, J. Cano, A. Teixido, I. Pujol, J. Gene, A. Castro, and P. Sarda. 2005. Use of random amplified microsatellites to type isolates from an outbreak of nosocomial aspergillosis in a general medical ward. Med. Mycol. 43:365-371. [DOI] [PubMed] [Google Scholar]
- 12.Hadrich, I., F. Makni, H. Sellami, F. Cheikhrouhou, A. Sellami, H. Bouaziz, S. Hdiji, M. Elloumi, and A. Ayadi. 4 June 2009. Invasive aspergillosis: epidemiology and environmental study in haematology patients (Sfax, Tunisia). Mycoses. [Epub ahead of print.] [DOI] [PubMed]
- 13.Heinemann, S., F. Symoens, B. Gordts, H. Jannes, and N. Nolard. 2004. Environmental investigations and molecular typing of Aspergillus flavus during an outbreak of postoperative infections. J. Hosp. Infect. 57:149-155. [DOI] [PubMed] [Google Scholar]
- 14.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James, M. J., B. A. Lasker, M. M. McNeil, M. Shelton, D. W. Warnock, and E. Reiss. 2000. Use of a repetitive DNA probe to type clinical and environmental isolates of Aspergillus flavus from a cluster of cutaneous infections in a neonatal intensive care unit. J. Clin. Microbiol. 38:3612-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klaassen, C. H. 2009. MLST versus microsatellites for typing Aspergillus fumigatus isolates. Med. Mycol. 47(Suppl. 1):S27-S33. [DOI] [PubMed] [Google Scholar]
- 17.Krishnan, S., E. K. Manavathu, and P. H. Chandrasekar. 2009. Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses 52:206-222. [DOI] [PubMed] [Google Scholar]
- 18.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moody, S. F., and B. M. Tyler. 1990. Use of nuclear DNA restriction fragment length polymorphisms to analyze the diversity of the Aspergillus flavus group: A. flavus, A. parasiticus, and A. nomius. Appl. Environ. Microbiol. 56:2453-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rath, P. M. 2001. Phenotypic and genotypic characterization of reference strains of the genus Aspergillus. Mycoses 44:65-72. [DOI] [PubMed] [Google Scholar]
- 21.Tran-Dinh, N., and D. Carter. 2000. Characterization of microsatellite loci in the aflatoxigenic fungi Aspergillus flavus and Aspergillus parasiticus. Mol. Ecol. 9:2170-2172. [PubMed] [Google Scholar]
- 22.van Belkum, A., W. van Leeuwen, S. Scherer, and H. Verbrugh. 1999. Occurrence and structure-function relationship of pentameric short sequence repeats in microbial genomes. Res. Microbiol. 150:617-626. [DOI] [PubMed] [Google Scholar]
- 23.Woo, P. C., C. C. Lau, K. T. Chong, H. Tse, D. N. Tsang, R. A. Lee, C. W. Tse, T. L. Que, L. M. Chung, A. H. Ngan, W. T. Hui, S. S. Wong, S. K. Lau, and K. Y. Yuen. 2007. MP1 homologue-based multilocus sequence system for typing the pathogenic fungus Penicillium marneffei: a novel approach using lineage-specific genes. J. Clin. Microbiol. 45:3647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]