Abstract
Three patients admitted to a Greek hospital were infected with Serratia marcescens isolates that exhibited reduced susceptibility to carbapenems and harbored Klebsiella pneumoniae carbapenemase (KPC) enzymes. In two of these cases, the patients were initially infected by carbapenem-susceptible S. marcescens isolates. Molecular typing and plasmid analysis suggested that all three patients had clonally indistinguishable isolates of S. marcescens that acquired a plasmid-mediated blaKPC-2 gene during the hospitalization.
The emergence of organisms producing class A β-lactamases of Klebsiella pneumoniae carbapenemase (KPC) types is a major clinical and public health concern (2, 10). They are typically transposon-encoded determinants and therefore have the potential to disseminate between plasmids and across bacterial species (9). K. pneumoniae remains the species most likely to harbor blaKPC genes (2, 8, 10, 13). Nevertheless, other species of Enterobacteriaceae, as well as species of nonfermenters, have been occasionally reported to exhibit this pattern of resistance (3, 15, 16, 19). In Serratia marcescens carbapenemase production is mostly attributed to class B metallo-β-lactamases (MBLs) as well as to the class A SME family of carbapenemases (14). Only recently has carbapenem-hydrolyzing activity in S. marcescens been attributed to the production of a KPC in China and the United States (3, 17, 23). We report the spread of three blaKPC-possessing S. marcescens isolates in a Greek intensive care unit and give in vitro and in vivo evidence of the potential acquisition of such plasmid-borne resistance genes.
In December 2008 a 77-year-old woman was admitted to the unit following a neurosurgical procedure. Ampicillin-sulbactam was administered postoperatively. Two months after her admission, the patient developed pneumonia, and bronchial lavage samples grew a S. marcescens isolate (S53) that exhibited reduced carbapenem susceptibility. The patient was successfully treated with tigecycline and inhaled colistin.
Approximately 5 months later, in April 2009, a 49-year-old man was admitted following the surgical removal of a subcranial hematoma. He remained febrile while receiving empirical prophylactic antibiotic treatment with ampicillin-sulbactam, vancomycin, and amikacin. Bronchial lavage samples produced a carbapenem-susceptible S. marcescens isolate (S51) and a carbapenem-resistant K. pneumoniae isolate (K72). Antibiotic therapy was changed to meropenem and colistin. A second episode of pneumonia occurred approximately 2 weeks later, and a new S. marcescens isolate (S54) with reduced susceptibility to carbapenems was recovered from the bronchial lavage cultures. The patient was successfully treated with tigecycline and colistin.
Finally, in April 2009 a 33-year-old woman was admitted following extensive surgery to the spine. The patient received ampicillin-sulbactam postoperatively. Approximately a week after her admission she presented with bacteremia due to a carbapenem-susceptible S. marcescens isolate (S52) and was treated with ciprofloxacin. Three weeks later the patient had an episode of pneumonia. Bronchial lavage sample cultures produced a new S. marcescens isolate (S55) that exhibited reduced susceptibility to carbapenems. Administration of ciprofloxacin in combination with gentamicin led to the successful treatment of this episode.
The isolates that were recovered from the aforementioned patients were evaluated. Species identification was performed with the Vitek 2 system (bioMérieux, Marcy l'Étoile, France) and confirmed with API 20E (bioMérieux). MICs for several β-lactams, aminoglycosides, ciprofloxacin, tigecycline, and colistin were further determined by agar dilution according to CLSI recommendations (4). The MBL Etest (AB Biodisk, Solna, Sweden) and the combined disk test with imipenem and EDTA (5) were used to screen for MBL production. The phenotypic detection of KPC-possessing isolates was evaluated with the boronic acid potentiation disk test using meropenem as an antibiotic substrate (20). Extended-spectrum β-lactamase (ESBL) production was tested with the CLSI confirmatory test and in the KPC-possessing isolates with the modified CLSI ESBL confirmatory test, using clavulanate in combination with boronic acid (21).
Isolates were screened for β-lactamase genes by PCR amplification using a panel of primers for the detection of all types of MBLs (6), KPCs (8), plasmid-mediated AmpCs in single PCRs for each gene (11), and ESBLs (22). PCR products were subjected to direct sequencing. Pulsed-field gel electrophoresis (PFGE) of SpeI- and of XbaI-digested genomic DNA of the S. marcescens isolates was performed with a CHEF-DRIII system (Bio-Rad, Hemel Hempstead, United Kingdom), and PFGE patterns were compared visually following previously described criteria (18). The potential for conjugational transfer of carbapenem resistance was examined in biparental matings using LB broth cultures and Escherichia coli 26R764 (lac+ Rifr) as the recipient strain. Transconjugant clones were screened on MacConkey agar plates containing rifampin (150 μg/ml) and amoxicillin (40 μg/ml) or ertapenem (0.5 μg/ml). MICs were determined by agar dilution (4). All β-lactamase genes were detected by PCR amplification. Plasmid extraction was performed by using an alkaline lysis protocol with E. coli 39R861 as a control strain (7).
The susceptibility patterns of the S. marcescens and K. pneumoniae isolates are shown in Table 1. S. marcescens isolates S51 and S52 were susceptible to all carbepenems and to most β-lactam antibiotics. Phenotypic tests were negative for carbapenemase and ESBL production, amplification of the β-lactamase genes confirmed the presence solely of the blaTEM gene, and DNA sequencing identified the gene in both isolates as blaTEM-1.
TABLE 1.
Antimicrobial susceptibility patterns of the study's clinical isolates, their transconjugants, and the recipient strain, E. coli 26R764
| Antibiotic(s) | MIC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|
| Clinical isolates |
Transconjugants |
E. coli 26R764 | |||||
| S. marcescens S51 and S52 | S. marcescens S53, S54, and S55 | K. pneumoniae K72 | E. coli 26R764 S53, S54, and S55 (pTEM-1) | E. coli 26R764 S54, and S55 (pKPC-2 and pTEM-1) | E. coli 26R764 K72 (pKPC-2/TEM-1) | ||
| Imipenem | 0.5 | 2-4 | 16 | 0.25 | 1-2 | 1 | 0.12 |
| Meropenem | 0.12 | 2-4 | 16 | 0.12 | 1 | 0.5 | 0.06 |
| Ertapenem | 0.12-0.25 | 8-16 | >32 | 0.12 | 2-4 | 0.5 | 0.06 |
| Aztreonam | 0.12 | 128-256 | >256 | 0.12 | 128 | 8 | 0.06 |
| Cefotaxime | 0.12 | >32 | >32 | 1 | 16 | 2 | 0.12 |
| Cefepime | 0.5 | 8-16 | 32 | 0.5 | 4-8 | 2 | 0.5 |
| Ceftazidime | 0.12 | 8-16 | 32 | 1 | 4-8 | 2 | 0.12 |
| Cefoxitin | 0.25-0.5 | 16-32 | 32 | 1 | 16 | 2 | 1 |
| Amoxicillin | >256 | >256 | >256 | >256 | >256 | >256 | 4 |
| Amoxicillin-clavulanate | 64-128 | 64 | >256 | 8 | 32-128 | 32 | 4 |
| Piperacillin-tazobactam | 2-4 | >256 | >256 | 2-4 | 128 | 32 | 2 |
| Tigecycline | 1-2 | 1 | 1 | 0.12 | 0.25 | 0.5 | 0.12 |
| Colistin | >16 | >16 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Ciprofloxacin | 0.12 | 0.06-0.12 | 16 | 0.03 | 0.03 | 0.015 | 0.03 |
| Amikacin | 2-8 | 16-32 | 32 | 16-32 | 16-32 | 2 | 2 |
| Tobramycin | 2-4 | 16-32 | 32 | 16 | 16 | 1 | 1 |
| Gentamicin | 1 | 1 | 2 | 1 | 2 | 1 | 1 |
S. marcescens isolates S53, S54, and S55 exhibited reduced susceptibility to imipenem and meropenem and were resistant to ertapenem. They were also resistant to various other β-lactam antibiotics and tobramycin; they also exhibited reduced susceptibility to amikacin but remained susceptible to gentamicin, ciprofloxacin, and tigecycline. The phenotypic tests were negative for MBL production, but the boronic acid potentiation disk test yielded a positive result for KPC production. The modified confirmatory test for ESBL production was negative. Amplification of the β-lactamase genes confirmed the presence of blaKPC and blaTEM genes, which were identified by sequencing analysis as blaKPC-2 and blaTEM-1, respectively.
K. pneumoniae isolate K72 was resistant to all carbapenems, ciprofloxacin, and tobramycin and exhibited reduced susceptibility to amikacin. The isolate remained susceptible to gentamicin, colistin, and tigecycline. Phenotypic tests showed KPC production. PCR assays and subsequent DNA sequencing confirmed the presence of blaKPC-2 and blaTEM-1 genes.
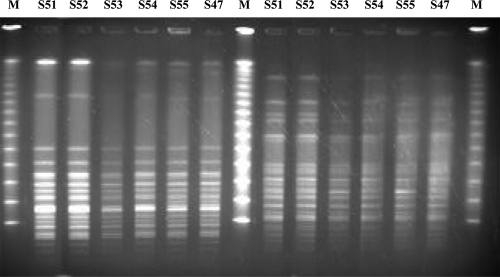
Analysis of the PFGE patterns of SpeI- and of XbaI-digested genomic DNA revealed that all five S. marcescens isolates, irrespective of their susceptibility patterns, were genetically indistinguishable (Fig. 1). For comparison we also tested five carbapenem-susceptible S. marcescens isolates that were recovered from separate patients hospitalized in the intensive care unit (ICU) before or during the period of the study (October 2008 to May 2009) and three historical S. marcescens isolates from our collections. All five isolates from the ICU belonged to the same pulsotype as the ones in our cases, whereas the historical isolates belonged to different pulsotypes (data not shown).
FIG. 1.
PFGE profiles of SpeI-digested genomic DNAs (left side) and of XbaI-digested genomic DNAs (right side) from S. marcescens isolates of the study (S51, S52, S53, S54, and S55) and a contemporary carbapenem-susceptible S. marcescens isolate (S47) from a patient hospitalized in the ICU before the study period. Lanes M, multimers of phage lambda DNA (48.5-kb) molecular mass markers.
Conjugation experiments using individual parental KPC-possessing S. marcescens isolates S53, S54, and S55 showed that resistance to β-lactams was self-transferable. Transconjugants received either solely the blaTEM gene or both blaKPC and blaTEM genes, thus presenting two distinctly different susceptibility patterns, depending on the resistance genes which they acquired (Table 1). Those positive for both blaKPC and blaTEM genes exhibited carbapenem MICs which were higher than those of the recipient strain. Reduced susceptibility to amikacin and resistance to tobramycin were transferred to all transconjugants. Plasmid analysis revealed that the parental isolates contained two separate plasmids with molecular sizes of approximately 85 kb and 35 kb, which were both transferable. The plasmid bands of the transconjugants were extracted from the gel and used as templates for the amplification of the blaKPC and blaTEM genes, the specific products of which were amplified from the larger and smaller plasmid, respectively, suggesting that the blaKPC gene was located on the 85-kb plasmid while the blaTEM gene was on the 35-kb one. EcoRI restriction analysis was performed on the purified 85-kb plasmids of the transconjugants. Restriction patterns were identical (data not shown), suggesting that all three S. marcescens isolates had acquired the same blaKPC-2-bearing plasmid.
Plasmid analysis of K. pneumoniae isolate K72 revealed the presence of only one plasmid of approximately 110 kb, which was transmissible and conferred resistance to β-lactams including carbapenems in the recipient strain. DNA extracted from plasmid bands of K72 and its E. coli transconjugant yielded positive results for both blaKPC and blaTEM genes. These transconjugants, however, presented a different susceptibility pattern in comparison to those which also contained both the blaKPC and blaTEM genes yet were derived from the S. marcescens isolates (Table 1). They were highly resistant to penicillin and combinations of penicillin with β-lactamase inhibitors and exhibited a lower increase in cephalosporin and aztreonam MICs, while MICs of imipenem, meropenem, and ertapenem were higher than those of the recipient strain yet lower than those of the S. marcescens transconjugants.
The present study documents the first report of KPC production in S. marcescens isolates in Europe and offers evidence of the potential for acquisition of these plasmid-borne enzymes during long-term hospitalization. These KPC-positive S. marcescens isolates were acquired while the patients were hospitalized in the ICU, following in two cases an initial infection by a non-carbapenemase-producing S. marcescens isolate. The isolates were genotypically indistinguishable, indicating the dissemination of the plasmid harboring the blaKPC-2 gene within the same clone.
KPC genes along with other resistance genes are typically located on mobile genetic elements (9, 12, 20). Previous studies have presented evidence which suggested the potential for horizontal dissemination of blaKPC genes between different clones of the same enterobacterial species (1) or even between different genera (3, 12, 15, 17). In one of our cases the patient was simultaneously infected with a K. pneumoniae isolate belonging to the clonal outbreak with which we are currently dealing in our hospital (data not shown). Our results, however, suggested that the blaKPC-2 gene of S. marcescens isolates was located on a different plasmid from that of the K. pneumoniae isolate. Therefore, it can be postulated that it was the plasmid-encoded blaKPC-2 gene of the index carbapenem-nonsusceptible S. marcescens isolate that disseminated. It cannot be excluded, however, that through a recombination event the blaKPC-2 gene was initially transferred from the K. pneumoniae clone to the carbapenem-susceptible S. marcescens clone. Although the initial source of infection could not be verified, all KPC-producing S. marcescens organisms were retrieved from bronchial lavage samples. It is therefore possible that the transfer of the plasmid-encoded KPC-2 enzyme occurred in the environment of the mixed bacterial flora typically found in bronchial secretions.
Our report provides evidence that S. marcescens has the ability to easily acquire KPC carbapenemases. Successfully combating infections caused by these carbapenem-resistant S. marcescens pathogens poses a difficult challenge, given their intrinsic resistance to last-resort antibiotics, such as colistin, and their potential for dissemination.
Acknowledgments
We declare no potential conflicts of interest.
Footnotes
Published ahead of print on 12 May 2010.
REFERENCES
- 1.Barbier, F., E. Ruppe, P. Giakkoupi, L. Wildenberg, J. C. Lucet, A. Vatopoulos, M. Wolff, and A. Andremont. 2010. Genesis of a KPC-producing Klebsiella pneumoniae after in vivo transfer from an imported Greek strain. Euro Surveill. 15:ii19457. [DOI] [PubMed] [Google Scholar]
- 2.Bratu, S., D. Landman, R. Haag, R. Recco, A. Eramo, M. Alam, and J. Quale. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 165:1430-1435. [DOI] [PubMed] [Google Scholar]
- 3.Cai, J. C., H. W. Zhou, R. Zhang, and G. X. Chen. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob. Agents Chemother. 52:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement. CLSI M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Franklin, C., L. Liolios, and A. Y. Peleg. 2006. Phenotypic detection of carbapenem-susceptible metallo-β-lactamase producing gram-negative bacilli in the clinical laboratory. J. Clin. Microbiol. 44:3139-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikonomidis, A., D. Tokatlidou, I. Kristo, D. Sofianou, A. Tsakris, P. Mantzana, S. Pournaras, and A. N. Maniatis. 2005. Outbreaks in distinct regions due to a single Klebsiella pneumoniae clone carrying a blaVIM-1 metallo-β-lactamase gene. J. Clin. Microbiol. 43:5344-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomaestro, B. M., E. H. Tobin, W. Shang, and T. Gootz. 2006. The spread of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae to upstate New York. Clin. Infect. Dis. 43:e26-e28. [DOI] [PubMed] [Google Scholar]
- 9.Naas, T., G. Cuzon, M. V. Villegas, M. F. Lartigue, J. P. Quinn, and P. Nordmann. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordmann, P., G. Cuzon, and T. Naas. 2009. The real threat of Klebsiella pneumoniae carbapenemase producing bacteria. Lancet Infect. Dis. 9:228-236. [DOI] [PubMed] [Google Scholar]
- 11.Pérez-Pérez, F. J., and N. D. Hanson. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrella, S., N. Ziental-Gelus, C. Mayer, M. Renard, V. Jarlier, and W. Sougakoff. 2008. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A β-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob. Agents Chemother. 52:3725-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pournaras, S., E. Protonotariou, E. Voulgari, I. Kristo, E. Dimitroulia, D. Vitti, M. Tsalidou, A. N. Maniatis, A. Tsakris, and D. Sofianou. 2009. Clonal spread of KPC-2 carbapenemase-producing Klebsiella pneumoniae strains in Greece. J. Antimicrob. Chemother. 64:348-352. [DOI] [PubMed] [Google Scholar]
- 14.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasheed, J. K., J. W. Biddle, K. F. Anderson, L. Washer, C. Chenoweth, J. Perrin, D. W. Newton, and J. B. Patel. 2008. Detection of the Klebsiella pneumoniae carbapenemase type 2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. J. Clin. Microbiol. 46:2066-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robledo, I. E., E. E. Aquino, M. I. Santé, J. L. Santana, D. M. Otero, C. F. León, and G. J. Vázquez. 2010. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 54:1354-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidjabat, H. E., F. P. Silveira, B. A. Potoski, K. M. Abu-Elmagd, J. M. Adams-Haduch, D. L. Paterson, and Y. Doi. 2009. Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin. Infect. Dis. 49:1736-1738. [DOI] [PubMed] [Google Scholar]
- 18.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tibbetts, R., J. G. Frye, J. Marschall, D. Warren, and W. Dunne. 2008. Detection of KPC-2 in a clinical isolate of Proteus mirabilis and first reported description of carbapenemase resistance caused by a KPC β-lactamase in P. mirabilis. J. Clin. Microbiol. 46:3080-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsakris, A., I. Kristo, A. Poulou, F. Markou, A. Ikonomidis, and S. Pournaras. 2008. First occurrence of KPC-2-possessing Klebsiella pneumoniae in a Greeek hospital and recommendation for detection with boronic acid disk tests. J. Antimicrob. Chemother. 62:1257-1260. [DOI] [PubMed] [Google Scholar]
- 21.Tsakris, A., A. Poulou, K. Themeli-Digalaki, E. Voulgari, T. Pittaras, D. Sofianou, S. Pournaras, and D. Petropoulou. 2009. Use of boronic acid disk tests to detect extended-spectrum β-lactamases in clinical isolates of KPC carbapenemase-possessing Enterobacteriaceae. J. Clin. Microbiol. 47:3420-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzelepi, E., C. Magana, E. Platsouka, D. Sofianou, O. Paniara, N. J. Legakis, A. C. Vatopoulos, and L. S. Tzouvelekis. 2003. Extended spectrum β-lactamase types in Klebsiella pneumoniae and Escherichia coli in two Greek hospitals. Int. J. Antimicrob. Agents 21:285-288. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, R., H.W. Zhou, J. C. Cai, and G. X. Chen. 2007. Plasmid-mediated carbapenem-hydrolysing β-lactamase KPC-2 in carbapenem-resistant Serratia marcescens isolates from Hangzhou, China. J. Antimicrob. Chemother. 59:574-576. [DOI] [PubMed] [Google Scholar]