Abstract
The genetic relatedness of Streptococcus milleri group isolates from the airways of cystic fibrosis patients was determined by using pulsed-field gel electrophoresis. This study reveals no evidence for patient-to-patient transmission in our patient population; however, within individual patients, complex inter- and intraspecies diversity and dynamics can be observed.
In addition to their role in purulent infections (3, 7, 11), members of the Streptococcus milleri group (SMG), also known as the Streptococcus anginosus group, comprised of Streptococcus constellatus, Streptococcus intermedius, and Streptococcus anginosus, have emerged as clinically relevant in chronic airway infections in cystic fibrosis (CF) patients and have been implicated as etiologic agents of pulmonary exacerbation (6, 10, 13, 14). We have recently described the isolation of a large number of SMG strains from a cohort of CF patients by using the semiselective medium McKay agar (12). This collection of SMG respiratory isolates was not recovered by conventional CF microbiology and enabled us to characterize the phenotypic properties of airway isolates and compare them to invasive strains (5). These results, in combination with analysis of the nucleotide sequence of the 16S rRNA gene of these strains, revealed clusters of isolates that included both CF and invasive isolates with indistinguishable phenotypic characteristics (5, 9).
In this study, we evaluated whether patient-to-patient transmission was occurring in our CF patient cohort. The molecular epidemiological relationship of the SMG isolates was determined by using pulsed-field gel electrophoresis (PFGE) (4, 8, 15).
PFGE was performed by modification of a protocol described by Bartie et al. (2). The isolates were cultured at 37°C for 48 h on brain heart infusion agar supplemented with colistin sulfate (10 μg/ml) and oxolinic acid (5 μg/ml) under anaerobic conditions. The cells were harvested and suspended to 20% transmittance (600 nm) in 100 mM Tris-HCl buffer (pH 7.6). Mutanolysin (100 U; Sigma-Aldrich, St. Louis, MO) was added to 500 μl of cell suspension before an equal volume of molten 1% SeaKem Gold agarose (Lonza, Rockland, ME) was added. Plugs were cast at room temperature and then transferred to 1.5 ml of lysis solution (0.25 M EDTA [pH 9.0], 0.5% Brij 58, 2 g/liter sodium deoxycholate, 5 g/liter lauroyl sarcosine, 100 U/ml mutanolysin) and incubated at 37°C for 2 h. The lysis solution was replaced with 1.5 ml ESP solution (0.25 M EDTA [pH 9.5], 1% sodium lauroyl sarcosine, 0.5 mg/ml proteinase K) and incubated at 55°C for 2 h. The plugs were rinsed with 1 ml of distilled water and then washed for 10-min intervals, once with distilled water and three times with 1× Tris-EDTA (TE; 10 mM Tris-Hcl, 1 mM EDTA [pH 8.0]) at room temperature.
For restriction digestion, the plugs were preincubated in 300 μl of 1× reaction buffer (Invitrogen, Carlsbad, CA) at room temperature for 15 min and then replaced with fresh 1× reaction buffer supplemented with 90 U of SmaI or ApaI (New England BioLabs, Beverly, MA). SmaI and ApaI digestion occurred at room temperature and 31°C, respectively, for 4 h. Following digestion, the plugs were briefly rinsed twice in 1× TE. Following a 5-min wash in 1× TE, the plugs were loaded into a 1% SeaKem Gold agarose gel, prepared in 0.5× TBE (1× TBE is 89 mM Tris-HCl [pH 7.4], 89 mM boric acid, 25 mM EDTA [pH 8.0]). The following parameters were used: gradient, 6.0 V/cm; run time, 22 h; included angle, 120°; initial switch time, 10 s; and final switch time, 35 s at 14°C.
A database of the PFGE profiles was developed with BioNumerics software (Applied Maths, Saint-Martens-Latem, Belgium). Dendrograms were generated using the unweighted pair group method using average linkages with a 1.0% position tolerance and the Dice coefficient correlation. The threshold required to justify analysis with an additional restriction enzyme was 90%.
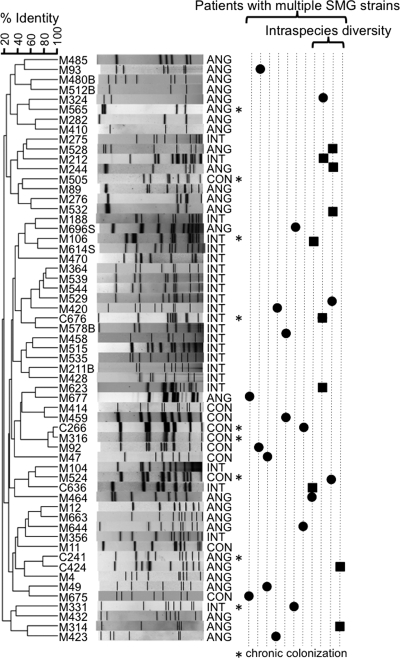
Fifty-nine unique profiles were observed from 76 SMG isolates cultured from expectorated sputum from 40 patients (Fig. 1). Such high genetic heterogeneity has been previously observed in the SMG (1). Four isolates from three patients were refractory to PFGE profiling.
FIG. 1.
Relationship between all of the unique PFGE profiles generated with SmaI from the SMG isolates used in this study. Chronically colonizing strains (recovered from the same patient on multiple occasions) are indicated with an asterisk. Patients with multiple SMG strains are represented as vertical dotted lines to the right of the gel profiles; SMG isolates of different species are shown by a solid black circle. In patients where intraspecies diversity was observed, strains of the same species are depicted with solid black squares. The strain and species (S. anginosus [ANG], S. intermedius [INT], and S. constellatus [CON]) are indicated to the left and right of the gel profiles, respectively.
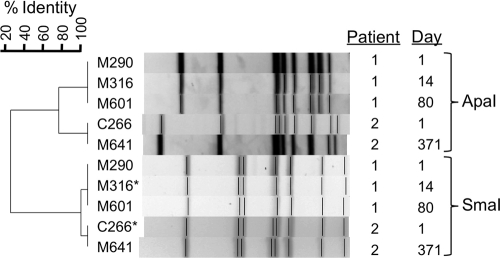
Notably, only two isolates (M316 and C266) recovered from different patients clustered above 90% identity by SmaI profiling (Fig. 1). To further resolve whether these isolates might represent patient-to-patient transmission, we tested all available S. constellatus isolates recovered longitudinally from these two patients (two isolates from the patient whose sputum yielded isolate M316 and three isolates from the patient whose sputum yielded isolate C266) with a secondary restriction enzyme, ApaI (Fig. 2). This analysis revealed that these strains were in fact more genetically divergent and clustered at less than 80% identity. Given that these strains represent distinct PFGE profiles and that these two patients had not attended the clinic on the same day or been inpatients at the same time, the two strains appear to represent different strains.
FIG. 2.
ApaI fingerprints of multiple isolates from two patients with closely related SmaI profiles reveal that the strains are genetically different enough to rule out patient-to-patient transmission. The multiple isolates were recovered at different times (shown in days to the right of the gel profile). The two isolates indicated with an asterisk are also represented in Fig. 1.
PFGE revealed a diverse SMG population. Multiple SMG isolates per patient (collected either longitudinally or cross-sectionally) were analyzed from a total of 15 patients. Multiple PFGE profiles were detected in 11 patients (Fig. 1). Four of the patients with multiple SMG strains revealed intraspecies diversity; as many as three distinct strains of the same species were represented in a single patient (Fig. 1).
Isolates were available for longitudinal analysis in 7 of the 11 patients (63.6%) with multiple SMG strains present. In six of these cases (85.7%), at least one of the strains was isolated on at least two occasions. It was possible to investigate longitudinal isolates by PFGE in four additional patients. In total, the same isolate was recovered at multiple time points in 9 of the 11 patients (81.8%) analyzed longitudinally. As previously reported, this supports the hypothesis that chronic colonization is common (2, 13), although it may not always be the case.
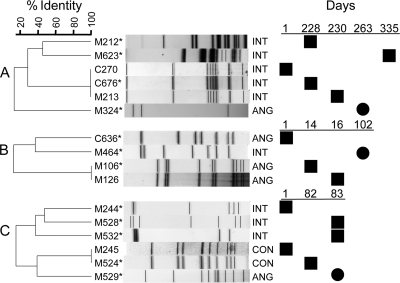
The complex dynamics of SMG populations within certain patients was noteworthy. Examples of SMG populations in three patients that illustrate both the diversity within the SMG at the species and strain level and the complex population dynamics that can occur over time are shown in Fig. 3.
FIG. 3.
Genetic relatedness of strains recovered from three different patients (A, B, and C) that demonstrate inter- and intraspecies diversity. Population dynamics of the SMG strains from longitudinal sampling are shown next to the corresponding PFGE profile. Strains of the same or different species are depicted with solid black squares or solid black circles, respectively. The strain and species (S. anginosus [ANG], S. intermedius [INT], and S. constellatus [CON]) are indicated to the left and right of the gel profiles, respectively. The isolates indicated with an asterisk are also represented in Fig. 1.
We have determined that SMG strains in our CF patient population are patient specific, and we have no evidence for the occurrence of patient-to-patient transmission. Intriguingly, this study reveals an unexpected level of complexity in SMG populations detected in sputum samples from certain CF patients. Moreover, it is important to consider that the intraspecies diversity we observe may still underrepresent the true population richness due to limited sampling. To fully appreciate the extent of genetic heterogeneity in chronic infections, deep sampling may be required. As we continue to investigate the CF airway microbiome, strain-level diversity and dynamics may be integral to developing predictive models of clinical perturbation.
Acknowledgments
We acknowledge the staff of the Southern Alberta CF Clinic for their contribution to patient care and for providing epidemiological data.
This work was supported by grants from the Canadian Cystic Fibrosis Foundation (CCFF) and the Canadian Institutes of Health Research (CIHR) to M.G.S. M.G.S. is supported as an Alberta Heritage Foundation for Medical Research (AHFMR) Scientist and Canada Research Chair in Microbial Gene Expression. An AHFMR studentship and a Canada Graduate Scholarship from CIHR support C.D.S. M.E.G. is supported by studentships from the CIHR, CCFF, and AHFMR.
Footnotes
Published ahead of print on 12 May 2010.
REFERENCES
- 1.Asmah, N., B. Eberspacher, T. Regnath, and M. Arvand. 2009. Prevalence of erythromycin and clindamycin resistance among clinical isolates of the Streptococcus anginosus group in Germany. J. Med. Microbiol. 58:222-227. [DOI] [PubMed] [Google Scholar]
- 2.Bartie, K. L., M. J. Wilson, D. W. Williams, and M. A. Lewis. 2000. Macrorestriction fingerprinting of “Streptococcus milleri” group bacteria by pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belko, J., D. A. Goldmann, A. Macone, and A. K. Zaidi. 2002. Clinically significant infections with organisms of the Streptococcus milleri group. Pediatr. Infect. Dis. J. 21:715-723. [DOI] [PubMed] [Google Scholar]
- 4.Bert, F., C. Branger, and N. Lambert-Zechovsky. 1997. Pulsed-field gel electrophoresis is more discriminating than multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for typing pyogenic streptococci. Curr. Microbiol. 34:226-229. [DOI] [PubMed] [Google Scholar]
- 5.Grinwis, M. E., C. D. Sibley, M. D. Parkins, C. S. Eshaghurshan, H. R. Rabin, and M. G. Surette. 2009. Characterization of Streptococcus milleri group isolates from expectorated sputum of adult cystic fibrosis patients. J. Clin. Microbiol. 48:395-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris, J. K., M. A. De Groote, S. D. Sagel, E. T. Zemanick, R. Kapsner, C. Penvari, H. Kaess, R. R. Deterding, F. J. Accurso, and N. R. Pace. 2007. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc. Natl. Acad. Sci. U. S. A. 104:20529-20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maskell, N. A., C. W. Davies, A. J. Nunn, E. L. Hedley, F. V. Gleeson, R. Miller, R. Gabe, G. L. Rees, T. E. Peto, M. A. Woodhead, D. J. Lane, J. H. Darbyshire, and R. J. Davies for the First Multicenter Intrapleural Sepsis Trial (MIST1) Group. 2005. U.K. controlled trial of intrapleural streptokinase for pleural infection. N. Engl. J. Med. 352:865-874. [DOI] [PubMed] [Google Scholar]
- 8.Moser, S. A., S. C. Mitchell, J. D. Ruby, S. Momeni, R. C. Osgood, J. Whiddon, and N. K. Childers. 2010. Repetitive extragenic palindromic PCR for study of Streptococcus mutans diversity and transmission in human populations. J. Clin. Microbiol. 48:599-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson, A. B., C. D. Sibley, L. Schmidt, M. A. Wilcox, M. G. Surette, and C. R. Corbett. 2010. Development of real-time PCR assays for detection of the Streptococcus milleri group from cystic fibrosis clinical specimens using targets for cpn60 and 16S rRNA genes. J. Clin. Microbiol. 48:1150-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkins, M. D., C. D. Sibley, M. G. Surette, and H. R. Rabin. 2008. The Streptococcus milleri group-an unrecognized cause of disease in cystic fibrosis: a case series and literature review. Pediatr. Pulmonol. 43:490-497. [DOI] [PubMed] [Google Scholar]
- 11.Prasad, K. N., A. M. Mishra, D. Gupta, N. Husain, M. Husain, and R. K. Gupta. 2006. Analysis of microbial etiology and mortality in patients with brain abscess. J. Infect. 53:221-227. [DOI] [PubMed] [Google Scholar]
- 12.Sibley, C. D., M. Grinwis, T. R. Field, M. D. Parkins, J. C. Noorgard, D. B. Gregson, H. R. Rabin, and M. Surette. 2010. McKay agar enables routine quantification of the Streptococcus milleri group in cystic fibrosis patients. J. Med. Microbiol. 59:534-540. [DOI] [PubMed] [Google Scholar]
- 13.Sibley, C. D., M. D. Parkins, H. R. Rabin, K. Duan, J. C. Norgaard, and M. G. Surette. 2008. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 105:15070-15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sibley, C. D., M. D. Parkins, H. R. Rabin, and M. G. Surette. 2009. The relevance of the polymicrobial nature of airway infection in the acute and chronic management of patients with cystic fibrosis. Curr. Opin. Investig. Drugs 10:787-794. [PubMed] [Google Scholar]
- 15.Soll, D. R., C. Pujol, and S. R. Lockhart. 2007. Laboratory procedures for epidemiological analysis of microorganisms, p. 129-151. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]