Abstract
This study used a diverse collection of epidemiologically unrelated Acinetobacter baumannii isolates to compare the robustness of a multilocus sequence typing (MLST) scheme, based on conserved regions of seven housekeeping genes, gltA, gdhB, recA, cpn60, rpoD, gyrB, and gpi, with that of sequence-based typing of blaOXA-51-like genes (SBT-blaOXA-51-like genes). The data obtained by analysis of MLST and SBT-blaOXA-51-like genes were compared to the data generated by pulsed-field gel electrophoresis (PFGE). The topologies of the phylogenetic trees generated for the gyrB and gpi genes showed evidence of recombination and were inconsistent with those of the trees generated for the other five genes. MLST identified 24 sequence types (STs), of which 19 were novel, and 5 novel alleles. Clonality was demonstrated by eBURST analysis and standardized index of association values of >1 (P < 0.001). MLST data revealed that all isolates harboring the major blaOXA-51-like alleles OXA-66, OXA-69, and OXA-71 fell within the three major European clonal lineages. However, the MLST data were not always in concordance with the PFGE data, and some isolates containing the same blaOXA-51-like allele demonstrated <50% relatedness by PFGE. It was concluded that the gyrB and gpi genes are not good candidates for use in MLST analysis and that a SBT-blaOXA-51-like gene scheme produced results comparable to those produced by MLST for the identification of the major epidemic lineages, with the advantage of having a significantly reduced sequencing cost and time. It is proposed that studies of A. baumannii epidemiology could involve initial screening of blaOXA-51-like alleles to identify isolates belonging to major epidemic lineages, followed by MLST analysis to categorize isolates from common lineages, with PFGE being reserved for fine-scale typing.
Acinetobacter baumannii is a Gram-negative bacterium that causes serious nosocomial infections, especially in critical care units (13, 14, 42). Several outbreaks have been caused by multidrug-resistant (MDR) strains of A. baumannii (23, 33, 43), and the rate of resistance to carbapenems, which have been the antibiotics of choice to treat infections caused by this pathogen, has increased considerably over the last decade (4, 32, 42, 46). In addition, the prevalence of A. baumannii in hospitals has increased worldwide (3, 25, 28, 29, 47), and, therefore, finding suitable molecular typing methods for A. baumannii is essential for epidemiological investigations and infection control studies. Many genomic typing methods have been used, including ribotyping (36), infrequent-restriction-site analysis (48), repetitive extragenic palindromic sequence-based PCR (rep-PCR) (18), random amplified polymorphic DNA (RAPD) analysis (21), amplified fragment length polymorphism (AFLP) analysis (39), and multilocus PCR and electrospray ionization mass spectrometry (PCR/ESI-MS) (6). Pulsed-field gel electrophoresis (PFGE) is still considered the “gold standard” for the typing of bacterial isolates (36), but it has drawbacks when its comes to interchanging data among laboratories for comparison purposes (35) and may lose its discriminatory power when isolates from geographically diverse areas are analyzed.
Multilocus sequence typing (MLST) schemes, which use several housekeeping genes, have already been used to type many pathogenic bacteria (15, 17, 20, 40), including A. baumannii (1, 31, 44), and MLST is emerging as an alternative to PFGE. MLST is used mainly for global epidemiology studies, but it has also been used successfully for short-term investigation of an outbreak of meningococcal disease (11). Although MLST has many advantages over other molecular typing methods, many questions remain to be answered, including whether several loci are required to obtain a robust scheme and whether the criteria for the selection of the housekeeping genes are sufficiently reliable to reveal the population structure of the strains analyzed.
blaOXA-51-like genes are unique to A. baumannii and may be used as markers for identification of this species (16). They have also successfully been used as one of three loci in a PCR-based typing scheme that is able to assign isolates of A. baumannii to sequence groups (SGs) that appear to correlate with the major epidemic lineages within the species (41). This raises the question of whether the blaOXA-51-like genes themselves could be utilized in a typing scheme. Accordingly, the aim of this work was to investigate the robustness of MLST in categorizing epidemiologically unrelated A. baumannii isolates from four continents and to evaluate the use of variations within the intrinsic blaOXA-51-like gene as a typing tool comparable to MLST.
MATERIALS AND METHODS
Bacterial isolates.
Forty-four isolates of A. baumannii from 22 countries, including three standard representatives of the endemic pan-European clonal lineages I, II, and III, were included in this study. All isolates were identified as A. baumannii as described previously (8) and by the presence of a blaOXA-51-like gene (45). All the isolates were recovered from cases of invasive disease in various hospitals between 1982 and 2006.
PFGE.
PFGE was performed essentially as described previously (27), but with minor modifications. The agarose-embedded bacterial genomic DNA was digested with ApaI (Promega, Southampton, United Kingdom) at 37°C overnight. Electrophoresis was performed on agarose (1%, wt/vol) gels with 0.5× Tris-borate-EDTA buffer. The following PFGE parameters were applied: voltage of 6 V/cm, initial switch time of 5 s, final switch time of 35 s, and run time of 24 h. Electrophoresis was performed in a CHEF DRII apparatus (Bio-Rad, Hemel Hempstead, United Kingdom). The stained gels were scanned using the Diversity Database software image-capturing system (Bio-Rad). The Dice coefficient was used to calculate similarities, and the unweighted-pair group method using average linkages (UPGMA) was used for cluster analysis with BioNumerics software, version 4.0 (Applied Maths, St-Martens-Latem, Belgium).
MLST analysis.
DNA was purified using a Puregene DNA purification system genomic DNA purification kit (Gentra Systems, Minneapolis, MN). PCRs for the seven housekeeping genes, gltA, gyrB, gdhB, recA, cpn60, gpi, and rpoD, were performed in 50-μl volumes containing 10 μl 5× Green GoTaq Flexi buffer, 1.5 mM MgCl2, 800 μM PCR nucleotide mix, and 1.25 U GoTaq DNA polymerase (Promega). The primers and PCR conditions were those described by Bartual et al. (1), except that the annealing temperatures were modified to 45°C for gyrB and rpoD, to 56°C for recA, and to 58°C for cpn60 and gpi.
A Px2 thermal cycler (Thermo Fisher Scientific, Waltham, MA) was used for all PCRs. Analysis of the PCR products was on agarose (1.5%, wt/vol) gels, followed by staining with ethidium bromide and visualization using the Diversity Database software image-capturing system (Bio-Rad). Following purification using a QIAquick PCR purification kit (Qiagen, Crawley, United Kingdom), the products were sequenced in both directions on a 3730 DNA analyzer (Applied Biosystems, Warrington, United Kingdom). The resulting sequences were analyzed using the online BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and MultAlin (http://bioinfo.genopole-toulouse.prd.fr/multalin/) software. The isolates were then assigned to sequence types (STs) using the tools on the A. baumannii MLST webpage (http://pubmlst.org/abaumannii/).
(i) Tree congruence.
Neighbor-joining trees for all identified alleles of the seven genes were constructed in Geneious (version 2.0.10) software using the Jukes-Cantor genetic distance model, with statistical support for the nodes being assessed via the bootstrap resampling method with 1,000 resamplings. Comparisons of the tree topologies were conducted visually, and differences were confirmed using the quartet measure of tree-to-tree distances implemented in COMPONENT (version 2.0) software (http://taxonomy.zoology.gla.ac.uk/rod/cpw/index.html).
(ii) PHI test.
The pairwise homoplasmy index (PHI) test was used to investigate the possibility of recombination among the alleles within the seven housekeeping genes (2) and was implemented in SplitsTree4 software (http://www.splitstree.org/).
(iii) Phylogenetic reconstruction.
The phylogeny for the 44 isolates was estimated via the neighbor-joining method using Geneious (version 2.0.10) software and the Jukes-Cantor genetic distance model, with statistical support for the nodes being assessed via the bootstrap resampling method with 1,000 resamplings. Concatenated sequences comprising the seven loci (gltA, gdhB, rpoD, cpn60, recA, gyrB, and gpi) and five loci (gltA, gdhB, rpoD, cpn60, and recA) were utilized (see Results).
(iv) eBURST analysis.
eBURST analysis was used to investigate the evolutionary relationships and clonal complexes (CCs) within the isolates, using the software on the eBURST website (http://eburst.mlst.net/v3/enter_data/single/), with statistical support for the complexes being assessed via the bootstrap resampling method with 1,000 resamplings (12). Analysis was performed by the use of both stringent (a minimum of six shared alleles) and relaxed (a minimum of five shared alleles) grouping parameters.
(v) IAS.
The standardized index of association (IAS) for the seven loci was calculated using START2 software (http://pubmlst.org/software/analysis/start2/) and 1,000 iterations.
Sequence-based typing of blaOXA-51-like genes (SBT-blaOXA-51-like genes).
Genomic DNA was extracted by boiling two to three colonies for 10 min in 50 μl sterile distilled water. The PCR mixtures contained 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, nuclease-free bovine serum albumin (0.1 mg/ml), 0.1% Triton X-100, 1.5 mM MgCl2, 800 μM PCR nucleotide mix, and 1.25 U of Pfu DNA polymerase (Promega) in a total volume of 50 μl. Primers OXA-69A and OXA-69B (16), external to the blaOXA-51-like gene, were used to amplify the entire sequence under the following conditions: 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, 48°C for 40 s, and 72°C for 3 min and then 72°C for 6 min. Primers were used at a final concentration of 0.25 μM, and reactions were performed with 0.5 μl crude DNA template. PCR product analysis and sequencing were performed as described above for the MLST products.
RESULTS
PFGE.
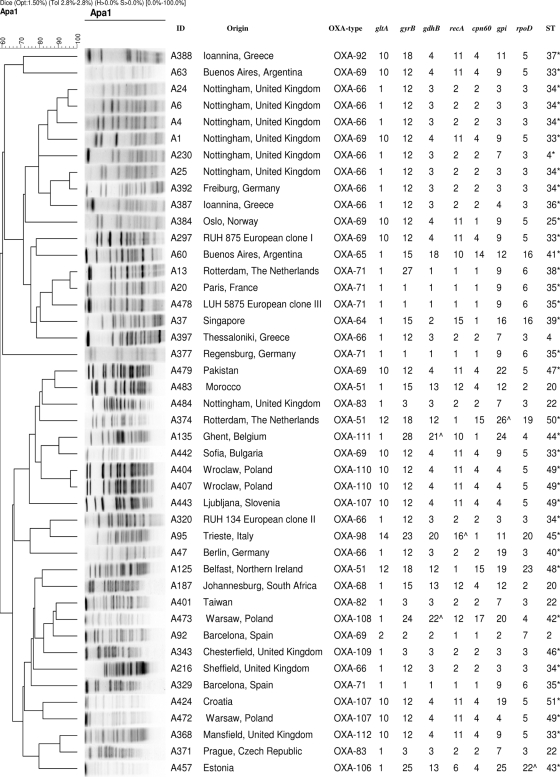
Six small clusters of isolates, each with a similarity of >80%, were identified (Fig. 1). The isolates forming each cluster were considered to belong to the same epidemic lineage. The remaining isolates had PFGE profiles with similarities of <80% and were considered to be unrelated.
FIG. 1.
Dendrogram constructed following determination of PFGE profiles using UPGMA. The OXA-51 type, the ST, and the MLST allelic profiles of seven housekeeping genes are shown for each isolate. ID, isolate identifier; ^, novel allelic profile; *, novel sequence.
MLST analysis.
Seven housekeeping genes, gltA, gdhB, recA, cpn60, rpoD, gyrB, and gpi, were amplified and sequenced for each isolate. Twenty-four different STs were identified, of which 19 (designated ST33 to ST51) were novel, and five novel alleles, officially named gdhB21, gdhB22, recA16, rpoD22, and gpi26, were identified (Fig. 1).
(i) Tree congruence.
Comparison of the tree topologies for all identified alleles of the seven genes revealed inconsistencies with the trees for gyrB and gpi in relation to one another and to the other five trees. The topologies of the trees for the gltA, cpn60, gdhB, recA, and rpoD sequences were broadly consistent, with the same isolates being grouped together in the same major clades with few exceptions. However, the gpi tree splits the 12 isolates usually grouped with isolate A297 (the representative of European clone I) across two major clades, while the gyrB tree splits the 17 isolates usually grouped with isolate A320 (the representative of European clone II) across two major clades. This observation was confirmed via the quartet measure of tree-to-tree distances (7). This method measures the similarity between two trees by breaking them down into quartets, subtrees containing just four isolates. By analyzing whether all possible quartets across both trees are resolved identically to or differently from one another, it can be determined whether the topologies of the two trees are similar or not. For the gyrB and gpi trees, the number of differently resolved trees was greater than the number of similarly resolved trees, in contrast to the results for the other five genes (Table 1), confirming the visual observation that the topologies of these two trees are inconsistent with those of the other five genes.
TABLE 1.
Quartet measures of tree-to-tree distances
| Alleles compared | No. of quartetsa |
|||
|---|---|---|---|---|
| Q | Same | Different | ||
| gyrB | cpn60 | 135,751 | 48,678 | 87,073 |
| gpi | cpn60 | 135,751 | 53,525 | 82,226 |
| gpi | gyrB | 135,751 | 54,176 | 81,575 |
| gltA | cpn60 | 135,751 | 72,781 | 62,970 |
| gltA | gyrB | 135,751 | 48,728 | 87,023 |
| gltA | gpi | 135,751 | 48,678 | 87,073 |
| gdhB | cpn60 | 135,751 | 81,976 | 53,775 |
| gdhB | gyrB | 135,751 | 66,570 | 69,181 |
| gdhB | gpi | 135,751 | 64,914 | 70,837 |
| gdhB | gltA | 135,751 | 79,804 | 55,947 |
| rpoD | cpn60 | 135,751 | 85,972 | 49,779 |
| rpoD | gyrB | 135,751 | 66,667 | 69,084 |
| rpoD | gpi | 135,751 | 62,436 | 73,315 |
| rpoD | gltA | 135,751 | 80,324 | 55,427 |
| rpoD | gdhB | 135,751 | 106,700 | 29,051 |
| recA | cpn60 | 135,751 | 101,700 | 34,051 |
| recA | gyrB | 135,751 | 51,342 | 84,409 |
| recA | gpi | 135,751 | 57,243 | 78,508 |
| recA | gltA | 135,751 | 81,937 | 53,814 |
| recA | gdhB | 135,751 | 93,872 | 41,879 |
| recA | rpoD | 135,751 | 92,481 | 43,270 |
Q, maximum number of resolved quartets; Same, number of quartets that were resolved and revealed to be the same; Different, number of quartets that were resolved and revealed to be different.
(ii) PHI test.
The PHI test utilizes DNA sequence data and infers whether patterns of nucleotide polymorphisms are consistent with a model of vertical transmission (clonal population structure) or not. Instances where they are not may indicate recombination events (2). Analysis of alleles for all seven genes with the PHI test detected statistically significant (P = 0.0036) evidence for recombination within the alleles for gyrB but not in gpi.
(iii) eBURST analysis.
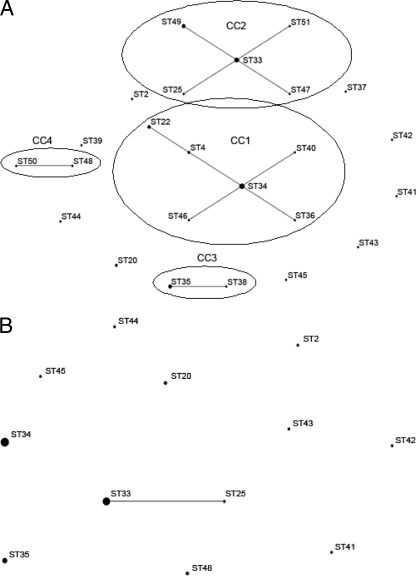
eBURST analysis of the allelic profiles for all seven loci by the use of both stringent and relaxed grouping parameters produced the same result (Fig. 2 A), revealing four CCs and nine singletons. The largest clonal complex, CC1, contained ST34, which was identified as a potential founder, with ST46, ST36, ST40, ST4, and ST22 radiating from it. A second major clonal complex, CC2, contained ST49, ST51, ST47, and ST25 surrounding ST33, which was identified as a potential founder for the complex. The other two minor clonal complexes identified contained ST35 and ST38 (CC3) and ST50 and ST48 (CC4), respectively. Analysis using only five loci (Fig. 2B) reduced the number of different allelic profiles such that all of the STs grouped in CC1 in Fig. 2A were identical to ST34; all of the STs, with the exception of ST25, grouped in CC2 were identical to ST33; and the two STs in each of CC3 and CC4 were also identical.
FIG. 2.
Results of eBURST analysis used to assign CCs within the 44 isolates of A. baumannii utilizing seven loci (A) and five loci (B). (A) The CCs are circled, and the predicted clonal ancestors for CC1 and CC2 are shown by the central points. (B) ST33 encompasses ST33, ST37, ST49, ST51, and ST47; ST34 encompasses ST34, ST4, ST36, ST40, ST46, and ST22; and ST35 includes ST35 and ST38, while ST48 covers ST48 and ST50. The sizes of the points are proportional to the number of isolates assigned to each ST.
(iv) IAS.
IAS is a measure of how clonal a population is and can be calculated using the allelic profiles generated by MLST. In a completely clonal population, alleles will be in linkage disequilibrium and the IAS approaches 1. Under panmixis, linkage equilibrium will be observed and IAS approaches 0. Values for IAS that differ significantly from 0 are considered to indicate a clonal population structure (26). Analysis of the entire data set of 44 isolates yielded an IAS value of 0.4907 (P < 0.001). This was decreased to 0.3569 (P < 0.001) when only one representative of each sequence type was included. Both values are significantly greater than 0 and indicate that the population is clonal.
(v) Phylogenetic reconstruction.
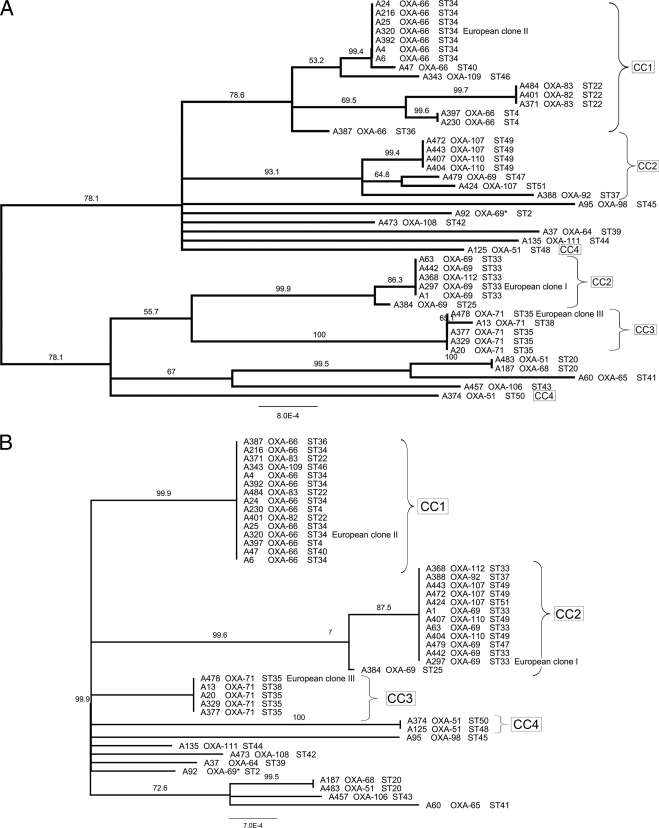
Neighbor-joining phylogenies for the 44 isolates were estimated using concatenated sequences for all seven loci (Fig. 3 A) and for five loci (gyrB and gpi were excluded) (Fig. 3B). The seven-locus phylogeny was not supported by very good percentage support values for the nodes, and isolates assigned to the same CCs by eBURST analysis were quite divergent and, in the case of CC2, occupied completely separate halves of the tree. As described above, there was evidence for horizontal gene transfer within the gyrB and gpi alleles and noncongruence between the individual locus trees. In order to assess the effect that the inclusion of the gyrB and gpi sequences was having on the tree topology, a neighbor-joining phylogeny for the 44 isolates was estimated using concatenated sequences for the other five genes only (Fig. 3B). The phylogeny was well supported with high percent support values for the nodes. The majority of isolates fell into one of three distinct and highly related clades: 14 isolates were grouped with the European clone II strain, 12 isolates were grouped with the European clone I strain, and 4 isolates were grouped with the European clone III strain. There was complete agreement between the CC groupings by eBURST analysis and the clades in the five-locus tree.
FIG. 3.
Neighbor-joining phylogenetic tree based on seven (A) and five (B) housekeeping genes. The corresponding OXA-51-like enzymes that the isolates encode and their sequence types are shown beside each isolate number. The CC that isolates were assigned to by eBURST analysis is indicated. Branches are labeled with percent support. OXA-69*, the OXA-69 enzyme encoded by a blaOXA-69 gene containing 5 silent nucleotide substitutions.
SBT-blaOXA-51-like genes.
Genes for 12 different OXA-51-like enzymes were identified, of which OXA-66 (n = 11), OXA-69 (n = 6), and OXA-71 (n = 5) constituted three major groups (Fig. 1). OXA-51 and OXA-107 were each found in three isolates, while OXA-83 was found in two isolates. OXA-68, OXA-82, OXA-92, OXA-109, and OXA-112 were found in a single isolate each (Fig. 1). As reported previously (8), representatives of the three major European clonal lineages were found to encode enzymes representative of the major groups, with clone I encoding OXA-69, clone II encoding OXA-66, and clone III encoding OXA-71.
Evaluation of SBT-blaOXA-51-like gene data and its correlation with MLST and PFGE data.
The SBT-blaOXA-51-like gene data correlated well with the MLST data, with respect to the identification of the major European lineages. The 14 isolates that grouped with the European clone II isolate each encoded closely related OXA-51-like enzymes of the OXA-66 group (Fig. 3A). Similarly, the 12 isolates that grouped with the European clone I isolate each encoded closely related members of the OXA-69 group of OXA-51-like enzymes, while the 4 isolates that grouped with the European clone III isolate each encoded OXA-71. A few exceptions were noticed in which isolate STs and OXA gene sequences were not consistent. Isolates A187 and A483, which had the same ST, ST20, encoded different OXAs, OXA-68 and OXA-51 (8 amino acid differences); and isolate A92, which had an OXA-69 enzyme, was not grouped with the other OXA-69-encoding isolates. However, the blaOXA-69 gene in A92 contained 5 silent nucleotide substitutions (G426 → A, C474 → A, C511 → T, G540 → A, and T801 → C), suggesting that this isolate may be quite different from the other OXA-69-encoding isolates. For the three major epidemic lineages, the SBT-blaOXA-51-like gene data were in concordance with the MLST-derived phylogeny for 93% of the isolates related to European clone I, 94% of the isolates related to European clone II, and 100% of the isolates related to European clone III. In contrast, typing by PFGE was not always in concordance with the SBT-blaOXA-51-like gene data or the MLST data. There were isolates with the same PFGE type (types A4, A6, and A24) that shared the same OXA-51-like gene, whereas others had the same ST but a different PFGE type (e.g., isolates A25 and A4 and isolates A6 and A24) (Fig. 1).
DISCUSSION
A. baumannii is becoming one of the most problematic organisms currently responsible for nosocomial infections, especially in intensive care units (9). Increasing antimicrobial resistance (42) and the ability of A. baumannii to survive on inanimate and dry surfaces (19) have been linked to the occurrence of outbreaks observed in various hospitals (5). MLST has emerged as the technique of choice for studying the population structure of many bacterial species (15, 17, 20, 30, 40), including A. baumannii (1, 10, 24, 29, 37, 38). However, profiles for only 93 isolates of A. baumannii are available in the current MLST database (http://pubmlst.org/abaumannii/), which is considerably less than the numbers deposited in databases for other bacteria (http://pubmlst.org/databases.shtml). Therefore, in order to investigate the MLST scheme thoroughly and validate the robustness of the MLST scheme, the present study investigated a previously characterized and diverse collection of epidemiologically unrelated A. baumannii isolates collected from 22 countries over four continents, Europe, Asia, Africa, and South America (8). In addition, because blaOXA-51-like genes are endogenous to A. baumannii, the study aimed to evaluate the use of these genes as the basis for a typing scheme.
The diversity of the isolates used in this study, compared with the limited diversity of the isolates in the A. baumannii database, meant that a considerable number of novel STs was expected. Indeed, 19 novel STs were detected, with this result being similar to that of Wisplinghoff et al. (44). Together with some earlier preliminary supporting evidence (1, 44), this indicates that A. baumannii is more diverse than originally thought, especially as A. baumannii tends to spread clonally during outbreaks (5, 34). Although MLST has advantages over many conventional methods for strain genotyping, it is not yet clear whether phylogenetic relationships based on several, often randomly selected, loci reflect the real phylogenies of the strains investigated or whether a smaller number of loci could provide results similar to those based on several loci (22).
The present work revealed that the tree topologies for all identified alleles of the seven genes were inconsistent, with the trees for gyrB and gpi being incongruent in relation to each other and to the trees for the other five genes. The incongruence observed between gene trees suggests that horizontal gene transfer has occurred at the gyrB and gpi loci with a sufficient frequency that their inclusion in the estimation of an A. baumannii phylogeny would distort it to such a degree that it would not be representative of the phylogenetic structure of the core genome. The only previous study (31) to have published the results of an in-depth analysis using the MLST scheme of Bartual et al. (1) appears to have had the same problems encountered here, with the trees for gyrB and gpi being nonconcordant with those for the other loci.
By omitting the gyrB and gpi genes and using only the remaining five genes, a well-supported phylogenetic tree was obtained. This poses two questions: (i) can fewer genes be utilized as effectively to estimate a phylogeny representative of the diversity of the core genome, and (ii) what are the criteria, if any, used to select the best housekeeping genes and/or other genes to reflect the core genome diversity? The present study revealed that only five genes (gltA, gdhB, recA, cpn60, and rpoD) were required to characterize A. baumannii and that these five genes were able to provide sufficient discriminatory power to separate the three major European clonal lineages, lineages I, II, and III, into three different groups. Furthermore, compared to MLST, SBT-blaOXA-51-like genes successfully identified 94% of these isolates as belonging to a major epidemic lineage, with only two results being discrepant. Members of the three major European lineages are responsible for the majority of outbreaks caused by A. baumannii worldwide and, as such, are currently of greater concern from an infection control perspective than other unrelated lineages belonging to this species. Therefore, it is very useful that isolates can quickly and easily be identified as belonging to one of the three major European epidemic lineages with a high degree of accuracy by SBT-blaOXA-51-like genes.
The IAS value for the entire data set was significantly greater than 0, indicating that the population structure is clonal. However, when only one representative of each ST was included in the analysis, the IAS value, while it was still significantly greater than 0, decreased from 0.4907 to 0.3569. This decrease is concordant with the epidemic nature of the European clonal lineages predominant in the data set, though it may also point toward there being a greater degree of recombination within more closely related lineages of A. baumannii. Identification of a clonal population structure by IAS analysis seems at odds with the detection of recombination at the gyrB and gpi loci. However, it has previously been shown that when IAS measures are used, the presence of recombination may be masked either due to an epidemic population structure or due to recombination occurring only between closely related lineages and not with those more distantly related. Analysis of a larger data set is required to determine which scenario is applicable within A. baumannii.
In contrast to a previous study (1), the PFGE profiles did not always correlate with the MLST results (Fig. 1). Therefore, it is probable that the swift increase in the genetic diversity indexed by PFGE has led to significant discrepancies in the ApaI DNA fragment patterns, such that these isolates are no longer recognized as being related to one another by the criteria utilized here.
With the exclusion of the gyrB and gpi loci, MLST analysis separated the representative isolates of European clones I (ST33), II (ST34), and III (ST35) and the isolates associated with these lineages into three major groups. Isolates A63 (ST33) and A479 (ST47), which grouped with clone I, are closely related (the two STs are single-locus variants at the gpi locus) and were recovered from Buenos Aires, Argentina, and Pakistan, respectively. Similarly, isolate A401 (ST22) from Taiwan grouped with clone II, ST34, though it differs at the gyrB and gpi loci. Thus, both of these clonal lineages are not confined solely to Europe. This is in agreement with other studies of worldwide A. baumannii isolates. The present study detected isolates belonging to the European clone III lineage only in European cities, though this does not preclude their spread and/or presence in other parts of the world.
Overall, typing of isolates by their blaOXA-51-like gene sequence yielded results that were broadly consistent with those obtained by MLST, with few exceptions. The data were considerably more consistent with the MLST data than with the PFGE data. The enzymes OXA-66, OXA-69, and OXA-71 are the predominant members of closely related OXA-51-like subgroups and are associated with particular epidemic lineages (8). Both SBT-blaOXA-51-like genes and MLST grouped the epidemic European clones I, II, and III into three different lineages. MLST shows clear applications for studying evolutionary relationships on a global scale and is the best state-of-the-art technique available to study population structures. However, the present data show that the gyrB and gpi loci should be treated with caution when the phylogeny of the core genome is estimated, particularly with respect to assigning isolates to the major epidemic lineages. Furthermore, the SBT-blaOXA-51-like gene scheme was found to be a useful tool for accurately identifying isolates belonging to the three major epidemic lineages within A. baumannii.
Acknowledgments
This work was supported by a grant (grant RA0119) from the Medical Research Council of the United Kingdom.
Footnotes
Published ahead of print on 26 April 2010.
REFERENCES
- 1.Bartual, S. G., H. Seifert, C. Hippler, M. A. D. Luzon, H. Wisplinghoff, and F. Rodríguez-Valera. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 45:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cisneros, J. M., J. Rodriguez-Bano, F. Fernandez-Cuenca, A. Ribera, J. Vila, A. Pascual, L. Martinez-Martinez, G. Bou, and J. Pachon. 2005. Risk-factors for the acquisition of imipenem-resistant Acinetobacter baumannii in Spain: a nationwide study. Clin. Microbiol. Infect. 11:874-879. [DOI] [PubMed] [Google Scholar]
- 4.Coelho, J. M., J. F. Turton, M. E. Kaufmann, J. Glover, N. Woodford, M. Warner, M. F. Palepou, R. Pike, T. L. Pitt, B. C. Patel, and D. M. Livermore. 2006. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and Southeast England. J. Clin. Microbiol. 44:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbella, X., A. Monteron, M. Pujol, M. A. Dominguez, J. Ayats, M. J. Argerich, F. Garrigosa, J. Ariza, and F. Gudiol. 2001. Emergence and rapid spread of carbapenem resistance during a large and sustained hospital outbreak of multiresistant Acinetobacter baumannii. J. Clin. Microbiol. 38:4086-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ecker, J. A., C. Massire, T. A. Hall, R. Ranken, T. D. Pennella, C. A. Ivy, L. B. Blyn, S. A. Hofstadler, T. P. Endy, P. T. Scott, L. Lindler, T. Hamilton, C. Gaddy, K. Snow, M. Pe, J. Fishbain, D. Craft, G. Deye, S. Riddel, E. Milstrey, B. Petruccelli, S. Brisse, V. Harpin, A. Schink, D. J. Ecker, R. Sampath, and M. W. Eshoo. 2006. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 44:2921-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estabrook, G. F., F. R. McMorris, and C. A. Meacham. 1985. Comparison of undirected phylogenetic trees based on subtrees of four evolutionary units. Syst. Zool. 34:193-200. [Google Scholar]
- 8.Evans, B. A., A. Hamouda, K. J. Towner, and S. G. B. Amyes. 2008. OXA-like β-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii. Clin. Microbiol. Infect. 14:268-275. [DOI] [PubMed] [Google Scholar]
- 9.Falagas, M. E., and E. A. Karveli. 2007. The changing global epidemiology of Acinetobacter baumannii infections: a development with major public health implications. Clin. Microbiol. Infect. 13:117-119. [DOI] [PubMed] [Google Scholar]
- 10.Fantana, C., M. Favaro, S. Minelli, M. C. Bossa, G. P. Testore, F. Leonardis, S. Natoli, and C. Favalli. 2008. Acinetobacter baumannii in intensive care unit: a novel system to study clonal relationship among the isolates. BMC Infect. Dis. 79:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feaver, I. M., S. J. Gray, R. Urwin, J. E. Russel, J. A. Bygraves, E. B. Kaczmarski, and M. C. Maiden. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 37:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier, P. E., and H. Richet. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42:692-699. [DOI] [PubMed] [Google Scholar]
- 14.Gaynes, R., J. R. Edwards, and the National Nosocomial Infections Surveillance System Division of Healthcare Quality Promotion, National Center for Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia. 2005. Overview of nosocomial infections caused by Gram-negative bacilli. Clin. Infect. Dis. 41:848-854. [DOI] [PubMed] [Google Scholar]
- 15.Harbottle, H., D. G. White, P. F. McDermott, R. D. Walker, and S. Zhao. 2006. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J. Clin. Microbiol. 44:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heym, B., M. Le Moal, L. Armand-Lefevre, and M. H. Nicolas-Chanoine. 2002. Multilocus sequence typing (MLST) shows that ‘Iberian’ clone of methicillin-resistant Staphylococcus aureus has spread to France and acquired reduced susceptibility to teicoplanin. J. Antimicrob. Chemother. 50:323-329. [DOI] [PubMed] [Google Scholar]
- 18.Huys, G., M. Cnockaert, A. Nemec, L. Dijkshoorn, S. Briss, M. Vaneechouette, and J. Swings. 2005. Repetitive-DNA-element PCR fingerprinting and antibiotic resistance of pan-European multi-resistant Acinetobacter baumannii clone III strains. J. Clin. Microbiol. 54:851-856. [DOI] [PubMed] [Google Scholar]
- 19.Jawad, A., H. Seifert, A. M. Snelling, J. Heritage, and P. M. Hawkey. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36:1938-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, K. J., S. M. Arduino, O. C. Stine, J. A. Johnson, and A. D. Harris. 2007. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J. Clin. Microbiol. 45:3707-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koeleman, J. G., J. Stoof, D. J. Biesmans, P. H. Savelkoul, and C. M. Vandenbroucke-Grauls. 1998. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J. Clin. Microbiol. 36:2522-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konstantinos, T. K., R. Alban, and T. M. James. 2006. Toward a more robust assessment of intraspecies diversity using fewer genetic markers. Appl. Environ. Microbiol. 72:7286-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraniotaki, E., R. Manganelli, E. Platsouka, A. Grossato, O. Paniara, and G. Palù. 2006. Molecular investigation of an outbreak of multidrug-resistant Acinetobacter baumannii, with characterization of class 1 integrons. Int. J. Antimicrob. Agents 28:193-199. [DOI] [PubMed] [Google Scholar]
- 24.Landman, D., M. Butnarius, S. Bratu, and J. Quale. 2008. Genetic relatedness of multidrug-resistant Acinetobacter baumannii endemic to New York City. Epidemiol. Infect. 137:174-180. [DOI] [PubMed] [Google Scholar]
- 25.Lolans, K., T. W. Rice, L. S. Munoz-Price, and J. P. Quinn. 2006. Multicity outbreaks of carbapenem-resistant Acinetobacter baumannii isolates producing the carbapenemase OXA-40. Antimicrob. Agents Chemother. 50:2941-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miranda, G., C. Kelly, F. Solorzano, B. Leanos, R. Coria, and J. E. Patterson. 1996. Use of pulsed-field gel electrophoresis typing to study an outbreak of infection due to Serratia marcescens in a neonatal intensive care unit. J. Clin. Microbiol. 34:3138-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naas, T., P. Bogaerts, C. Bauraing, Y. Degheldre, Y. Glupczynski, and P. Nordmann. 2006. Emergence of PER and VEB extended-spectrum beta-lactamases in Acinetobacter baumannii in Belgium. J. Antimicrob. Chemother. 58:178-182. [DOI] [PubMed] [Google Scholar]
- 29.Nemec, A., L. Kíová, M. Maixnerová, L. Diancourt, T. J. K. van der Reijden, S. Brisse, P. van den Broek, and L. Dijkshoorn. 2008. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J. Antimicrob. Chemother. 62:484-489. [DOI] [PubMed] [Google Scholar]
- 30.Nemoy, L., M. Kottetishvili, J. Tigno, A. Keefer-Norris, A. D. Harris, E. N. Perencevich, J. A. Johnson, D. Torpey, A. Sulakvelidze, J. G. Morris, and O. C. Stine. 2005. Multilocus sequence typing versus pulsed-field gel electrophoresis for characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates. J. Clin. Microbiol. 43:1776-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park, Y. K., S. Jung, K. Park, H. S. Cheong, K. R. Peck, J. Song, and K. S. Ko. 2009. Independent emergence of colistin-resistant Acinetobacter spp. isolates from Korea. Diagn. Microbiol. Infect. Dis. 64:43-52. [DOI] [PubMed] [Google Scholar]
- 32.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, S. A., R. Findlay, and S. D. R. Lang. 2001. Investigation of an outbreak of multi-drug resistant Acinetobacter baumannii in an intensive care burns unit. J. Hosp. Infect. 48:228-232. [DOI] [PubMed] [Google Scholar]
- 34.Schulte, B., C. Goerke, P. Weyrich, S. Grobner, C. Bahrs, C. Wolz, I. B. Autenrieth, and S. Borgmann. 2005. Clonal spread of meropenem-resistant Acinetobacter baumannii strains in hospitals in the Mediterranean region and transmission to south-west Germany. J. Hosp. Infect. 61:356-357. [DOI] [PubMed] [Google Scholar]
- 35.Seifert, H., L. Dolzani, R. Bressan, T. Van der Reijden, B. van Strijen, D. Stefanik, H. Heersma, and L. Dijkshoorn. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifert, H., and P. Gerner-Smidt. 1995. Comparison of ribotyping and pulsed-field gel electrophoresis for molecular typing of Acinetobacter isolates. J. Clin. Microbiol. 33:1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seok-Mo, H., H. M. Elaine, J. L. Alan, R. G. Steven, and A. S. Frank. 2008. Genetic relationships between respiratory pathogens isolated from dental plaque and bronchoalveolar lavage fluid from patients in the intensive care unit undergoing mechanical ventilation. Clin. Infect. Dis. 47:1562-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shelburne, S. A., K. V. Singh, A. Clinton-White, L. Byrne, A. Carmer, C. Austin, E. Graviss, C. Stager, B. E. Murray, and R. L. Atmar. 2008. Sequential outbreaks of infections by distinct Acinetobacter baumannii strains in a public teaching hospital in Houston, Texas. J. Clin. Microbiol. 46:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spence, R. P., T. J. K. van der Reijden, L. Dijkshoorn, and K. J. Towner. 2004. Comparison of Acinetobacter baumannii isolates from United Kingdom hospitals with predominant Northern European genotypes by amplified-fragment length polymorphism analysis. J. Clin. Microbiol. 42:832-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanabe, Y., F. Kasai, and M. M. Watanabe. 2007. Multilocus sequence typing reveals high genetic diversity and clonal population structure of the toxic cyanobacterium Microcystis aeruginosa. Microbiology 153:3695-3703. [DOI] [PubMed] [Google Scholar]
- 41.Turton, J. F., S. N. Gabriel, C. Valderrey, M. E. Kaufmann, and T. L. Pitt. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineage of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807-815. [DOI] [PubMed] [Google Scholar]
- 42.Van Looveren, M., H. Goossens, and the ARPAC Steering Group. 2004. Antimicrobial resistance of Acinetobacter spp. in Europe. Clin. Microbiol. Infect. 10:684-704. [DOI] [PubMed] [Google Scholar]
- 43.Villegas, M. V., and A. I. Hartstein. 2003. Acinetobacter outbreaks, 1977-2000. Infect. Control Hosp. Epidemiol. 24:284-295. [DOI] [PubMed] [Google Scholar]
- 44.Wisplinghoff, H., C. Hippler, S. G. Bartual, C. Haefs, D. Stefanik, P. G. Higgins, and H. Seifert. 2008. Molecular epidemiology of clinical Acinetobacter baumannii and Acinetobacter genomic species 13TU isolates using a multilocus sequencing typing scheme. Clin. Microbiol. Infect. 14:708-715. [DOI] [PubMed] [Google Scholar]
- 45.Woodford, N., M. J. Ellington, J. M. Coelho, J. F. Turton, M. E. Ward, S. Brown, S. G. B. Amyes, and D. M. Livermore. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351-353. [DOI] [PubMed] [Google Scholar]
- 46.Ying, C. M., T. K. W. Ling, C. C. Lee, and J. M. Ling. 2006. Characterization of carbapenem-resistant Acinetobacter baumannii in Shanghai and Hong Kong. J. Med. Microbiol. 55:799-802. [DOI] [PubMed] [Google Scholar]
- 47.Yong, D., J. H. Shin, S. Kim, Y. Lim, J. H. Yum, K. Lee, Y. Chong, and A. Bauernfeind. 2003. High prevalence of PER-1 extended-spectrum β-lactamase-producing Acinetobacter spp. in Korea. Antimicrob. Agents Chemother. 47:1749-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo, J. H., J. H. Choi, W. S. Shin, D. H. Huh, Y. K. Cho, K. M. Kim, M. Y. Kim, and M. W. Kang. 1999. Application of infrequent-restriction-site PCR to clinical isolates of Acinetobacter baumannii and Serratia marcescens. J. Clin. Microbiol. 37:3108-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]